Natural Compounds in Liposomal Nanoformulations of Potential Clinical Application in Glioblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Compounds and Reagents
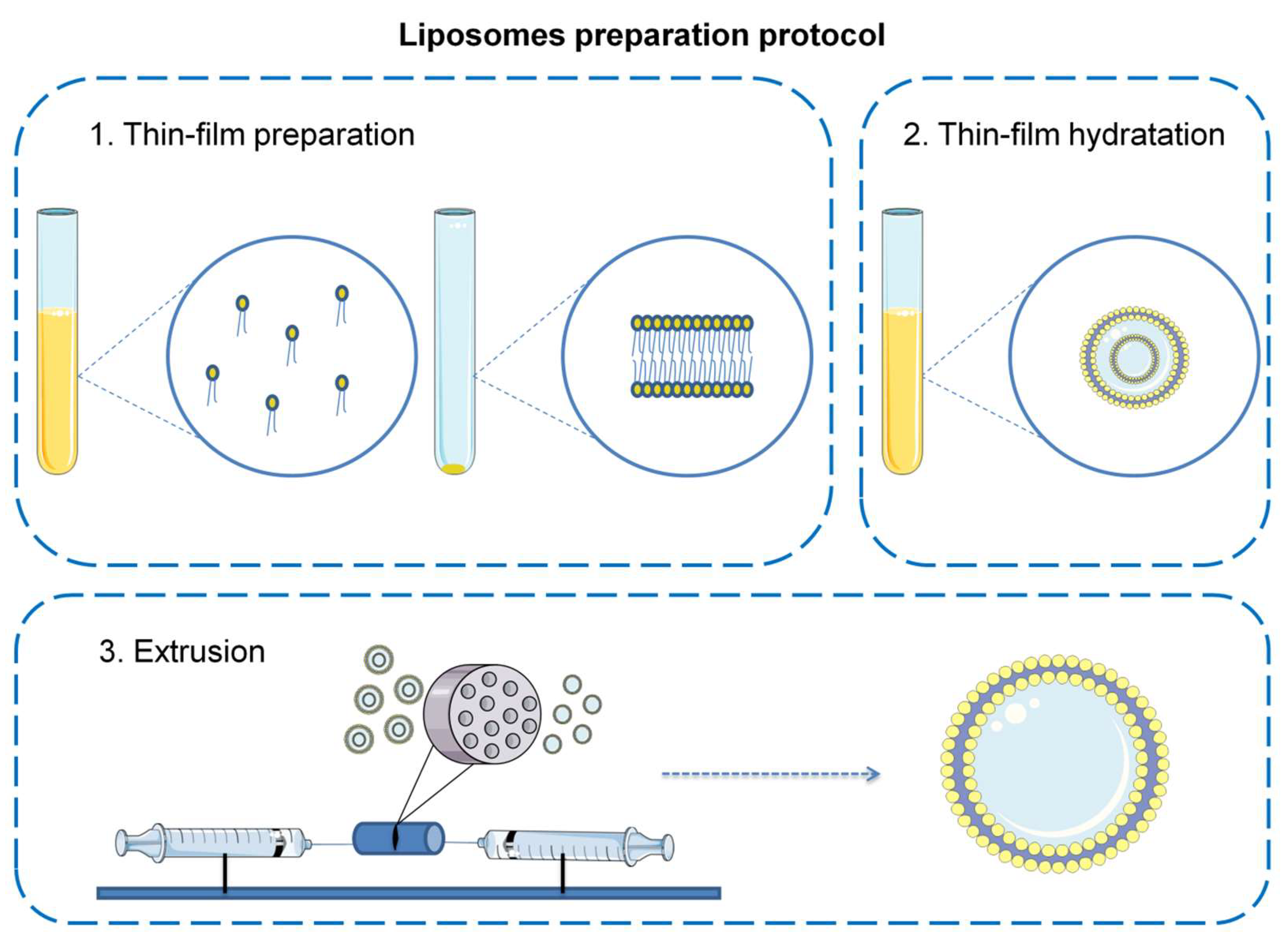
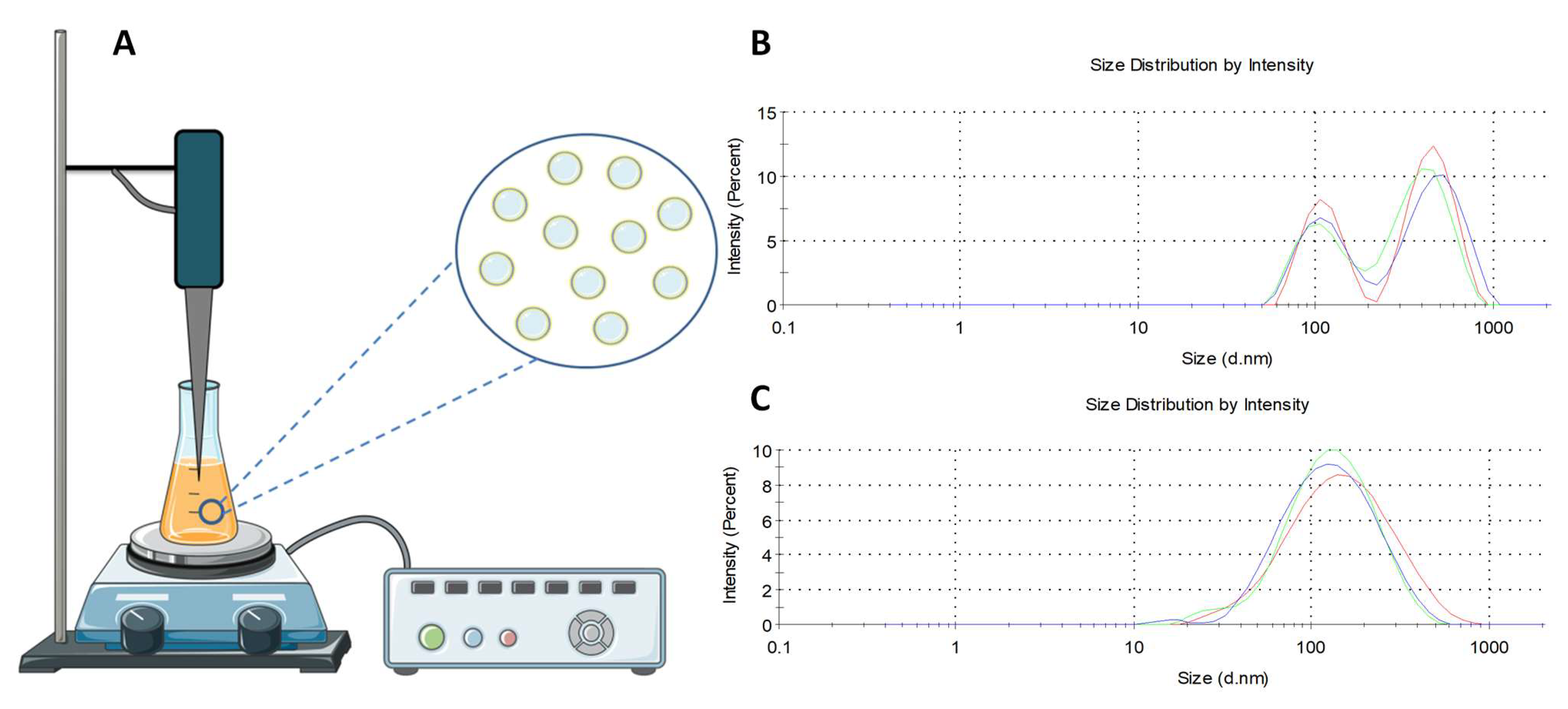
2.2. Liposome Preparation
2.3. Liposome Size, Polydispersity Index, and Zeta Potential Measurement
2.4. HPLC Analysis and Encapsulation Efficiency
2.5. NMR and EPR Measurements
2.6. Biological Activity Assessment (MTT Test)
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Liposome Size, Encapsulation, Polydispersity Index, and Zeta Potential Measurement
3.2. NMR Measurements
3.3. EPR Measurements
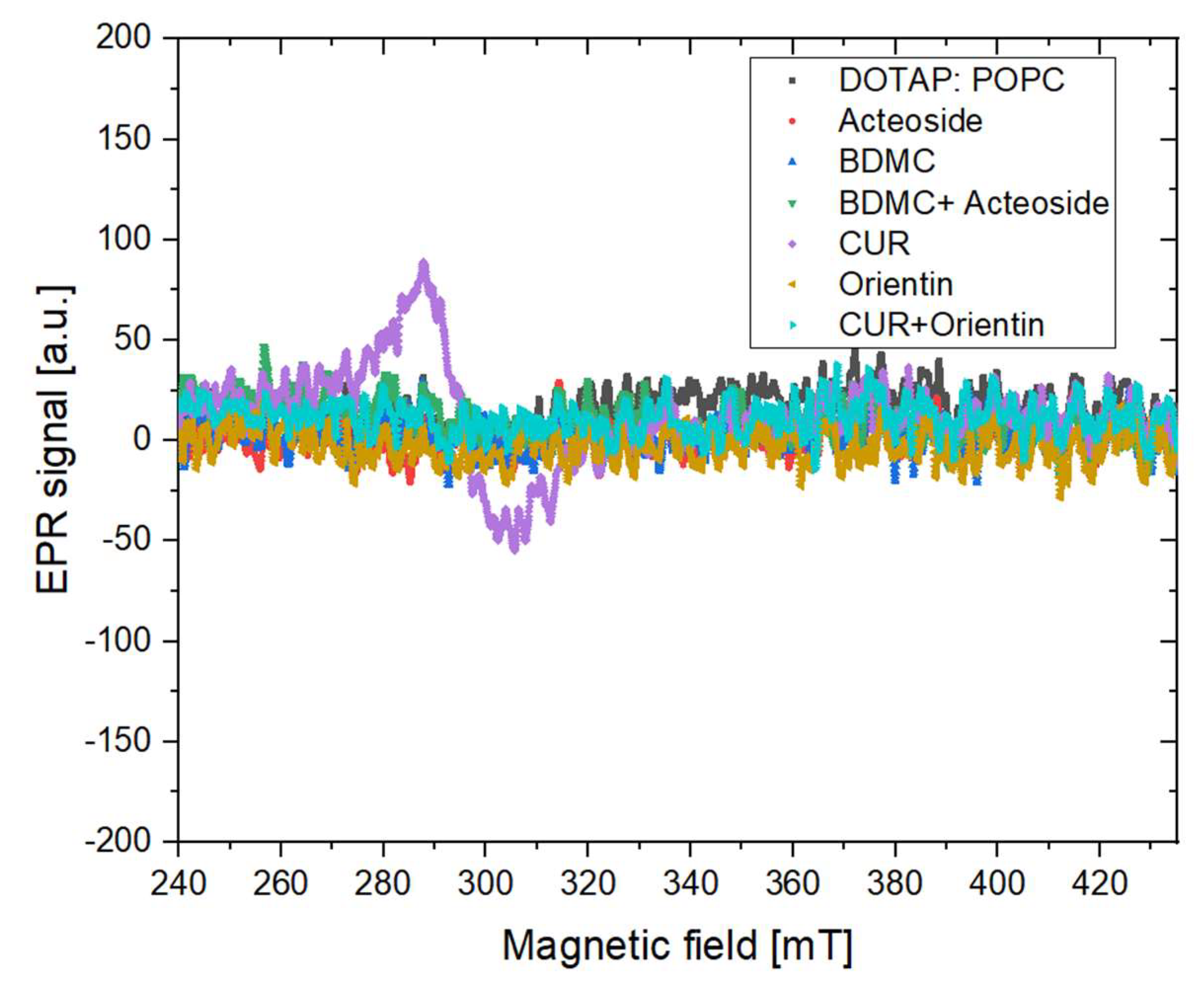
3.3.1. X Band Measurement for the Presence of Stable Free Radicals
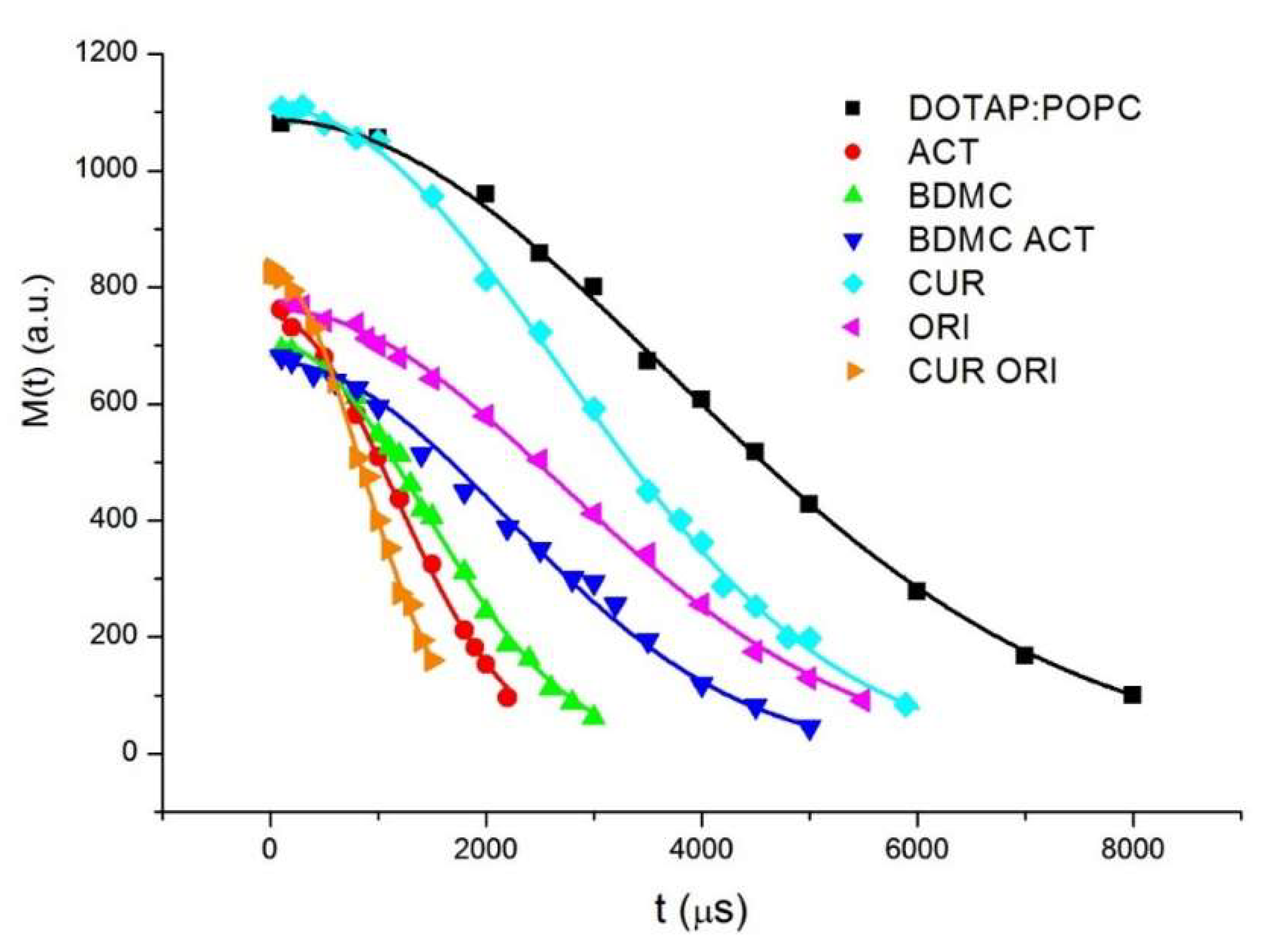
3.3.2. L Band Free Radical Scavenging
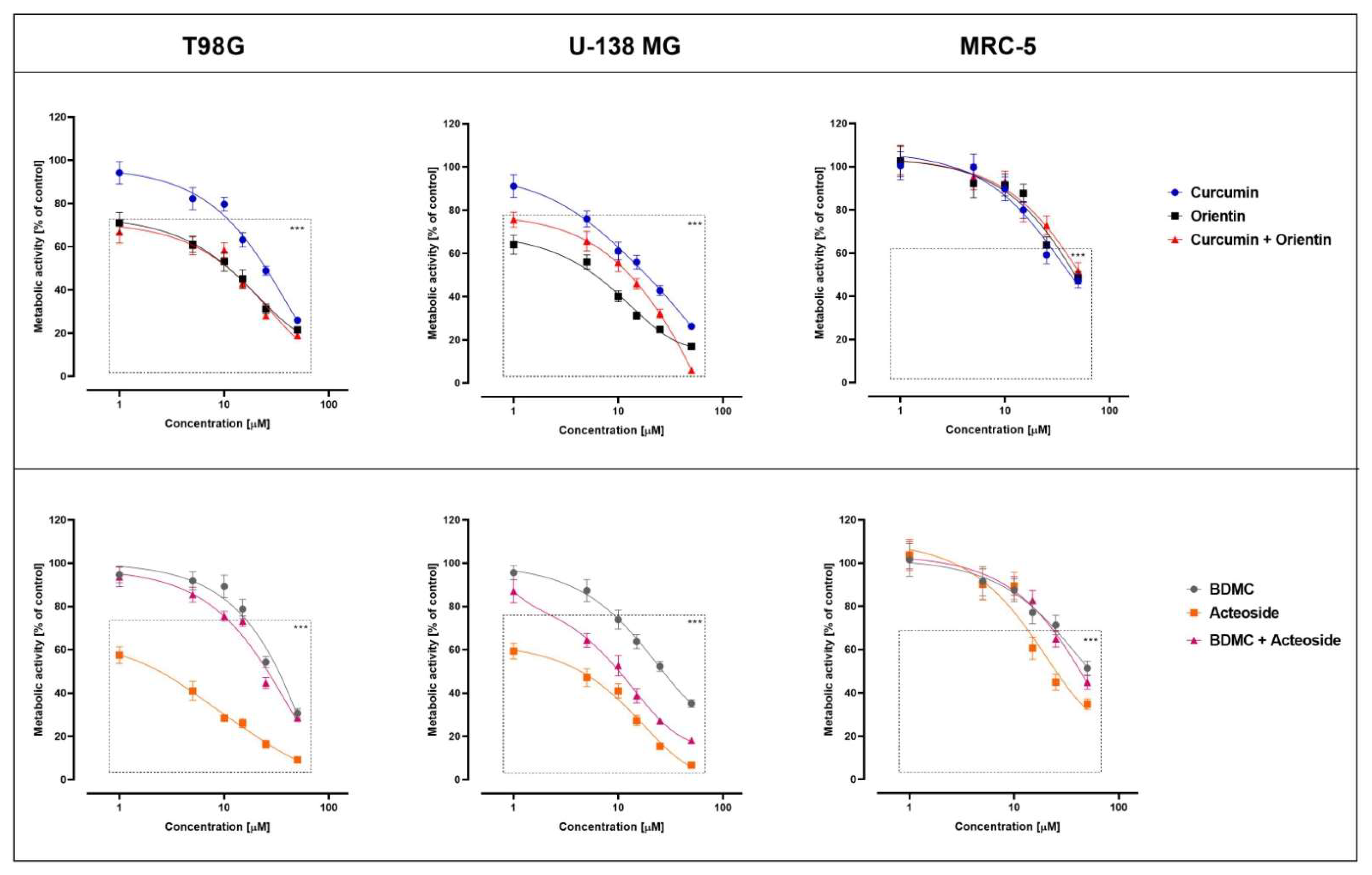
3.4. Biological Activity
3.5. The Alternative Method of Liposome Extrusion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Kapatia, G.; Salunke, P.; Ahuja, C.K.; Singh, V. Intraoperative consultation in the diagnosis of posterior fossa brain tumors following the 2016 WHO update. Cytopathology 2021, 32, 459–471. [Google Scholar] [CrossRef]
- McNamara, C.; Mankad, K.; Thust, S.; Dixon, L.; Limback-Stanic, C.; D’Arco, F.; Jacques, T.S.; Löbel, U. 2021 WHO classification of tumours of the central nervous system: A review for the neuroradiologist. Neuroradiology 2022, 64, 1919–1950. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Reuss, D.E.; Kratz, A.; Sahm, F.; Capper, D.; Schrimpf, D.; Koelsche, C.; Hovestadt, V.; Bewerunge-Hudler, M.; Jones, D.T.W.; Schittenhelm, J.; et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015, 130, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Hasselblatt, M.; Jaber, M.; Reuss, D.; Grauer, O.; Bibo, A.; Terwey, S.; Schick, U.; Ebel, H.; Niederstadt, T.; Stummer, W.; et al. Diffuse Astrocytoma, IDH-Wildtype: A Dissolving Diagnosis. J. Neuropathol. Exp. Neurol. 2018, 77, 422–425. [Google Scholar] [CrossRef] [Green Version]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014-2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Delgado-López, P.D.; Corrales-García, E.M. Survival in glioblastoma: A review on the impact of treatment modalities. Clin. Transl. Oncol. 2016, 18, 1062–1071. [Google Scholar] [CrossRef]
- Ghani, U. Chapter three—Polyphenols. In Alpha-Glucosidase Inhibitors; Ghani, U., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 61–100. ISBN 978-0-08-102779-0. [Google Scholar]
- Sandur, S.K.; Pandey, M.K.; Sung, B.; Ahn, K.S.; Murakami, A.; Sethi, G.; Limtrakul, P.; Badmaev, V.; Aggarwal, B.B. Curcumin, demethoxycurcumin, bisdemethoxycurcumin, tetrahydrocurcumin and turmerones differentially regulate anti-inflammatory and anti-proliferative responses through a ROS-independent mechanism. Carcinogenesis 2007, 28, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tayyem, R.F.; Heath, D.D.; Al-Delaimy, W.K.; Rock, C.L. Curcumin content of turmeric and curry powders. Nutr. Cancer 2006, 55, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Yodkeeree, S.; Chaiwangyen, W.; Garbisa, S.; Limtrakul, P. Curcumin, demethoxycurcumin and bisdemethoxycurcumin differentially inhibit cancer cell invasion through the down-regulation of MMPs and uPA. J. Nutr. Biochem. 2009, 20, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Lu, H.-F.; Chen, Y.-H.; Chen, J.-C.; Chou, W.-H.; Huang, H.-C. Curcumin, demethoxycurcumin, and bisdemethoxycurcumin induced caspase-dependent and –independent apoptosis via Smad or Akt signaling pathways in HOS cells. BMC Complement. Med. Ther. 2020, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hu, X.-P.; Zeng, Y.; Li, Y.; Wu, H.-Q.; Qiu, R.-Z.; Ma, W.-J.; Li, T.; Li, C.-Y.; He, Z.-D. Advanced research on acteoside for chemistry and bioactivities. J. Asian Nat. Prod. Res. 2011, 13, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ren, Q.; Wu, L. The pharmacokinetic property and pharmacological activity of acteoside: A review. Biomed. Pharmacother. 2022, 153, 113296. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, Z.; Huo, Z.; Xu, Z.; Zhong, Y.; Wang, X.; Yang, Y.; Wang, Z. Osmanthus fragrans Flower Aqueous Extract and its Enriched Acteoside inhibit Melanogenesis and Ultraviolet-induced Pigmentation. Nat. Prod. Commun. 2018, 13, 1934578X1801300515. [Google Scholar] [CrossRef]
- Verbascoside: Identification, Quantification, and Potential Sensitization of Colorectal Cancer Cells to 5-FU by Targeting PI3K/AKT Pathway | Scientific Reports. Available online: https://www.nature.com/articles/s41598-018-35083-2 (accessed on 2 August 2022).
- Mulani, S.K.; Guh, J.-H.; Mong, K.-K.T. A general synthetic strategy and the anti-proliferation properties on prostate cancer cell lines for natural phenylethanoid glycosides. Org. Biomol. Chem. 2014, 12, 2926–2937. [Google Scholar] [CrossRef]
- Lam, K.Y.; Ling, A.P.K.; Koh, R.Y.; Wong, Y.P.; Say, Y.H. A Review on Medicinal Properties of Orientin. Adv. Pharmacol. Sci. 2016, 2016, 4104595. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Tong, M.; Li, Z.; Huang, W.; Jin, Y.; Cao, Q.; Zhou, X.; Tong, G. The Effects of Orientin on Proliferation and Apoptosis of T24 Human Bladder Carcinoma Cells Occurs Through the Inhibition of Nuclear Factor-kappaB and the Hedgehog Signaling Pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 9547–9554. [Google Scholar] [CrossRef]
- Abu Lila, A.S.; Ishida, T. Liposomal Delivery Systems: Design Optimization and Current Applications. Biol. Pharm. Bull. 2017, 40, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Du, C.; Guo, N.; Teng, Y.; Meng, X.; Sun, H.; Li, S.; Yu, P.; Galons, H. Composition design and medical application of liposomes. Eur. J. Med. Chem. 2019, 164, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 601, 120571. [Google Scholar] [CrossRef] [PubMed]
- Large, D.E.; Abdelmessih, R.G.; Fink, E.A.; Auguste, D.T. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021, 176, 113851. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Lu, Y.; Qi, J.; Zhu, Q.; Chen, Z.; Wu, W. Adapting liposomes for oral drug delivery. Acta Pharm. Sin. B 2019, 9, 36. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, U.S. Liposomes in drug delivery: Progress and limitations. Int. J. Pharm. 1997, 154, 123–140. [Google Scholar] [CrossRef]
- Ishida, T.; Harashima, H.; Kiwada, H. Liposome clearance. Biosci. Rep. 2002, 22, 197–224. [Google Scholar] [CrossRef]
- Oku, N.; Namba, Y. Long-circulating liposomes. Crit. Rev. Ther. Drug Carrier Syst. 1994, 11, 231–270. [Google Scholar]
- Schwendener, R.A.; Lagocki, P.A.; Rahman, Y.E. The effects of charge and size on the interaction of unilamellar liposomes with macrophages. Biochim. Biophys. Acta BBA-Biomembr. 1984, 772, 93–101. [Google Scholar] [CrossRef]
- Oussoren, C.; Zuidema, J.; Crommelin, D.J.; Storm, G. Lymphatic uptake and biodistribution of liposomes after subcutaneous injection. II. Influence of liposomal size, lipid compostion and lipid dose. Biochim. Biophys. Acta 1997, 1328, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Pröhl, M.; Bus, T.; Czaplewska, J.A.; Traeger, A.; Deicke, M.; Weiss, H.; Weigand, W.; Schubert, U.S.; Gottschaldt, M. Synthesis and in vitro Toxicity of d-Glucose and d-Fructose Conjugated Curcumin–Ruthenium Complexes. Eur. J. Inorg. Chem. 2016, 2016, 5197–5204. [Google Scholar] [CrossRef]
- Budzianowska, A.; Skrzypczak, L.; Budzianowski, J. Phenylethanoid Glucosides from in vitro Propagated Plants and Callus Cultures of Plantago lanceolata. Planta Med. 2004, 70, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Budzianowski, J.; Pakulski, G.; Robak, J. Studies on antioxidative activity of some C-glycosylflavones. Pol. J. Pharmacol. Pharm. 1991, 43, 395–401. [Google Scholar] [PubMed]
- Piwowarczyk, L.; Kucinska, M.; Tomczak, S.; Mlynarczyk, D.T.; Piskorz, J.; Goslinski, T.; Murias, M.; Jelinska, A. Liposomal Nanoformulation as a Carrier for Curcumin and pEGCG—Study on Stability and Anticancer Potential. Nanomaterials 2022, 12, 1274. [Google Scholar] [CrossRef]
- European Medicines Agency (EMA). Q 2 (R1) Validation of Analytical Procedures: Text and Methodology; June 1995 CPMP/ICH/381/95; European Medicines Agency (EMA): Amsterdam, The Netherlands, 2006; p. 15. [Google Scholar]
- Hudiyanti, D.; Al Khafiz, M.F.; Anam, K.; Siahaan, P.; Christa, S.M. In Vitro Evaluation of Curcumin Encapsulation in Gum Arabic Dispersions under Different Environments. Molecules 2022, 27, 3855. [Google Scholar] [CrossRef]
- Baranowski, M.; Woźniak-Braszak, A.; Jurga, K. High homogeneity B(1) 30.2 MHz Nuclear Magnetic Resonance Probe for off-resonance relaxation times measurements. J. Magn. Reson. 2011, 208, 163–166. [Google Scholar] [CrossRef]
- Czechowski, T.; Baranowski, M.; Woźniak-Braszak, A.; Jurga, K.; Jurga, J.; Kędzia, P. The Instrument Set for Generating Fast Adiabatic Passage. Appl. Magn. Reson. 2012, 43, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Margaret Cheng, H.-L.; Stikov, N.; Ghugre, N.R.; Wright, G.A. Practical medical applications of quantitative MR relaxometry. J. Magn. Reson. Imaging 2012, 36, 805–824. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Zielińska-Przyjemska, M.; Wierzchowski, M.; Kleszcz, R.; Studzińska-Sroka, E.; Kaczmarek, M.; Paluszczak, J.; Cielecka-Piontek, J.; Krajka-Kuźniak, V. Methoxy-stilbenes downregulate the transcription of Wnt/β-catenin-dependent genes and lead to cell cycle arrest and apoptosis in human T98G glioblastoma cells. Adv. Med. Sci. 2021, 66, 6–20. [Google Scholar] [CrossRef]
- Jain, A.K.; Thareja, S. In vitro and in vivo characterization of pharmaceutical nanocarriers used for drug delivery. Artif. Cells Nanomedicine Biotechnol. 2019, 47, 524–539. [Google Scholar] [CrossRef] [Green Version]
- Doane, T.L.; Chuang, C.-H.; Hill, R.J.; Burda, C. Nanoparticle ζ-Potentials. Acc. Chem. Res. 2012, 45, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Crist, R.M.; Clogston, J.D.; McNeil, S.E. Zeta potential: A case study of cationic, anionic, and neutral liposomes. Anal. Bioanal. Chem. 2017, 409, 5779–5787. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Marioli, M.; Zhang, K. Analytical characterization of liposomes and other lipid nanoparticles for drug delivery. J. Pharm. Biomed. Anal. 2021, 192, 113642. [Google Scholar] [CrossRef] [PubMed]
- Lasic, D.D. Magnetic Resonance Methods in The Studies of Liposomes. Bull. Magn. Reason. 1991, 13, 3–13. [Google Scholar]
- The Principles of Nuclear Magnetism—A. Abragam—Google Książki. Available online: https://books.google.pl/books/about/The_Principles_of_Nuclear_Magnetism.html?id=9M8U_JK7K54C&redir_esc=y (accessed on 16 August 2022).
- A Stochastic Theory of Line- Shape and Relaxation|CiNii Research. Available online: https://cir.nii.ac.jp/crid/1571135649070600576 (accessed on 16 August 2022).
- Smith, I.C.P.; Stockton, G.W.; Tulloch, A.P.; Polnaszek, C.F.; Johnson, K.G. Deuterium NMR and spin label ESR as probes of membrane organization. J. Colloid Interface Sci. 1977, 58, 439–451. [Google Scholar] [CrossRef]
- Duda, M.; Cygan, K.; Wisniewska-Becker, A. Effects of Curcumin on Lipid Membranes: An EPR Spin-label Study. Cell Biochem. Biophys. 2020, 78, 139–147. [Google Scholar] [CrossRef]
- Baranowski, M.; Gonet, M.; Czechowski, T.; Kucinska, M.; Plewinski, A.; Szczepanik, P.; Murias, M. Dynamic Electron Paramagnetic Resonance Imaging: Modern Technique for Biodistribution and Pharmacokinetic Imaging. J. Phys. Chem. C 2020, 124, 19743–19752. [Google Scholar] [CrossRef]
- Tadyszak, K.; Boś-Liedke, A.; Jurga, J.; Baranowski, M.; Mrówczyński, R.; Chlewicki, W.; Jurga, S.; Czechowski, T. Overmodulation of projections as signal-to-noise enhancement method in EPR imaging. Magn. Reson. Chem. 2016, 54, 136–142. [Google Scholar] [CrossRef]
- Gonet, M.; Baranowski, M.; Czechowski, T.; Kucinska, M.; Plewinski, A.; Szczepanik, P.; Jurga, S.; Murias, M. Multiharmonic electron paramagnetic resonance imaging as an innovative approach for in vivo studies. Free Radic. Biol. Med. 2020, 152, 271–279. [Google Scholar] [CrossRef]
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front. Pharmacol. 2018, 9, 1300. [Google Scholar] [CrossRef] [Green Version]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, Z.-Y.; Wang, P.-Y.; Li, Y.-J.; Wang, R.-R.; Xie, S.-Y. Nanoplatform-based natural products co-delivery system to surmount cancer multidrug-resistant. J. Control. Release Off. J. Control. Release Soc. 2021, 336, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, W.; Chen, J.; Yuan, S.; Hu, J.; Han, B.; Huang, Y.; Zhou, W. Abnormal saccharides affecting cancer multi-drug resistance (MDR) and the reversal strategies. Eur. J. Med. Chem. 2021, 220, 113487. [Google Scholar] [CrossRef] [PubMed]
- Piwowarczyk, L.; Stawny, M.; Mlynarczyk, D.T.; Muszalska-Kolos, I.; Goslinski, T.; Jelińska, A. Role of Curcumin and (−)-Epigallocatechin-3-O-Gallate in Bladder Cancer Treatment: A Review. Cancers 2020, 12, 1801. [Google Scholar] [CrossRef]
- Tseng, Y.-H.; Chiou, S.-S.; Weng, J.-P.; Lin, P.-C. Curcumin and tetrahydrocurcumin induce cell death in Ara-C-resistant acute myeloid leukemia. Phytother. Res. 2019, 33, 1199–1207. [Google Scholar] [CrossRef] [Green Version]
- Bisdemethoxycurcumin Sensitizes Cisplatin-Resistant Lung Cancer Cells to Chemotherapy by Inhibition of CA916798 and PI3K/AKT Signaling|SpringerLink. Available online: https://link.springer.com/article/10.1007/s10495-017-1395-x (accessed on 3 August 2022).
- Mahinfar, P.; Baradaran, B.; Davoudian, S.; Vahidian, F.; Cho, W.C.-S.; Mansoori, B. Long Non-Coding RNAs in Multidrug Resistance of Glioblastoma. Genes 2021, 12, 455. [Google Scholar] [CrossRef]
- Mahinfar, P.; Mansoori, B.; Rostamzadeh, D.; Baradaran, B.; Cho, W.C.; Mansoori, B. The Role of microRNAs in Multidrug Resistance of Glioblastoma. Cancers 2022, 14, 3217. [Google Scholar] [CrossRef]
- Di Nunno, V.; Franceschi, E.; Tosoni, A.; Di Battista, M.; Gatto, L.; Lamperini, C.; Minichillo, S.; Mura, A.; Bartolini, S.; Brandes, A.A. Treatment of recurrent glioblastoma: State-of-the-art and future perspectives. Expert Rev. Anticancer Ther. 2020, 20, 785–795. [Google Scholar] [CrossRef]
- Hwang, T.W.; Kim, D.H.; Kim, D.B.; Jang, T.W.; Kim, G.-H.; Moon, M.; Yoon, K.A.; Choi, D.E.; Park, J.H.; Kim, J.-J. Synergistic anticancer effect of acteoside and temozolomide-based glioblastoma chemotherapy. Int. J. Mol. Med. 2019, 43, 1478–1486. [Google Scholar] [CrossRef]
- Jia, W.-Q.; Zhu, J.-W.; Yang, C.-Y.; Ma, J.; Pu, T.-Y.; Han, G.-Q.; Zou, M.-M.; Xu, R.-X. Verbascoside inhibits progression of glioblastoma cells by promoting Let-7g-5p and down-regulating HMGA2 via Wnt/beta-catenin signalling blockade. J. Cell. Mol. Med. 2020, 24, 2901–2916. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-J.; Pham, T.-H.; Bak, Y.; Ryu, H.-W.; Oh, S.-R.; Yoon, D.-Y. Orientin inhibits invasion by suppressing MMP-9 and IL-8 expression via the PKCα/ ERK/AP-1/STAT3-mediated signaling pathways in TPA-treated MCF-7 breast cancer cells. Phytomedicine Int. J. Phytother. Phytopharm. 2018, 50, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, F.; Shafiee, M.; Banikazemi, Z.; Pourhanifeh, M.H.; Khanbabaei, H.; Shamshirian, A.; Amiri Moghadam, S.; ArefNezhad, R.; Sahebkar, A.; Avan, A.; et al. Curcumin inhibits NF-kB and Wnt/β-catenin pathways in cervical cancer cells. Pathol. Res. Pract. 2019, 215, 152556. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.P.; Kim, H.G.; Choi, J.H.; Park, B.H.; Jeong, M.H.; Jeong, T.C.; Jeong, H.G. Acteoside inhibits PMA-induced matrix metalloproteinase-9 expression via CaMK/ERK- and JNK/NF-κB-dependent signaling. Mol. Nutr. Food Res. 2011, 55, S103–S116. [Google Scholar] [CrossRef]
- Zlokovic, B.V.; Apuzzo, M.L. Cellular and molecular neurosurgery: Pathways from concept to reality--part II: Vector systems and delivery methodologies for gene therapy of the central nervous system. Neurosurgery 1997, 40, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Thangaraj, K.; Balasubramanian, B.; Park, S.; Natesan, K.; Liu, W.; Manju, V. Orientin Induces G0/G1 Cell Cycle Arrest and Mitochondria Mediated Intrinsic Apoptosis in Human Colorectal Carcinoma HT29 Cells. Biomolecules 2019, 9, 418. [Google Scholar] [CrossRef] [Green Version]
- Rizeq, B.; Gupta, I.; Ilesanmi, J.; AlSafran, M.; Rahman, M.M.; Ouhtit, A. The Power of Phytochemicals Combination in Cancer Chemoprevention. J. Cancer 2020, 11, 4521–4533. [Google Scholar] [CrossRef]
- Jhaveri, A.; Deshpande, P.; Pattni, B.; Torchilin, V. Transferrin-targeted, resveratrol-loaded liposomes for the treatment of glioblastoma. J. Control. Release Off. J. Control. Release Soc. 2018, 277, 89–101. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Kleszcz, R.; Stasiłowicz-Krzemień, A.; Cielecka-Piontek, J. Sodium Butyrate Enhances Curcuminoids Permeability through the Blood-Brain Barrier, Restores Wnt/β-Catenin Pathway Antagonists Gene Expression and Reduces the Viability of Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 11285. [Google Scholar] [CrossRef]
- An, F.; Wang, S.; Tian, Q.; Zhu, D. Effects of orientin and vitexin from Trollius chinensis on the growth and apoptosis of esophageal cancer EC-109 cells. Oncol. Lett. 2015, 10, 2627–2633. [Google Scholar] [CrossRef]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. In Liposomes: Methods and Protocols; D’Souza, G.G.M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 17–22. ISBN 978-1-4939-6591-5. [Google Scholar]







| Compound | Particle Size (±SD) [nm] | PDI | Zeta Potential [mV] |
|---|---|---|---|
| CUR | 168.2 ± 7.5 | 0.07 | +38.8 |
| ORI | 163.2 ± 2.6 | 0.07 | +37.0 |
| CUR + ORI | 172.5 ± 0.06 | 0.09 | +37.6 |
| BDMC | 163.9 ± 5.5 | 0.07 | +37.3 |
| ACT | 187.5 ± 3.4 | 0.11 | +37.3 |
| BDMC + ACT | 166.7 ± 2.7 | 0.08 | +38.1 |
| Compound | T1 (s) | T2 (ms) |
|---|---|---|
| DOTAP:POPC | 2.7 | 5.3 |
| ACT | 2.8 | 1.6 |
| BDMC | 2.6 | 2.0 |
| BDMC + ACT | 2.7 | 3.0 |
| CUR | 2.7 | 3.7 |
| ORI | 2.6 | 3.8 |
| CUR + ORI | 2.3 | 1.2 |
| Parameters | DOTAP:POPC | ACT | BDMC + ACT | CUR | CUR + ORI | ORI |
|---|---|---|---|---|---|---|
| A [a.u.] | 0.047 | 0.053 | 0.067 | 0.111 | 0.020 | 0.029 |
| T [s] | 21.25 | 22.2 | 23.1 | 240.6 | 123.29 | 52.3 |
| y0 [a.u.] | 0.953 | 0.946 | 0.932 | 0.89 | 0.98 | 0.97 |
| IC50 [µM] | ||
|---|---|---|
| Compound | Cell Line | |
| T98G | U-138 MG | |
| CUR | 47.5 ± 3.1 | 19.0 ± 3.6 |
| ORI | nd | nd |
| CUR + ORI | nd | 24.0 ± 2.3 |
| BDMC | 39.0 ± 2.7 | 40.0 ± 3.9 |
| ACT | 85.0 ± 4.3 | 44.0 ± 4.1 |
| BDMC + ACT | nd | 69.0 ± 5.2 |
| IC50 [µM] | ||
|---|---|---|
| Compounds in Liposomes | Cell Line | |
| T98G | U-138 MG | |
| CUR | 24.0 ± 2.1 | 19.5 ± 2.4 |
| ORI | 12.0 ± 1.7 | 7.0 ± 1.5 |
| CUR + ORI | 12.5 ± 2.6 | 13.0 ± 1.4 |
| BDMC | 29.0 ± 2.2 | 28.0 ± 1.9 |
| ACT | 2.9 ± 0.9 | 4.0 ± 1.1 |
| BDMC + ACT | 23.0 ± 1.8 | 11.0 ± 1.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piwowarczyk, L.; Mlynarczyk, D.T.; Krajka-Kuźniak, V.; Majchrzak-Celińska, A.; Budzianowska, A.; Tomczak, S.; Budzianowski, J.; Woźniak-Braszak, A.; Pietrzyk, R.; Baranowski, M.; et al. Natural Compounds in Liposomal Nanoformulations of Potential Clinical Application in Glioblastoma. Cancers 2022, 14, 6222. https://doi.org/10.3390/cancers14246222
Piwowarczyk L, Mlynarczyk DT, Krajka-Kuźniak V, Majchrzak-Celińska A, Budzianowska A, Tomczak S, Budzianowski J, Woźniak-Braszak A, Pietrzyk R, Baranowski M, et al. Natural Compounds in Liposomal Nanoformulations of Potential Clinical Application in Glioblastoma. Cancers. 2022; 14(24):6222. https://doi.org/10.3390/cancers14246222
Chicago/Turabian StylePiwowarczyk, Ludwika, Dariusz T. Mlynarczyk, Violetta Krajka-Kuźniak, Aleksandra Majchrzak-Celińska, Anna Budzianowska, Szymon Tomczak, Jaromir Budzianowski, Aneta Woźniak-Braszak, Rafał Pietrzyk, Mikołaj Baranowski, and et al. 2022. "Natural Compounds in Liposomal Nanoformulations of Potential Clinical Application in Glioblastoma" Cancers 14, no. 24: 6222. https://doi.org/10.3390/cancers14246222
APA StylePiwowarczyk, L., Mlynarczyk, D. T., Krajka-Kuźniak, V., Majchrzak-Celińska, A., Budzianowska, A., Tomczak, S., Budzianowski, J., Woźniak-Braszak, A., Pietrzyk, R., Baranowski, M., Goslinski, T., & Jelinska, A. (2022). Natural Compounds in Liposomal Nanoformulations of Potential Clinical Application in Glioblastoma. Cancers, 14(24), 6222. https://doi.org/10.3390/cancers14246222









