Chemokine Receptor-Targeted Therapies: Special Case for CCR8
Abstract
:Simple Summary
Abstract
1. Introduction
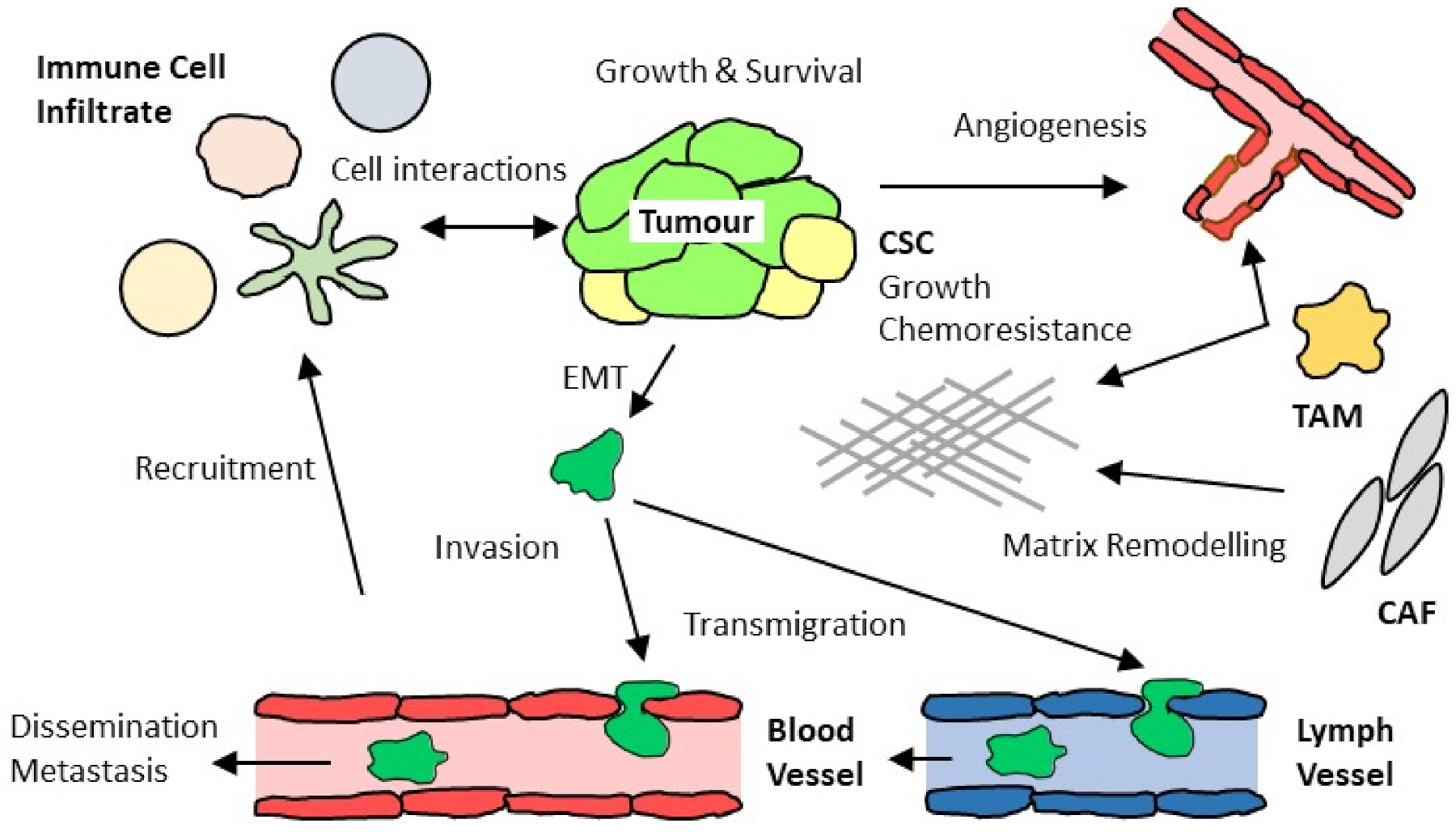
2. Complex Interactions of Chemokines with Tumor-Associated Immune and Stromal Cells
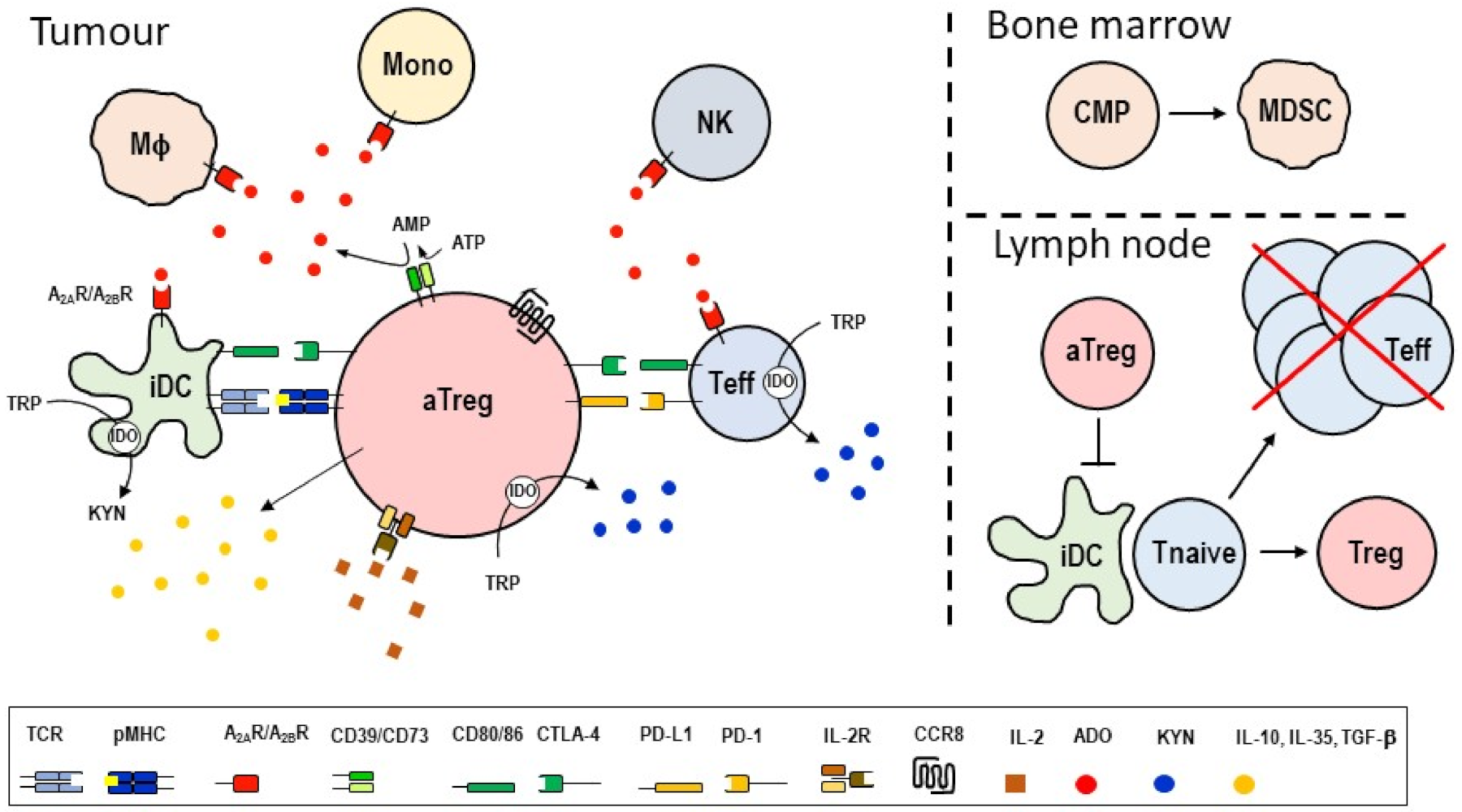
3. Treg Cell-Targeted Therapies
4. Brief History of CC Chemokine Receptor 8 (CCR8)
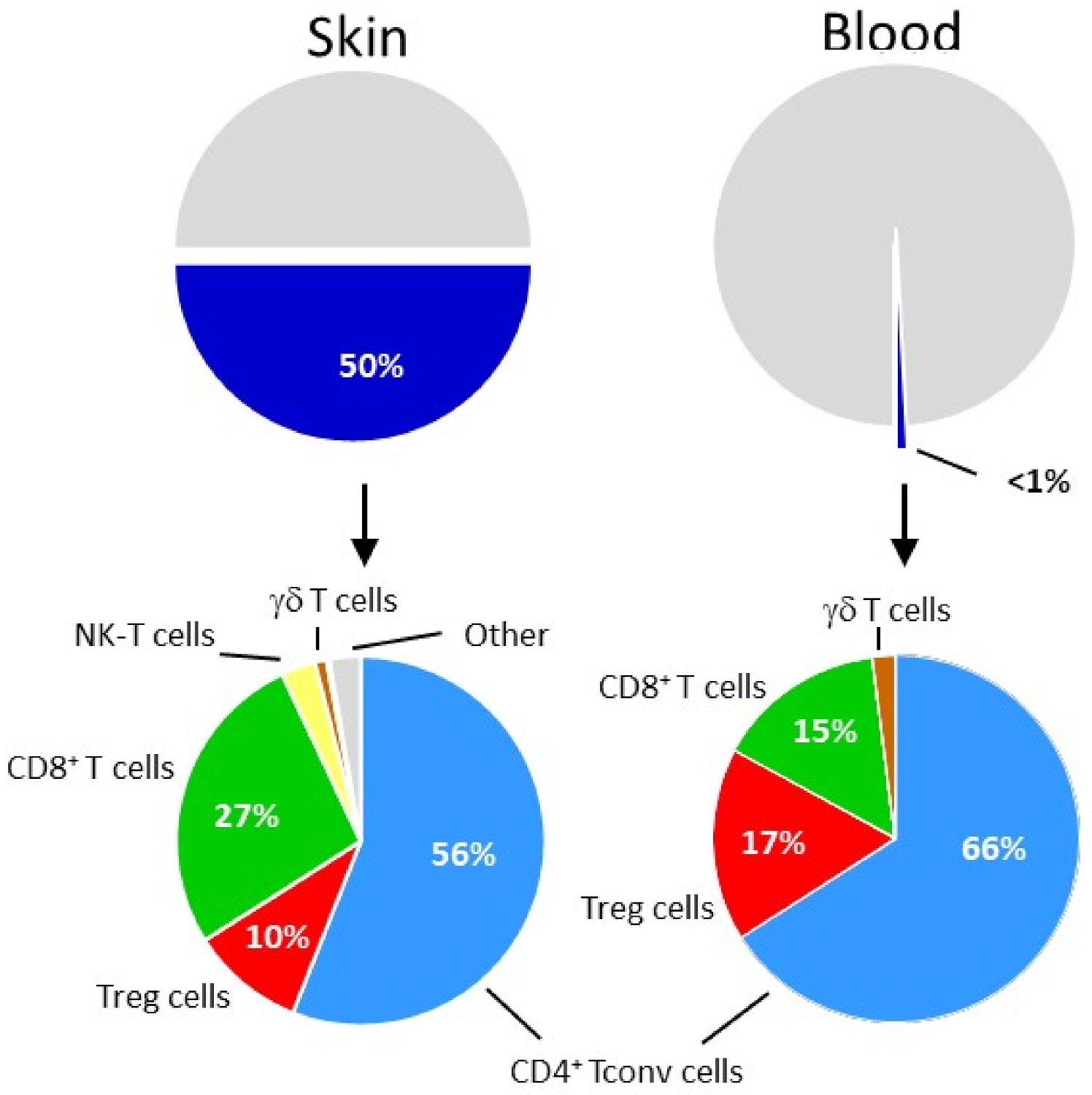
5. Relationship between Humans Skin and Tumor Treg Cells
6. CCR8 Marks Activated Intratumoral Treg Cells
7. Safety Concerns Related to CCR8-Targeted Therapies
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CCR8 | CC chemokine receptor 8 |
| CTLA-4 | cytotoxic T lymphocyte associated protein-4 |
| PD-1/PD-L1 | program death receptor-1/ program death receptor-1 ligand-1 |
| irAEs | immune-related adverse events |
| CBIs | checkpoint blockade inhibitors |
| Treg/aTreg | regulatory T cells/activated regulatory T cells |
| Tconv | conventional T cell |
| Teff | effector T cell |
| NK | natural killer cell |
| DC/iDC | dendritic cell/inhibitory dendritic cell |
| MDSC | myeloid derived suppressor cell |
| Mf | macrophage |
| CAF | cancer associated fibroblast |
| TAM | tumor associated macrophage |
| IFN-g | interferon gamma |
| TNF-a | tumor necrosis factor alpha |
| TGF-b | transforming growth factor beta |
| TRP | tryptophane |
| KYN | N-formyl-kynurenine |
| IDO | indoleamine 2,3-dioxygenase |
| EMT | epithelial mesenchymal transition |
| CSC | cancer stem cell |
References
- Moser, B.; Loetscher, P. Lymphocyte traffic control by chemokines. Nat. Immunol. 2001, 2, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and Chemokine Receptors: Positioning Cells for Host Defense and Immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCully, M.L.; Kouzeli, A.; Moser, B. Peripheral Tissue Chemokines: Homeostatic Control of Immune Surveillance T Cells. Trends Immunol. 2018, 39, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Bachelerie, F.; Ben-Baruch, A.; Burkhardt, A.M.; Combadiere, C.; Farber, J.M.; Graham, G.J.; Horuk, R.; Sparre-Ulrich, A.H.; Locati, M.; Luster, A.D.; et al. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2013, 66, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Amarandi, R.M.; Hjorto, G.M.; Rosenkilde, M.M.; Karlshoj, S. Probing Biased Signaling in Chemokine Receptors. Methods Enzymol. 2016, 570, 155–186. [Google Scholar] [CrossRef]
- Moser, B.; Wolf, M.; Walz, A.; Loetscher, P. Chemokines: Multiple levels of leukocyte migration control. Trends Immunol. 2004, 25, 75–84. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The Chemokine Superfamily Revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef] [Green Version]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Galdiero, M.R.; Marone, G.; Mantovani, A. Cancer Inflammation and Cytokines. Cold Spring Harb. Perspect. Biol. 2018, 10, a028662. [Google Scholar] [CrossRef] [Green Version]
- Shalapour, S.; Karin, M. Pas de Deux: Control of Anti-tumor Immunity by Cancer-Associated Inflammation. Immunity 2019, 51, 15–26. [Google Scholar] [CrossRef]
- Mollica Poeta, V.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef] [Green Version]
- Vilgelm, A.E.; Richmond, A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019, 10, 333. [Google Scholar] [CrossRef] [Green Version]
- Morein, D.; Erlichman, N.; Ben-Baruch, A. Beyond Cell Motility: The Expanding Roles of Chemokines and Their Receptors in Malignancy. Front. Immunol. 2020, 11, 952. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, X.Y.; Jiang, P. Cytokine and Chemokine Signals of T-Cell Exclusion in Tumors. Front. Immunol. 2020, 11, 594609. [Google Scholar] [CrossRef]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Geraud, A.; Gougis, P.; Vozy, A.; Anquetil, C.; Allenbach, Y.; Romano, E.; Funck-Brentano, E.; Moslehi, J.J.; Johnson, D.B.; Salem, J.E. Clinical Pharmacology and Interplay of Immune Checkpoint Agents: A Yin-Yang Balance. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 85–112. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Guc, E.; Pollard, J.W. Redefining macrophage and neutrophil biology in the metastatic cascade. Immunity 2021, 54, 885–902. [Google Scholar] [CrossRef]
- Sakaguchi, S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat. Immunol. 2005, 6, 345–352. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression-implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Wing, J.B.; Tanaka, A.; Sakaguchi, S. Human FOXP3(+) Regulatory T Cell Heterogeneity and Function in Autoimmunity and Cancer. Immunity 2019, 50, 302–316. [Google Scholar] [CrossRef] [Green Version]
- Raffin, C.; Vo, L.T.; Bluestone, J.A. Treg cell-based therapies: Challenges and perspectives. Nat. Rev. Immunol. 2020, 20, 158–172. [Google Scholar] [CrossRef]
- Nussing, S.; Trapani, J.A.; Parish, I.A. Revisiting T Cell Tolerance as a Checkpoint Target for Cancer Immunotherapy. Front. Immunol. 2020, 11, a589641. [Google Scholar] [CrossRef]
- Sivakumar, S.; Abu-Shah, E.; Ahern, D.J.; Arbe-Barnes, E.H.; Jainarayanan, A.K.; Mangal, N.; Reddy, S.; Rendek, A.; Easton, A.; Kurz, E.; et al. Activated Regulatory T-Cells, Dysfunctional and Senescent T-Cells Hinder the Immunity in Pancreatic Cancer. Cancers 2021, 13, 1776. [Google Scholar] [CrossRef]
- Berendt, M.J.; North, R.J. T-cell-mediated suppression of anti-tumor immunity. An explanation for progressive growth of an immunogenic tumor. J. Exp. Med. 1980, 151, 69–80. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar]
- Khattri, R.; Cox, T.; Yasayko, S.A.; Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003, 4, 337–342. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [Green Version]
- Ohue, Y.; Nishikawa, H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019, 110, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Mikami, N.; Wing, J.B.; Tanaka, A.; Ichiyama, K.; Ohkura, N. Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 2020, 38, 541–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delacher, M.; Imbusch, C.D.; Hotz-Wagenblatt, A.; Mallm, J.P.; Bauer, K.; Simon, M.; Riegel, D.; Rendeiro, A.F.; Bittner, S.; Sanderink, L.; et al. Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity 2020, 52, 295–312.e211. [Google Scholar] [CrossRef] [PubMed]
- Miragaia, R.J.; Gomes, T.; Chomka, A.; Jardine, L.; Riedel, A.; Hegazy, A.N.; Whibley, N.; Tucci, A.; Chen, X.; Lindeman, I.; et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 2019, 50, 493–504.e497. [Google Scholar] [CrossRef] [Green Version]
- Savas, P.; Virassamy, B.; Ye, C.; Salim, A.; Mintoff, C.P.; Caramia, F.; Salgado, R.; Byrne, D.J.; Teo, Z.L.; Dushyanthen, S.; et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018, 24, 986–993. [Google Scholar] [CrossRef]
- Clarke, J.; Panwar, B.; Madrigal, A.; Singh, D.; Gujar, R.; Wood, O.; Chee, S.J.; Eschweiler, S.; King, E.V.; Awad, A.S.; et al. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J. Exp. Med. 2019, 216, 2128–2149. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Pasetto, A.; Jia, L.; Deniger, D.C.; Stevanovic, S.; Robbins, P.F.; Rosenberg, S.A. Tumor-infiltrating human CD4(+) regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci. Immunol. 2019, 4, eaao4310. [Google Scholar] [CrossRef]
- Klicznik, M.M.; Morawski, P.A.; Hollbacher, B.; Varkhande, S.R.; Motley, S.J.; Kuri-Cervantes, L.; Goodwin, E.; Rosenblum, M.D.; Long, S.A.; Brachtl, G.; et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 2019, 4, eaav8995. [Google Scholar] [CrossRef]
- Fonseca, R.; Beura, L.K.; Quarnstrom, C.F.; Ghoneim, H.E.; Fan, Y.; Zebley, C.C.; Scott, M.C.; Fares-Frederickson, N.J.; Wijeyesinghe, S.; Thompson, E.A.; et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 2020, 21, 412–421. [Google Scholar] [CrossRef]
- Brunner, M.C.; Chambers, C.A.; Chan, F.K.; Hanke, J.; Winoto, A.; Allison, J.P. CTLA-4-Mediated inhibition of early events of T cell proliferation. J. Immunol. 1999, 162, 5813–5820. [Google Scholar]
- Takahashi, T.; Kuniyasu, Y.; Toda, M.; Sakaguchi, N.; Itoh, M.; Iwata, M.; Shimizu, J.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: Induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 1998, 10, 1969–1980. [Google Scholar] [CrossRef] [Green Version]
- Ohta, A.; Sitkovsky, M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature 2001, 414, 916–920. [Google Scholar] [CrossRef] [Green Version]
- Sundstrom, P.; Stenstad, H.; Langenes, V.; Ahlmanner, F.; Theander, L.; Ndah, T.G.; Fredin, K.; Borjesson, L.; Gustavsson, B.; Bastid, J.; et al. Regulatory T Cells from Colon Cancer Patients Inhibit Effector T-cell Migration through an Adenosine-Dependent Mechanism. Cancer Immunol. Res. 2016, 4, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Newton, R.C.; Friedman, S.M.; Scherle, P.A. Indoleamine 2,3-dioxygenase, an emerging target for anti-cancer therapy. Curr. Cancer Drug Targets 2009, 9, 938–952. [Google Scholar] [CrossRef]
- Yokosuka, T.; Takamatsu, M.; Kobayashi-Imanishi, W.; Hashimoto-Tane, A.; Azuma, M.; Saito, T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 2012, 209, 1201–1217. [Google Scholar] [CrossRef] [Green Version]
- Melero, I.; Berman, D.M.; Aznar, M.A.; Korman, A.J.; Perez Gracia, J.L.; Haanen, J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat. Rev. Cancer 2015, 15, 457–472. [Google Scholar] [CrossRef]
- Kubli, S.P.; Berger, T.; Araujo, D.V.; Siu, L.L.; Mak, T.W. Beyond immune checkpoint blockade: Emerging immunological strategies. Nat. Rev. Drug Discov. 2021, 20, 899–919. [Google Scholar] [CrossRef]
- Strauss, L.; Mahmoud, M.A.A.; Weaver, J.D.; Tijaro-Ovalle, N.M.; Christofides, A.; Wang, Q.; Pal, R.; Yuan, M.; Asara, J.; Patsoukis, N.; et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci. Immunol. 2020, 5, eaay1863. [Google Scholar] [CrossRef]
- De Simone, M.; Arrigoni, A.; Rossetti, G.; Gruarin, P.; Ranzani, V.; Politano, C.; Bonnal, R.J.P.; Provasi, E.; Sarnicola, M.L.; Panzeri, I.; et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity 2016, 45, 1135–1147. [Google Scholar] [CrossRef] [Green Version]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e1316. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Simons, D.L.; Lu, X.; Tu, T.Y.; Solomon, S.; Wang, R.; Rosario, A.; Avalos, C.; Schmolze, D.; Yim, J.; et al. Connecting blood and intratumoral Treg cell activity in predicting future relapse in breast cancer. Nat. Immunol. 2019, 20, 1220–1230. [Google Scholar] [CrossRef]
- Alvisi, G.; Brummelman, J.; Puccio, S.; Mazza, E.M.; Tomada, E.P.; Losurdo, A.; Zanon, V.; Peano, C.; Colombo, F.S.; Scarpa, A.; et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J. Clin. Investig. 2020, 130, 3137–3150. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhou, Q.; Zeng, H.; Zhang, H.; Liu, Z.; Shao, J.; Wang, Z.; Xiong, Y.; Wang, J.; Bai, Q.; et al. CCR8 blockade primes anti-tumor immunity through intratumoral regulatory T cells destabilization in muscle-invasive bladder cancer. Cancer Immunol. Immunother. 2020, 69, 1855–1867. [Google Scholar] [CrossRef]
- Villarreal, D.O.; L’Huillier, A.; Armington, S.; Mottershead, C.; Filippova, E.V.; Coder, B.D.; Petit, R.G.; Princiotta, M.F. Targeting CCR8 induces protective antitumor immunity and enhances vaccine-induced responses in colon cancer. Cancer Res. 2018, 78, 5340–5348. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, H.; Dombrecht, B.; Kiss, M.; Roose, H.; Allen, E.; Van Overmeire, E.; Kancheva, D.; Martens, L.; Murgaski, A.; Bardet, P.M.R.; et al. Therapeutic depletion of CCR8(+) tumor-infiltrating regulatory T cells elicits antitumor immunity and synergizes with anti-PD-1 therapy. J. Immunother. Cancer 2021, 9, e001749. [Google Scholar] [CrossRef]
- Campbell, J.R.; McDonald, B.R.; Mesko, P.B.; Siemers, N.O.; Singh, P.B.; Selby, M.; Sproul, T.W.; Korman, A.J.; Vlach, L.M.; Houser, J.; et al. Fc-Optimized Anti-CCR8 Antibody Depletes Regulatory T Cells in Human Tumor Models. Cancer Res. 2021, 81, 2983–2994. [Google Scholar] [CrossRef]
- Whiteside, S.K.; Grant, F.M.; Gyori, D.S.; Conti, A.G.; Imianowski, C.J.; Kuo, P.; Nasrallah, R.; Sadiyah, F.; Lira, S.A.; Tacke, F.; et al. CCR8 marks highly suppressive Treg cells within tumours but is dispensable for their accumulation and suppressive function. Immunology 2021, 163, 512–520. [Google Scholar] [CrossRef]
- Bhatt, D.; Kang, B.; Sawant, D.; Zheng, L.; Perez, K.; Huang, Z.; Sekirov, L.; Wolak, D.; Huang, J.Y.; Liu, X.; et al. STARTRAC analyses of scRNAseq data from tumor models reveal T cell dynamics and therapeutic targets. J. Exp. Med. 2021, 218, e20201329. [Google Scholar] [CrossRef]
- Tiffany, H.L.; Lautens, L.L.; Gao, J.L.; Pease, J.; Locati, M.; Combadiere, C.; Modi, W.; Bonner, T.I.; Murphy, P.M. Identification of CCR8: A human monocyte and thymus receptor for the CC chemokine I-309. J. Exp. Med. 1997, 186, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Roos, R.S.; Loetscher, M.; Legler, D.F.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. Identification of CCR8, the receptor for the human CC chemokine I-309. J. Biol. Chem. 1997, 272, 17251–17254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.D.; Hata, S.; De Waal Malefyt, R.; Krangel, M.S. A novel polypeptide secreted by activated human T lymphocytes. J. Immunol. 1989, 143, 2907–2916. [Google Scholar] [PubMed]
- Islam, S.A.; Ling, M.F.; Leung, J.; Shreffler, W.G.; Luster, A.D. Identification of human CCR8 as a CCL18 receptor. J. Exp. Med. 2013, 210, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Goya, I.; Gutiérrez, J.; Varona, R.; Kremer, L.; Zaballos, A.; Márquez, G. Identification of CCR8 as the specific receptor for the human b-chemokine I-309: Cloning and molecular characterization of murine CCR8 as the receptor for TCA-3. J. Immunol. 1998, 160, 1975–1981. [Google Scholar]
- Burd, P.R.; Freeman, G.J.; Wilson, S.D.; Berman, M.; DeKruyff, R.; Billings, P.R.; Dorf, M.E. Cloning and characterization of a novel T cell activation gene. J. Immunol. 1987, 139, 3126–3131. [Google Scholar]
- Islam, S.A.; Chang, D.S.; Colvin, R.A.; Byrne, M.H.; McCully, M.L.; Moser, B.; Lira, S.A.; Charo, I.F.; Luster, A.D. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat. Immunol. 2011, 12, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Chensue, S.W.; Lukacs, N.W.; Yang, T.Y.; Shang, X.; Frait, K.A.; Kunkel, S.L.; Kung, T.; Wiekowski, M.T.; Hedrick, J.A.; Cook, D.N.; et al. Aberrant in vivo T helper type 2 cell response and impaired eosinophil recruitment in CC chemokine receptor 8 knockout mice. J. Exp. Med. 2001, 193, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Goya, I.; Villares, R.; Zaballos, A.; Gutierrez, J.; Kremer, L.; Gonzalo, J.A.; Varona, R.; Carramolino, L.; Serrano, A.; Pallares, P.; et al. Absence of CCR8 does not impair the response to ovalbumin-induced allergic airway disease. J. Immunol. 2003, 170, 2138–2146. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.D.; Kuo, F.; Kumer, J.; Motani, A.S.; Lawrence, C.E.; Henderson, W.R., Jr.; Venkataraman, C. CCR8 is not essential for the development of inflammation in a mouse model of allergic airway disease. J. Immunol. 2003, 170, 581–587. [Google Scholar] [CrossRef] [Green Version]
- Gombert, M.; Dieu-Nosjean, M.C.; Winterberg, F.; Bunemann, E.; Kubitza, R.C.; Da Cunha, L.; Haahtela, A.; Lehtimaki, S.; Muller, A.; Rieker, J.; et al. CCL1-CCR8 interactions: An axis mediating the recruitment of T cells and Langerhans-type dendritic cells to sites of atopic skin inflammation. J. Immunol. 2005, 174, 5082–5091. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Perez, F.; Kato, Y.; Gonzalez-Menendez, I.; Laino, J.; Ohbayashi, M.; Burggraf, M.; Krause, M.; Kirberg, J.; Iwakura, Y.; Martella, M.; et al. CCR8 leads to eosinophil migration and regulates neutrophil migration in murine allergic enteritis. Sci. Rep. 2019, 9, 9608. [Google Scholar] [CrossRef]
- Qu, C.; Edwards, E.W.; Tacke, F.; Angeli, V.; Llodra, J.; Sanchez-Schmitz, G.; Garin, A.; Haque, N.S.; Peters, W.; Van Rooijen, N.; et al. Role of CCR8 and other chemokine pathways in the migration of monocyte-derived dendritic cells to lymph nodes. J. Exp. Med. 2004, 200, 1231–1241. [Google Scholar] [CrossRef]
- Nagao, K.; Kobayashi, T.; Moro, K.; Ohyama, M.; Adachi, T.; Kitashima, D.Y.; Ueha, S.; Horiuchi, K.; Tanizaki, H.; Kabashima, K.; et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 2012, 13, 744–752. [Google Scholar] [CrossRef] [Green Version]
- Yabe, R.; Shimizu, K.; Shimizu, S.; Azechi, S.; Choi, B.I.; Sudo, K.; Kubo, S.; Nakae, S.; Ishigame, H.; Kakuta, S.; et al. CCR8 regulates contact hypersensitivity by restricting cutaneous dendritic cell migration to the draining lymph nodes. Int. Immunol. 2015, 27, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Sokol, C.L.; Camire, R.B.; Jones, M.C.; Luster, A.D. The Chemokine Receptor CCR8 Promotes the Migration of Dendritic Cells into the Lymph Node Parenchyma to Initiate the Allergic Immune Response. Immunity 2018, 49, 449–463. [Google Scholar] [CrossRef]
- Ebert, L.M.; Meuter, S.; Moser, B. Homing and function of human skin gammadelta T cells and NK cells: Relevance for tumor surveillance. J. Immunol. 2006, 176, 4331–4336. [Google Scholar] [CrossRef]
- Schaerli, P.; Ebert, L.; Willimann, K.; Blaser, A.; Roos, R.S.; Loetscher, P.; Moser, B. A Skin-selective Homing Mechanism for Human Immune Surveillance T Cells. J. Exp. Med. 2004, 199, 1265–1275. [Google Scholar] [CrossRef]
- McCully, M.L.; Ladell, K.; Hakobyan, S.; Mansel, R.E.; Price, D.A.; Moser, B. Epidermis instructs skin homing receptor expression in human T cells. Blood 2012, 120, 4591–4598. [Google Scholar] [CrossRef] [Green Version]
- McCully, M.L.; Ladell, K.; Andrews, R.; Jones, R.E.; Miners, K.L.; Roger, L.; Baird, D.M.; Cameron, M.J.; Jessop, Z.M.; Whitaker, I.S.; et al. CCR8 Expression Defines Tissue-Resident Memory T Cells in Human Skin. J. Immunol. 2018, 200, 1639–1650. [Google Scholar] [CrossRef] [Green Version]
- Iellem, A.; Mariani, M.; Lang, R.; Recalde, H.; Panina-Bordignon, P.; Sinigaglia, F.; D’Ambrosio, D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 2001, 194, 847–853. [Google Scholar] [CrossRef] [Green Version]
- D’Ambrosio, D.; Iellem, A.; Bonecchi, R.; Mazzeo, D.; Sozzani, S.; Mantovani, A.; Sinigaglia, F. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J. Immunol. 1998, 161, 5111–5115. [Google Scholar]
- Colantonio, L.; Iellem, A.; Sinigaglia, F.; D’Ambrosio, D. Skin-homing CLA+ T cells and regulatory CD25+ T cells represent major subsets of human peripheral blood memory T cells migrating in response to CCL1/I-309. Eur. J. Immunol. 2002, 32, 3506–3514. [Google Scholar] [CrossRef]
- Sebastiani, S.; Allavena, P.; Albanesi, C.; Nasorri, F.; Bianchi, G.; Traidl, C.; Sozzani, S.; Girolomoni, G.; Cavani, A. Chemokine receptor expression and function in CD4+ T lymphocytes with regulatory activity. J. Immunol. 2001, 166, 996–1002. [Google Scholar] [CrossRef] [Green Version]
- Soler, D.; Chapman, T.R.; Poisson, L.R.; Wang, L.; Cote-Sierra, J.; Ryan, M.; McDonald, A.; Badola, S.; Fedyk, E.; Coyle, A.J.; et al. CCR8 expression identifies CD4 memory T cells enriched for FOXP3+ regulatory and Th2 effector lymphocytes. J. Immunol. 2006, 177, 6940–6951. [Google Scholar] [CrossRef] [Green Version]
- Mutalithas, K.; Guillen, C.; Raport, C.; Kolbeck, R.; Soler, D.; Brightling, C.E.; Pavord, I.D.; Wardlaw, A.J. Expression of CCR8 is increased in asthma. Clin. Exp. Allergy 2010, 40, 1175–1185. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.D.; Krangel, M.S. The human cytokine I-309 is a monocyte chemoattractant. Proc. Natl. Acad. Sci. USA 1992, 89, 2950–2954. [Google Scholar] [CrossRef] [Green Version]
- Lüttichau, H.R.; Stine, J.; Boesen, T.P.; Johnsen, A.H.; Chantry, D.; Gerstoft, J.; Schwartz, T.W. A highly selective CC chemokine receptor (CCR)8 antagonist encoded by the poxvirus molluscum contagiosum. J. Exp. Med. 2000, 191, 171–179. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Masopust, D. Tissue-resident memory T cells. Immunity 2014, 41, 886–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.O.; Kupper, T.S. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 2015, 21, 688–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, S.N.; Mackay, L.K. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 2016, 16, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Zaid, A.; Hor, J.L.; Christo, S.N.; Groom, J.R.; Heath, W.R.; Mackay, L.K.; Mueller, S.N. Chemokine Receptor-Dependent Control of Skin Tissue-Resident Memory T Cell Formation. J. Immunol. 2017, 199, 2451–2459. [Google Scholar] [CrossRef] [Green Version]
- Inngjerdingen, M.; Damaj, B.; Maghazachi, A.A. Expression and regulation of chemokine receptors in human natural killer cells. Blood 2001, 97, 367–375. [Google Scholar] [CrossRef]
- Inngjerdingen, M.; Damaj, B.; Maghazachi, A.A. Human NK cells express CC chemokine receptors 4 and 8 and respond to thymus and activation-regulated chemokine, macrophage-derived chemokine, and I-309. J. Immunol. 2000, 164, 4048–4054. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Tiffany, H.L.; King, L.; Murphy, P.M.; Golding, H.; Zaitseva, M.B. CCR8 on human thymocytes functions as a human immunodeficiency virus type 1 coreceptor. J. Virol. 2000, 74, 6946–6952. [Google Scholar] [CrossRef] [Green Version]
- Annunziato, F.; Cosmi, L.; Liotta, F.; Lazzeri, E.; Manetti, R.; Vanini, V.; Romagnani, P.; Maggi, E.; Romagnani, S. Phenotype, localization, and mechanism of suppression of CD4(+)CD25(+) human thymocytes. J. Exp. Med. 2002, 196, 379–387. [Google Scholar] [CrossRef] [Green Version]
- Cosmi, L.; Liotta, F.; Lazzeri, E.; Francalanci, M.; Angeli, R.; Mazzinghi, B.; Santarlasci, V.; Manetti, R.; Vanini, V.; Romagnani, P.; et al. Human CD8+CD25+ thymocytes share phenotypic and functional features with CD4+CD25+ regulatory thymocytes. Blood 2003, 102, 4107–4114. [Google Scholar] [CrossRef]
- Haque, N.S.; Fallon, J.T.; Pan, J.J.; Taubman, M.B.; Harpel, P.C. Chemokine receptor-8 (CCR8) mediates human vascular smooth muscle cell chemotaxis and metalloproteinase-2 secretion. Blood 2004, 103, 1296–1304. [Google Scholar] [CrossRef]
- Haque, N.S.; Fallon, J.T.; Taubman, M.B.; Harpel, P.C. The chemokine receptor CCR8 mediates human endothelial cell chemotaxis induced by I-309 and Kaposi sarcoma herpesvirus-encoded vMIP-I and by lipoprotein(a)-stimulated endothelial cell conditioned medium. Blood 2001, 97, 39–45. [Google Scholar] [CrossRef] [Green Version]
- Ruckes, T.; Saul, D.; Van Snick, J.; Hermine, O.; Grassmann, R. Autocrine antiapoptotic stimulation of cultured adult T-cell leukemia cells by overexpression of the chemokine I-309. Blood 2001, 98, 1150–1159. [Google Scholar] [CrossRef] [Green Version]
- McCully, M.L.; Collins, P.J.; Hughes, T.R.; Thomas, C.P.; Billen, J.; O’Donnell, V.B.; Moser, B. Skin Metabolites Define a New Paradigm in the Localization of Skin Tropic Memory T Cells. J. Immunol. 2015, 195, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Agace, W.W.; Persson, E.K. How vitamin A metabolizing dendritic cells are generated in the gut mucosa. Trends Immunol. 2012, 33, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Urry, Z.; Chambers, E.S.; Xystrakis, E.; Dimeloe, S.; Richards, D.F.; Gabrysova, L.; Christensen, J.; Gupta, A.; Saglani, S.; Bush, A.; et al. The role of 1alpha,25-dihydroxyvitamin D3 and cytokines in the promotion of distinct Foxp3+ and IL-10+ CD4+ T cells. Eur. J. Immunol. 2012, 42, 2697–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakdash, G.; Vogelpoel, L.T.; van Capel, T.M.; Kapsenberg, M.L.; de Jong, E.C. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. 2015, 8, 265–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Aar, A.M.; Sibiryak, D.S.; Bakdash, G.; van Capel, T.M.; van der Kleij, H.P.; Opstelten, D.J.; Teunissen, M.B.; Kapsenberg, M.L.; de Jong, E.C. Vitamin D3 targets epidermal and dermal dendritic cells for induction of distinct regulatory T cells. J. Allergy Clin. Immunol. 2011, 127, 1532–1540.e1537. [Google Scholar] [CrossRef]
- Porcuna, J.; Menendez-Gutierrez, M.P.; Ricote, M. Molecular control of tissue-resident macrophage identity by nuclear receptors. Curr. Opin. Pharmacol. 2020, 53, 27–34. [Google Scholar] [CrossRef]
- Barsheshet, Y.; Wildbaum, G.; Levy, E.; Vitenshtein, A.; Akinseye, C.; Griggs, J.; Lira, S.A.; Karin, N. CCR8+FOXp3+ Treg cells as master drivers of immune regulation. Proc. Natl. Acad. Sci. USA 2017, 114, 6086–6091. [Google Scholar] [CrossRef] [Green Version]
- Greenhough, A.; Smartt, H.J.; Moore, A.E.; Roberts, H.R.; Williams, A.C.; Paraskeva, C.; Kaidi, A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Porter, C.D.; Parkar, M.H.; Levinsky, R.J.; Collins, M.K.L.; Kinnon, C. X-linked chronic granulomatous disease: Correction of NADPH oxidase defect by retrovirus-mediated expression of gp91-phox. Blood 1993, 82, 2196–2202. [Google Scholar] [CrossRef]
- Eruslanov, E.; Stoffs, T.; Kim, W.J.; Daurkin, I.; Gilbert, S.M.; Su, L.M.; Vieweg, J.; Daaka, Y.; Kusmartsev, S. Expansion of CCR8(+) inflammatory myeloid cells in cancer patients with urothelial and renal carcinomas. Clin. Cancer Res. 2013, 19, 1670–1680. [Google Scholar] [CrossRef] [Green Version]
- Kuehnemuth, B.; Piseddu, I.; Wiedemann, G.M.; Lauseker, M.; Kuhn, C.; Hofmann, S.; Schmoeckel, E.; Endres, S.; Mayr, D.; Jeschke, U.; et al. CCL1 is a major regulatory T cell attracting factor in human breast cancer. BMC Cancer 2018, 18, 1278. [Google Scholar] [CrossRef]
- Fujikawa, M.; Koma, Y.I.; Hosono, M.; Urakawa, N.; Tanigawa, K.; Shimizu, M.; Kodama, T.; Sakamoto, H.; Nishio, M.; Shigeoka, M.; et al. Chemokine (C-C Motif) Ligand 1 Derived from Tumor-Associated Macrophages Contributes to Esophageal Squamous Cell Carcinoma Progression via CCR8-Mediated Akt/Proline-Rich Akt Substrate of 40 kDa/Mammalian Target of Rapamycin Pathway. Am. J. Pathol. 2021, 191, 686–703. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R. A revisited concept. Tumors: Wounds that do not heal. Crit. Rev. Oncol. Hematol. 2018, 128, 65–69. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef] [Green Version]
- Delacher, M.; Simon, M.; Sanderink, L.; Hotz-Wagenblatt, A.; Wuttke, M.; Schambeck, K.; Schmidleithner, L.; Bittner, S.; Pant, A.; Ritter, U.; et al. Single-cell chromatin accessibility landscape identifies tissue repair program in human regulatory T cells. Immunity 2021, 54, 702–720.e717. [Google Scholar] [CrossRef]
- Vasanthakumar, A.; Moro, K.; Xin, A.; Liao, Y.; Gloury, R.; Kawamoto, S.; Fagarasan, S.; Mielke, L.A.; Afshar-Sterle, S.; Masters, S.L.; et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 2015, 16, 276–285. [Google Scholar] [CrossRef]
- Cipolletta, D.; Feuerer, M.; Li, A.; Kamei, N.; Lee, J.; Shoelson, S.E.; Benoist, C.; Mathis, D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012, 486, 549–553. [Google Scholar] [CrossRef]
- Feuerer, M.; Herrero, L.; Cipolletta, D.; Naaz, A.; Wong, J.; Nayer, A.; Lee, J.; Goldfine, A.B.; Benoist, C.; Shoelson, S.; et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009, 15, 930–939. [Google Scholar] [CrossRef]
- Ali, N.; Zirak, B.; Rodriguez, R.S.; Pauli, M.L.; Truong, H.A.; Lai, K.; Ahn, R.; Corbin, K.; Lowe, M.M.; Scharschmidt, T.C.; et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017, 169, 1119–1129.e1111. [Google Scholar] [CrossRef] [Green Version]
- Nosbaum, A.; Prevel, N.; Truong, H.A.; Mehta, P.; Ettinger, M.; Scharschmidt, T.C.; Ali, N.H.; Pauli, M.L.; Abbas, A.K.; Rosenblum, M.D. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J. Immunol. 2016, 196, 2010–2014. [Google Scholar] [CrossRef]
- Li, A.; Herbst, R.H.; Canner, D.; Schenkel, J.M.; Smith, O.C.; Kim, J.Y.; Hillman, M.; Bhutkar, A.; Cuoco, M.S.; Rappazzo, C.G.; et al. IL-33 Signaling Alters Regulatory T Cell Diversity in Support of Tumor Development. Cell Rep. 2019, 29, 2998–3008.e2998. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, H.; Saito, Y.; Ohuchida, K.; Kawakami, E.; Fujiki, S.; Watanabe, T.; Ono, R.; Kaneko, A.; Takagi, S.; Najima, Y.; et al. Deregulated Mucosal Immune Surveillance through Gut-Associated Regulatory T Cells and PD-1(+) T Cells in Human Colorectal Cancer. J. Immunol. 2018, 200, 3291–3303. [Google Scholar] [CrossRef] [Green Version]
- Magnuson, A.M.; Kiner, E.; Ergun, A.; Park, J.S.; Asinovski, N.; Ortiz-Lopez, A.; Kilcoyne, A.; Paoluzzi-Tomada, E.; Weissleder, R.; Mathis, D.; et al. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc. Natl. Acad. Sci. USA 2018, 115, E10672–E10681. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, A.P.; Pinto, M.L.; Castro, F.; Costa, A.M.; Marques-Magalhaes, A.; Canha-Borges, A.; Cruz, T.; Velho, S.; Oliveira, M.J. The immunosuppressive and pro-tumor functions of CCL18 at the tumor microenvironment. Cytokine Growth Factor Rev. 2021, 60, 107–119. [Google Scholar] [CrossRef]
- Kumar, P.; Saini, S.; Prabhakar, B.S. Cancer immunotherapy with check point inhibitor can cause autoimmune adverse events due to loss of Treg homeostasis. Semin. Cancer Biol. 2020, 64, 29–35. [Google Scholar] [CrossRef]
- Walsh, M.J.; Dougan, M. Checkpoint blockade toxicities: Insights into autoimmunity and treatment. Semin. Immunol. 2021, 52, 101473. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, R. Immunotherapy-related adverse events (irAEs): Extraction from FDA drug labels and comparative analysis. JAMIA Open 2019, 2, 173–178. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Dissecting the mechanisms of immune checkpoint therapy. Nat. Rev. Immunol. 2020, 20, 75–76. [Google Scholar] [CrossRef]
- Yoshie, O.; Fujisawa, R.; Nakayama, T.; Harasawa, H.; Tago, H.; Izawa, D.; Hieshima, K.; Tatsumi, Y.; Matsushima, K.; Hasegawa, H.; et al. Frequent expression of CCR4 in adult T-cell leukemia and human T-cell leukemia virus type 1-transformed T cells. Blood 2002, 99, 1505–1511. [Google Scholar] [CrossRef] [Green Version]
- Imai, T.; Baba, M.; Nishimura, M.; Kakizaki, M.; Takagi, S.; Yoshie, O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J. Biol. Chem. 1997, 272, 15036–15042. [Google Scholar] [CrossRef] [Green Version]
- Yoshie, O.; Matsushima, K. CCR4 and its ligands: From bench to bedside. Int. Immunol. 2015, 27, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Blackmon, A.L.; Pinter-Brown, L. Spotlight on Mogamulizumab-Kpkc for Use in Adults with Relapsed or Refractory Mycosis Fungoides or Sezary Syndrome: Efficacy, Safety, and Patient Selection. Drug Des. Dev. Ther. 2020, 14, 3747–3754. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, J.P.; Albrecht, J.D.; Alberti-Violetti, S.; Berti, E. CCR4 in cutaneous T-cell lymphoma: Therapeutic targeting of a pathogenic driver. Eur. J. Immunol. 2021, 51, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Kasamon, Y.L.; Chen, H.; de Claro, R.A.; Nie, L.; Ye, J.; Blumenthal, G.M.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Mogamulizumab-kpkc for Mycosis Fungoides and Sezary Syndrome. Clin. Cancer Res. 2019, 25, 7275–7280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Tissue/Cells | Comments | Refs |
|---|---|---|
| Blood Tregs cells | Subset of CD4+CD25+ T cells (IHC 1) that co-express CCR4 and CLA FC 2 data with CCL1-AF or antibody reagents confirm that 40% of blood FoxP3+ Treg cells express CCR8 CCR8+ Treg cells co-express CCR4 and skin-homing receptor CLA In vitro chemotaxis to CCL1 | [77,80,82,84] |
| Skin Treg cells | Majority (>90%) of cutaneous FoxP3+ Treg cells express CCR8 and co-express CCR4 and CLA | [79] |
| Blood Tconv cells | Original data with activated Th2 cells (NB 3) FC data with CCL1-AF or antibody reagents demonstrate minor (<20%) fraction of Tconv cells that express CCR8 (FC) CCR8+CD4+ Tconv cells outnumber CCR8+CD8+ T cells by 4:1 (FC) CCR8 expressed on minor fraction of Vδ1+ γδ T cells, whereas dominant fraction of Vδ2+ γδ T cells lack CCR8 (FC) | [76,77,78,81,84] |
| Skin Tconv cells | Half of all cutaneous Tconv cells express CCR8; of these, CD4+ T cells outnumber CD8+ T cells by 2:1 (FC) Co-expression of CCR4, CXCR3 and skin homing receptor CLA (FC) Diverse cytokine expression profile by CCR8+ Tconv cells (FC) Vδ1+ γδ T cells express CCR8 (Vδ2+ γδ T cells are not present in skin) (FC) | [76,77,78,79,84] |
| Blood NK cells | Activated NK lines express CCR8 (NB, FC) and respond to CCL1 (chemotaxis) CCR8 expression rare on blood NK cells (FC) | [76,84,92,93] |
| Skin NK cells | CCR8 expression on cutaneous CD56+CD16− NK cells (FC); CD56+CD16+ fraction of NK cells, which predominate in blood, lack CCR8 (FC) | [76,79] |
| Thymocytes | CD4+CD25+ thymocytes express CCR8 (NB, FC) and migrate in response to CCL1 | [94,95,96] |
| Endothelial cells | Aortic endothelial cells express CCR8 (IHC) Human vascular endothelial cell line migrates in response to CCL1 | [97,98] |
| Cancer | Human adult T cell leukemia express CCR8 (NB) and migrate in response to CCL1 Activated Treg cells in solid tumors express CCR8 (RNASeq 4, FC), and migrate in response to CCL1 | [49,50,51,52,53,54,99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moser, B. Chemokine Receptor-Targeted Therapies: Special Case for CCR8. Cancers 2022, 14, 511. https://doi.org/10.3390/cancers14030511
Moser B. Chemokine Receptor-Targeted Therapies: Special Case for CCR8. Cancers. 2022; 14(3):511. https://doi.org/10.3390/cancers14030511
Chicago/Turabian StyleMoser, Bernhard. 2022. "Chemokine Receptor-Targeted Therapies: Special Case for CCR8" Cancers 14, no. 3: 511. https://doi.org/10.3390/cancers14030511






