Kras Gene Analysis Using Liquid-Based Cytology Specimens Predicts Therapeutic Responses and Prognosis in Patients with Pancreatic Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
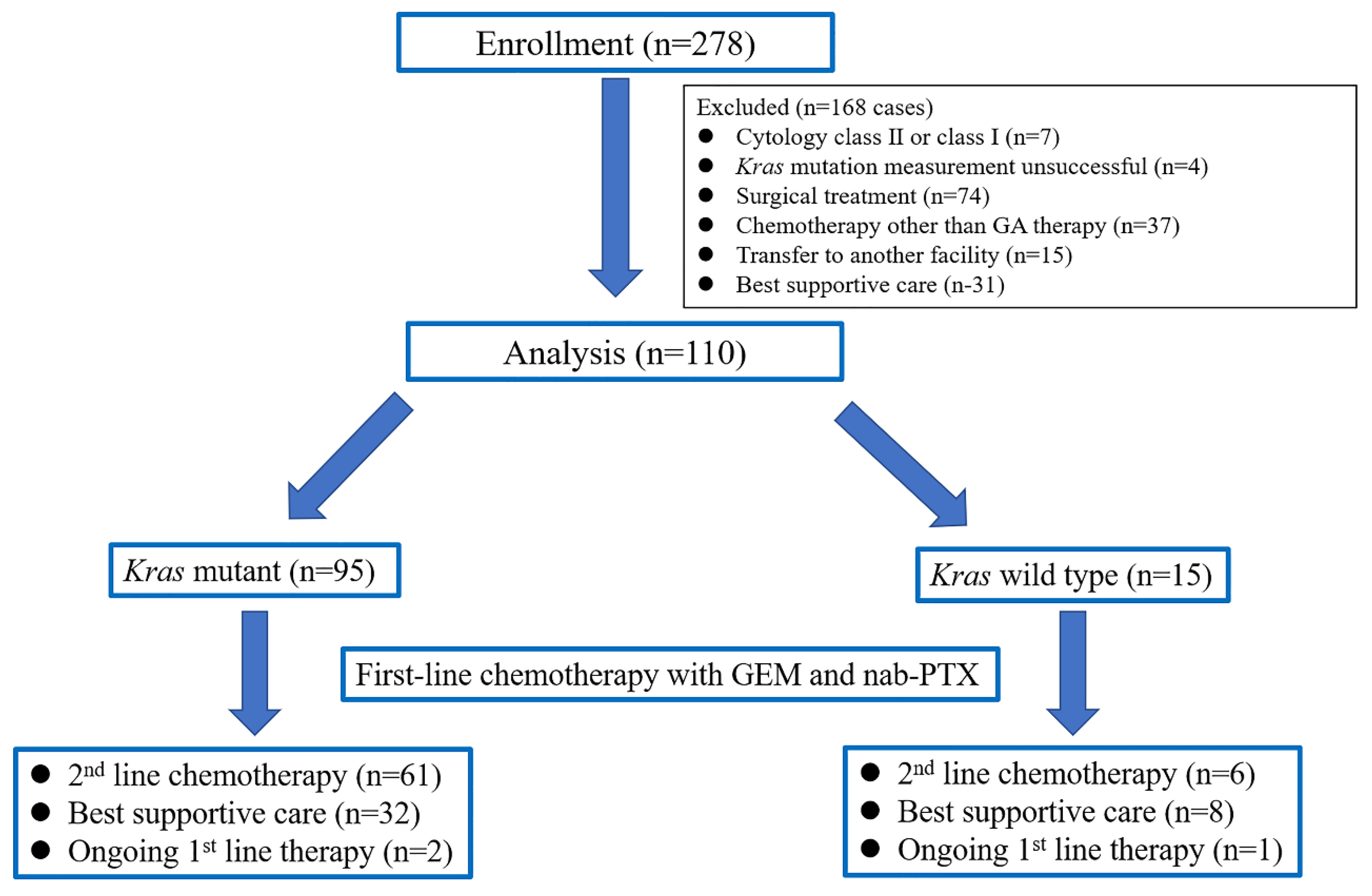
2. Materials and Methods
2.1. Study Design
2.2. Patients
2.3. EUS-FNA Procedure and Specimen Processing
2.4. DNA Extraction and Kras Mutation Analysis
2.5. Definitions
2.6. Chemotherapy
2.7. Statistical Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luchini, C.; Capelli, P.; Scarpa, A. Pancreatic Ductal Adenocarcinoma and Its Variants. Surg. Pathol. Clin. 2016, 9, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef] [Green Version]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Linder, J. Recent advances in thin-layer cytology. Diagn. Cytopathol. 1998, 18, 24–32. [Google Scholar] [CrossRef]
- Itonaga, M.; Murata, S.I.; Hatamaru, K.; Tamura, T.; Nuta, J.; Kawaji, Y.; Maekita, T.; Iguchi, M.; Kato, J.; Kojima, F.; et al. Diagnostic efficacy of smear plus liquid-based cytology for EUS-FNA of solid pancreatic lesions: A propensity-matched study. Medicine 2019, 98, e15575. [Google Scholar] [CrossRef]
- Chun, J.W.; Lee, K.; Lee, S.H.; Kim, H.; You, M.S.; Hwang, Y.J.; Paik, W.H.; Ryu, J.K.; Kim, Y.T. Comparison of liquid-based cytology with conventional smear cytology for EUS-guided FNA of solid pancreatic masses: A prospective randomized noninferiority study. Gastrointest. Endosc. 2020, 91, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K.; Kato, H.; Nouso, K.; Ako, S.; Kinugasa, H.; Horiguchi, S.; Saragai, Y.; Takada, S.; Yabe, S.; Muro, S.; et al. Evaluation of Local Recurrence of Pancreatic Cancer by KRAS Mutation Analysis Using Washes from Endoscopic Ultrasound-Guided Fine-Needle Aspiration. Dig. Dis. Sci. 2020, 65, 2907–2913. [Google Scholar] [CrossRef]
- Sekita-Hatakeyama, Y.; Nishikawa, T.; Takeuchi, M.; Morita, K.; Takeda, M.; Hatakeyama, K.; Nakai, T.; Uchiyama, T.; Itami, H.; Fujii, T.; et al. K-ras mutation analysis of residual liquid-based cytology specimens from endoscopic ultrasound-guided fine needle aspiration improves cell block diagnosis of pancreatic ductal adenocarcinoma. PLoS ONE 2018, 13, e0193692. [Google Scholar] [CrossRef]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733. [Google Scholar] [CrossRef] [Green Version]
- Tao, L.Y.; Zhang, L.F.; Xiu, D.R.; Yuan, C.H.; Ma, Z.L.; Jiang, B. Prognostic significance of K-ras mutations in pancreatic cancer: A meta-analysis. World J. Surg. Oncol. 2016, 14, 146. [Google Scholar] [CrossRef] [Green Version]
- Haas, M.; Ormanns, S.; Baechmann, S.; Remold, A.; Kruger, S.; Westphalen, C.B.; Siveke, J.T.; Wenzel, P.; Schlitter, A.M.; Esposito, I.; et al. Extended RAS analysis and correlation with overall survival in advanced pancreatic cancer. Br. J. Cancer 2017, 116, 1462–1469. [Google Scholar] [CrossRef] [Green Version]
- Akagi, K.; Arai, Y. Tm analysis method using a quenching probe is a simple and rapid way to simultaneously detect KRAS and BRAF mutations. Rinsho. Byori. 2011, 59, 757–762. [Google Scholar] [PubMed]
- Suzuki, S.; Komori, M.; Hirai, M.; Ureshino, N.; Kimura, S. Development of a novel, fully-automated genotyping system: Principle and applications. Sensors 2012, 12, 16614–16627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windon, A.L.; Loaiza-Bonilla, A.; Jensen, C.E.; Randall, M.; Morrissette, J.J.D.; Shroff, S.G. A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma. J. Gastrointest. Oncol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hingorani, S.R.; Tuveson, D.A. Ras redux: Rethinking how and where Ras acts. Curr. Opin. Genet. Dev. 2003, 13, 6–13. [Google Scholar] [CrossRef]
- Shimizu, K.; Nishiyama, T.; Hori, Y. Gemcitabine Enhances Kras-MEK-Induced Matrix Metalloproteinase-10 Expression Via Histone Acetylation in Gemcitabine-Resistant Pancreatic Tumor-initiating Cells. Pancreas 2017, 46, 268–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.W.; Lee, J.E.; Jung, K.H.; Son, M.K.; Shin, S.M.; Kim, S.J.; Fang, Z.; Yan, H.H.; Park, J.H.; Han, B.; et al. KRAS targeting antibody synergizes anti-cancer activity of gemcitabine against pancreatic cancer. Cancer Lett. 2018, 438, 174–186. [Google Scholar] [CrossRef]
- Ryu, W.J.; Han, G.; Lee, S.H.; Choi, K.Y. Suppression of Wnt/β-catenin and RAS/ERK pathways provides a therapeutic strategy for gemcitabine-resistant pancreatic cancer. Biochem. Biophys. Res. Commun. 2021, 549, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Schlitter, A.M.; Segler, A.; Steiger, K.; Michalski, C.W.; Jäger, C.; Konukiewitz, B.; Pfarr, N.; Endris, V.; Bettstetter, M.; Kong, B.; et al. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci. Rep. 2017, 7, 41064. [Google Scholar] [CrossRef] [Green Version]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; Klempner, S.J.; Hendifar, A.; Milind, J.M.; Golan, T.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted with Existing Drugs or Used as Biomarkers. Gastroenterology 2019, 156, 2242–2253. [Google Scholar] [CrossRef] [Green Version]
- Luchini, C.; Paolino, G.; Mattiolo, P.; Piredda, M.L.; Cavaliere, A.; Gaule, M.; Melisi, D.; Salvia, R.; Malleo, G.; Shin, J.I.; et al. KRAS wild-type pancreatic ductal adenocarcinoma: Molecular pathology and therapeutic opportunities. J. Exp. Clin. Cancer Res. 2020, 39, 227. [Google Scholar] [CrossRef]
- Luchini, C.; Brosens, L.A.A.; Wood, L.D.; Chatterjee, D.; Shin, J.I.; Sciammarella, C.; Fiadone, G.; Malleo, G.; Salvia, R.; Kryklyva, V.; et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: Histology, molecular pathology and clinical implications. Gut 2021, 70, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Yokose, T.; Kitago, M.; Matsuda, S.; Sasaki, Y.; Masugi, Y.; Nakamura, Y.; Shinoda, M.; Yagi, H.; Abe, Y.; Oshima, G.; et al. Combination of KRAS and SMAD4 mutations in formalin-fixed paraffin-embedded tissues as a biomarker for pancreatic cancer. Cancer Sci. 2020, 111, 2174–2182. [Google Scholar] [CrossRef]
- Kwon, H.; Kim, W.G.; Eszlinger, M.; Paschke, R.; Song, D.E.; Kim, M.; Park, S.; Jeon, M.J.; Kim, T.Y.; Shong, Y.K.; et al. Molecular Diagnosis Using Residual Liquid-Based Cytology Materials for Patients with Nondiagnostic or Indeterminate Thyroid Nodules. Endocrinol. Metab. 2016, 31, 586–591. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Qiu, T.; Guo, H.; Ying, J.; Li, J.; Zhang, Z. Detection of EGFR and KRAS gene mutations using suspension liquid-based cytology specimens in metastatic lung adenocarcinoma. Oncotarget 2017, 8, 106685–106692. [Google Scholar] [CrossRef] [Green Version]
- Park, J.K.; Lee, J.H.; Noh, D.H.; Park, J.K.; Lee, K.T.; Lee, J.K.; Lee, K.H.; Jang, K.T.; Cho, J. Factors of Endoscopic Ultrasound-Guided Tissue Acquisition for Successful Next-Generation Sequencing in Pancreatic Ductal Adenocarcinoma. Gut Liver 2020, 14, 387–394. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, M.; Irie, H.; Takagi, T.; Suzuki, R.; Konno, N.; Asama, H.; Sato, Y.; Nakamura, J.; Takasumi, M.; Hashimoto, M.; et al. Efficacy of EUS-guided FNB using a Franseen needle for tissue acquisition and microsatellite instability evaluation in unresectable pancreatic lesions. BMC Cancer 2020, 20, 1094. [Google Scholar] [CrossRef]
- Suzuki, S.; Matsusaka, S.; Hirai, M.; Shibata, H.; Takagi, K.; Mizunuma, N.; Hatake, K. A novel approach to detect KRAS/BRAF mutation for colon cancer: Highly sensitive simultaneous detection of mutations and simple pre-treatment without DNA extraction. Int. J. Oncol. 2015, 47, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indini, A.; Rijavec, E.; Ghidini, M.; Cortellini, A.; Grossi, F. Targeting KRAS in Solid Tumors: Current Challenges and Future Opportunities of Novel KRAS Inhibitors. Pharmaceutics 2021, 13, 653. [Google Scholar] [CrossRef] [PubMed]


| n = 110 | |
|---|---|
| Sex, male/female | 47/73 |
| Patient age, y, mean (range) | 67.8 (44–82) |
| Performance status 0/1 | 104/6 |
| Lesion size, mm, mean (range) | 28.8 (9.8–72) |
| Location of lesion | |
| head/body or tail | 54/56 |
| Disease status | |
| local/ metastatic | 30/80 |
| CA19-9, mg/mL | |
| <37/≥37 | 21/89 |
| Kras mutation status | |
| mutant/wild type | 95/15 |
| Amount of extracted DNA, ng/µL, mean (range) | 28.2 (1.6–217.0) |
| Second line chemotherapy * | |
| absent/present | 38/69 |
| Kras, Mutant | Kras, Wild Type | p-Value | |
|---|---|---|---|
| (n = 95) | (n = 15) | ||
| Sex, male/female, n (%) | 55/42 (57.9/42.1) | 9/6 (60/40) | 1.000 |
| Patient age, y, mean | 68.4 | 64.3 | 0.102 |
| Performance status 0/1, n (%) | 88/9 (92.6/7.4) | 14/1 (93.3/6.7) | 1.000 |
| Lesion size, mm, mean | 29.1 | 27.1 | 0.524 |
| Location of lesion | |||
| head/body or tail, n | 42/53 (44.2/55.8) | 12/3 (80/20) | 0.012 |
| Disease status | |||
| local/metastatic, n (%) | 24/71 (25.3/74.7) | 6/9 (40/60) | 0.348 |
| CA19-9, mg/mL | |||
| <37/≥37, n (%) | 19/78 (20/80) | 4/11 (26.7/73.3) | 0.480 |
| Amount of extracted DNA, ng/µL, mean | 29.4 | 20.5 | 0.384 |
| Second line chemotherapy | |||
| absent/present, n (%) | 32/61 (52.5/47.5) | 6/8 (40/60) | 0.560 |
| t | Kras, Mutant | Kras, Wild Type | p-Value | |
|---|---|---|---|---|
| (n = 95) | (n = 15) | |||
| Response, no. (%) | Complete response | 0 (0) | 0 (0) | |
| Partial response | 15 (15.8) | 3 (20) | ||
| Stable disease | 40 (42.1) | 10 (66.7) | ||
| Progressive disease | 40 (42.1) | 2 (13.3) | ||
| Rate of objective response *, no. (%) | 15 (15.8) | 3 (20) | 0.701 | |
| Rate of disease control **, no. (%) | 55 (57.9) | 13 (86.7) | 0.044 | |
| Progression-Free Survival | |||||
|---|---|---|---|---|---|
| Univariate | Multivariate | ||||
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| Sex | Female/male | 0.99 (0.67–1.45) | 0.992 | 0.94 (0.60–1.46) | 0.769 |
| Patient age, y | >70/≦70 | 0.99 (0.67–1.45) | 0.946 | 1.04 (0.68–1.60) | 0.842 |
| Performance status | 1/0 | 1.46 (0.64–3.34) | 0.368 | 1.60 (0.65–3.96) | 0.309 |
| Lesion size, mm | >20/≦20 | 1.19 (0.77–1.85) | 0.424 | 1.12 (0.69–1.81) | 0.639 |
| Location of lesion | Body or tail /head | 1.09 (0.74–1.59) | 0.665 | 1.03 (0.69–1.81) | 0.886 |
| Disease status | Local/metastatic | 0.83 (0.54–1.28) | 0.399 | 0.98 (0.61–1.57) | 0.924 |
| Kras status | Wild type/mutant | 0.56 (0.31–0.99) | 0.049 | 0.53 (0.28–0.99) | 0.045 |
| CA19-9, mg/mL | ≥37/<37 | 0.95 (0.59–1.55) | 0.850 | 0.89 (0.53–1.49) | 0.658 |
| Overall Survival | |||||
|---|---|---|---|---|---|
| Univariate | Multivariate | ||||
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| Sex | Female/male | 0.87 (0.58–1.31) | 0.560 | 1.10 (0.69–1.77) | 0.462 |
| Patient age, y | >70/≦70 | 0.93 (0.62–1.40) | 0.735 | 0.97 (0.61–1.52) | 0.853 |
| Performance status | 1/0 | 1.66 (0.72–3.81) | 0.230 | 1.33 (0.54–3.25) | 0.491 |
| Lesion size, mm | >20/≦20 | 1.26 (0.82–1.83) | 0.313 | 1.10 (0.67–1.81) | 0.860 |
| Location of lesion | Body or tail/head | 0.82 (0.55–1.83) | 0.323 | 1.16 (0.75–1.79) | 0.424 |
| Disease status | Local/metastatic | 0.60 (0.37–0.96) | 0.026 | 0.57 (0.36–0.92) | 0.048 |
| Kras status | Wild type /mutant | 0.50 (0.26–0.97) | 0.026 | 0.35 (0.16–0.74) | 0.007 |
| CA19-9, mg/mL | ≥37/<37 | 1.02 (0.60–1.74) | 0.932 | 0.62 (0.34–1.10) | 0.104 |
| Second line chemotherapy | Present /absent | 0.44 (0.29–0.68) | <0.001 | 0.20 (0.20–0.50) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Itonaga, M.; Ashida, R.; Murata, S.-I.; Yamashita, Y.; Hatamaru, K.; Tamura, T.; Kawaji, Y.; Kayama, Y.; Emori, T.; Kawai, M.; et al. Kras Gene Analysis Using Liquid-Based Cytology Specimens Predicts Therapeutic Responses and Prognosis in Patients with Pancreatic Cancer. Cancers 2022, 14, 551. https://doi.org/10.3390/cancers14030551
Itonaga M, Ashida R, Murata S-I, Yamashita Y, Hatamaru K, Tamura T, Kawaji Y, Kayama Y, Emori T, Kawai M, et al. Kras Gene Analysis Using Liquid-Based Cytology Specimens Predicts Therapeutic Responses and Prognosis in Patients with Pancreatic Cancer. Cancers. 2022; 14(3):551. https://doi.org/10.3390/cancers14030551
Chicago/Turabian StyleItonaga, Masahiro, Reiko Ashida, Shin-Ichi Murata, Yasunobu Yamashita, Keiichi Hatamaru, Takashi Tamura, Yuki Kawaji, Yuudai Kayama, Tomoya Emori, Manabu Kawai, and et al. 2022. "Kras Gene Analysis Using Liquid-Based Cytology Specimens Predicts Therapeutic Responses and Prognosis in Patients with Pancreatic Cancer" Cancers 14, no. 3: 551. https://doi.org/10.3390/cancers14030551
APA StyleItonaga, M., Ashida, R., Murata, S. -I., Yamashita, Y., Hatamaru, K., Tamura, T., Kawaji, Y., Kayama, Y., Emori, T., Kawai, M., Yamaue, H., Matsuzaki, I., Nagai, H., Kinoshita, Y., Wan, K., Shimokawa, T., & Kitano, M. (2022). Kras Gene Analysis Using Liquid-Based Cytology Specimens Predicts Therapeutic Responses and Prognosis in Patients with Pancreatic Cancer. Cancers, 14(3), 551. https://doi.org/10.3390/cancers14030551








