Predicting Glioblastoma Cellular Motility from In Vivo MRI with a Radiomics Based Regression Model
Abstract
:Simple Summary
Abstract
1. Introduction
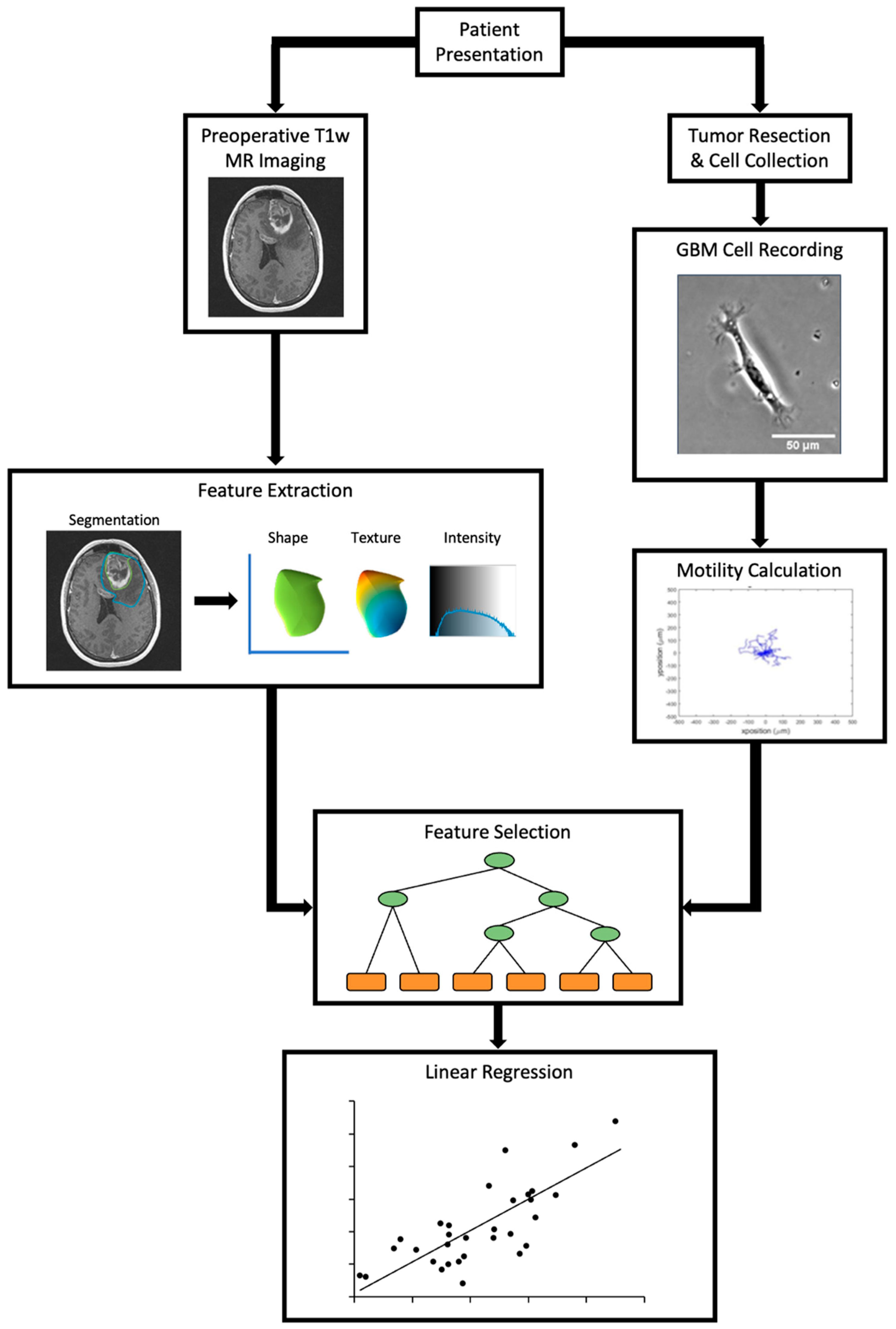
2. Materials and Methods
2.1. Patients
2.2. Live Cell Imaging and Motility Calculations
2.3. Image Feature Extraction
2.4. Feature Selection
2.5. Regression
2.6. Model Validation
3. Results
3.1. Demographics
3.2. Feature Selection
3.3. Regression
3.4. Permutation Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, B.M.; Cloughesy, T.F. Adult Glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef] [PubMed]
- Campos, B.; Olsen, L.R.; Urup, T.; Poulsen, H.S. A Comprehensive Profile of Recurrent Glioblastoma. Oncogene 2016, 35, 5819–5825. [Google Scholar] [CrossRef] [PubMed]
- Noch, E.K.; Ramakrishna, R.; Magge, R. Challenges in the Treatment of Glioblastoma: Multisystem Mechanisms of Therapeutic Resistance. World Neurosurg. 2018, 116, 505–517. [Google Scholar] [CrossRef]
- Daubon, T.; Magaut, R.; Bikfalvi, A. Tumor Cell Invasion and the Tumor Microenvironment: Special Focus on Brain Tumors. Adv. Exp. Med. Biol. 2021, 1329, 109–121. [Google Scholar]
- Lefranc, F.; Brotchi, J.; Kiss, R. Possible Future Issues in the Treatment of Glioblastomas: Special Emphasis on Cell Migration and the Resistance of Migrating Glioblastoma Cells to Apoptosis. J. Clin. Oncol. 2005, 23, 2411–2422. [Google Scholar] [CrossRef]
- Klank, R.L.; Decker Grunke, S.A.; Bangasser, B.L.; Forster, C.L.; Price, M.A.; Odde, T.J.; SantaCruz, K.S.; Rosenfeld, S.S.; Canoll, P.; Turley, E.A.; et al. Biphasic Dependence of Glioma Survival and Cell Migration on CD44 Expression Level. Cell Rep. 2017, 18, 23–31. [Google Scholar] [CrossRef]
- Fulbert, C.; Gaude, C.; Sulpice, E.; Chabardès, S. Moderate Hypothermia Inhibits Both Proliferation and Migration of Human Glioblastoma Cells. J. Neuro 2019, 144, 489–499. [Google Scholar] [CrossRef]
- Lefranc, F.; Le Rhun, E.; Kiss, R.; Weller, M. Glioblastoma Quo Vadis: Will Migration and Invasiveness Reemerge as Therapeutic Targets? Cancer Treat. Rev. 2018, 68, 145–154. [Google Scholar] [CrossRef]
- Xu, T.J.; Qiu, P.; Zhang, Y.B.; Yu, S.Y.; Xu, G.M.; Yang, W. MiR-148a Inhibits the Proliferation and Migration of Glioblastoma by Targeting ITGA9. Hum. Cell 2019, 32, 548–556. [Google Scholar] [CrossRef]
- Pace, K.R.; Dutt, R.; Galileo, D.S. Exosomal L1CAM Stimulates Glioblastoma Cell Motility, Proliferation, and Invasiveness. Int. J. Mol. Sci. 2019, 20, 3982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mega, A.; Hartmark Nilsen, M.; Leiss, L.W.; Tobin, N.P.; Miletic, H.; Sleire, L.; Strell, C.; Nelander, S.; Krona, C.; Hägerstrand, D.; et al. Astrocytes Enhance Glioblastoma Growth. Glia 2020, 68, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Stuelten, C.H.; Parent, C.A.; Montell, D.J. Cell Motility in Cancer Invasion and Metastasis: Insights from Simple Model Organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Panopoulos, A.; Howell, M.; Fotedar, R.; Margolis, R.L. Glioblastoma Motility Occurs in the Absence of Actin Polymer. Mol. Biol. Cell 2011, 22, 2212–2220. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019, 9, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Nael, K.; Bauer, A.H.; Hormigo, A.; Lemole, M.; Germano, I.M.; Puig, J.; Stea, B. Multiparametric MRI for Differentiation of Radiation Necrosis From Recurrent Tumor in Patients With Treated Glioblastoma. AJR Am. J. Roentgenol. 2018, 210, 18–23. [Google Scholar] [CrossRef]
- Soike, M.H.; McTyre, E.R.; Shah, N.; Puchalski, R.B.; Holmes, J.A.; Paulsson, A.K.; Miller, L.D.; Cramer, C.K.; Lesser, G.J.; Strowd, R.E.; et al. Glioblastoma Radiomics: Can Genomic and Molecular Characteristics Correlate with Imaging Response Patterns? Neuroradiology 2018, 60, 1043–1051. [Google Scholar] [CrossRef]
- Osman, A.F.I. A Multi-Parametric MRI-Based Radiomics Signature and a Practical ML Model for Stratifying Glioblastoma Patients Based on Survival Toward Precision Oncology. Front. Comput. Neurosci. 2019, 13, 58. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lu, D.; Gao, P.; Tian, Q.; Lu, H.; Xu, X.; He, X.; Liu, Y. Survival-Relevant High-Risk Subregion Identification for Glioblastoma Patients: The MRI-Based Multiple Instance Learning Approach. Eur. Radiol. 2020, 30, 5602–5610. [Google Scholar] [CrossRef] [PubMed]
- Bangasser, B.L.; Shamsan, G.A.; Chan, C.E.; Opoku, K.N.; Tüzel, E.; Schlichtmann, B.W.; Kasim, J.A.; Fuller, B.J.; McCullough, B.R.; Rosenfeld, S.S.; et al. Shifting the Optimal Stiffness for Cell Migration. Nat. Commun. 2017, 8, 15313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, K.J.; Kimes, R.V. Empirical Characterization of Random Forest Variable Importance Measures. Comput. Stat. Data Anal. 2008, 52, 2249–2260. [Google Scholar] [CrossRef]
- Zou, H. The Adaptive Lasso and Its Oracle Properties. J. Am. Stat. Assoc. 2006, 101, 1418–1429. [Google Scholar] [CrossRef] [Green Version]
- Phipson, B.; Smyth, G.K. Permutation P-Values Should Never Be Zero: Calculating Exact P-Values When Permutations Are Randomly Drawn. Stat. Appl. Genet. Mol. Biol. 2010, 9. [Google Scholar] [CrossRef]
- Ernst, M.D. Permutation Methods: A Basis for Exact Inference. Stat. Sci. 2004, 19, 676–685. [Google Scholar] [CrossRef]
- Wirsching, H.-G.; Galanis, E.; Weller, M. Glioblastoma. Handb. Clin. Neurol. 2016, 134, 381–397. [Google Scholar]
- Cohen, A.L.; Colman, H. Glioma Biology and Molecular Markers. Cancer Treat. Res. 2015, 163, 15–30. [Google Scholar]
- Piao, Y.; Lu, L.; de Groot, J. AMPA Receptors Promote Perivascular Glioma Invasion via β1 Integrin–dependent Adhesion to the Extracellular Matrix. Neuro-Oncology 2009, 11, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Chow, D.; Chang, P.; Weinberg, B.D.; Bota, D.A.; Grinband, J.; Filippi, C.G. Imaging Genetic Heterogeneity in Glioblastoma and Other Glial Tumors: Review of Current Methods and Future Directions. AJR Am. J. Roentgenol. 2018, 210, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Eder, K.; Kalman, B. Molecular Heterogeneity of Glioblastoma and Its Clinical Relevance. Pathol. Oncol. Res. 2014, 20, 777–787. [Google Scholar] [CrossRef] [PubMed]



| Variable | Number of Patients |
|---|---|
| Sex (female/male) | 13/18 |
| Age–Median (Range) | 57 (25–75) |
| De Novo GBM (Recurrent GBM) | 25 (6) |
| IDH Mutation | 2 (6.5%) |
| MGMT Promoter Methylation | 13 (42.0%) |
| Location | |
| Frontal Lobe | 13 (42.0%) |
| Temporal Lobe | 11 (35.5%) |
| Parietal Lobe | 5 (16.0%) |
| Occipital Lobe | 2 (6.5%) |
| Median (range) | |
| Tumor Volume (cm3) | 47.9 (0.8–118.7) |
| Variable | Bootstrap Forest Rank | Adaptive Lasso Estimate | p-Value |
|---|---|---|---|
| First-Order 10th Percentile | 1 | 1.671 | <0.0001 |
| First-Order Minimum | 2 | −4.230 | <0.0001 |
| GLRLM Gray Level Non-Uniformity | 3 | −0.00028 | <0.0001 |
| GLRLM Long-Run Low Gray-Level Emphasis | 4 | 0.00395 | <0.0001 |
| GLSZM Grey-Level Variance | 5 | 0 | 1.00 |
| GLRLM Run Percentage | 6 | 0 | 1.00 |
| GLDM High Gray-Level Emphasis | 7 | 0 | 1.00 |
| GLSZM Zone Entropy | 8 | 0 | 1.00 |
| GLCM Joint Average | 9 | 0 | 1.00 |
| GLDM Dependance Non-Uniformity | 10 | 0 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulford, K.; McMahon, M.; Gardeck, A.M.; Hunt, M.A.; Chen, C.C.; Odde, D.J.; Wilke, C. Predicting Glioblastoma Cellular Motility from In Vivo MRI with a Radiomics Based Regression Model. Cancers 2022, 14, 578. https://doi.org/10.3390/cancers14030578
Mulford K, McMahon M, Gardeck AM, Hunt MA, Chen CC, Odde DJ, Wilke C. Predicting Glioblastoma Cellular Motility from In Vivo MRI with a Radiomics Based Regression Model. Cancers. 2022; 14(3):578. https://doi.org/10.3390/cancers14030578
Chicago/Turabian StyleMulford, Kellen, Mariah McMahon, Andrew M. Gardeck, Matthew A. Hunt, Clark C. Chen, David J. Odde, and Christopher Wilke. 2022. "Predicting Glioblastoma Cellular Motility from In Vivo MRI with a Radiomics Based Regression Model" Cancers 14, no. 3: 578. https://doi.org/10.3390/cancers14030578






