Advances in and the Applicability of Machine Learning-Based Screening and Early Detection Approaches for Cancer: A Primer
Abstract
:Simple Summary
Abstract
1. Introduction
2. A Brief Introduction to Machine Learning
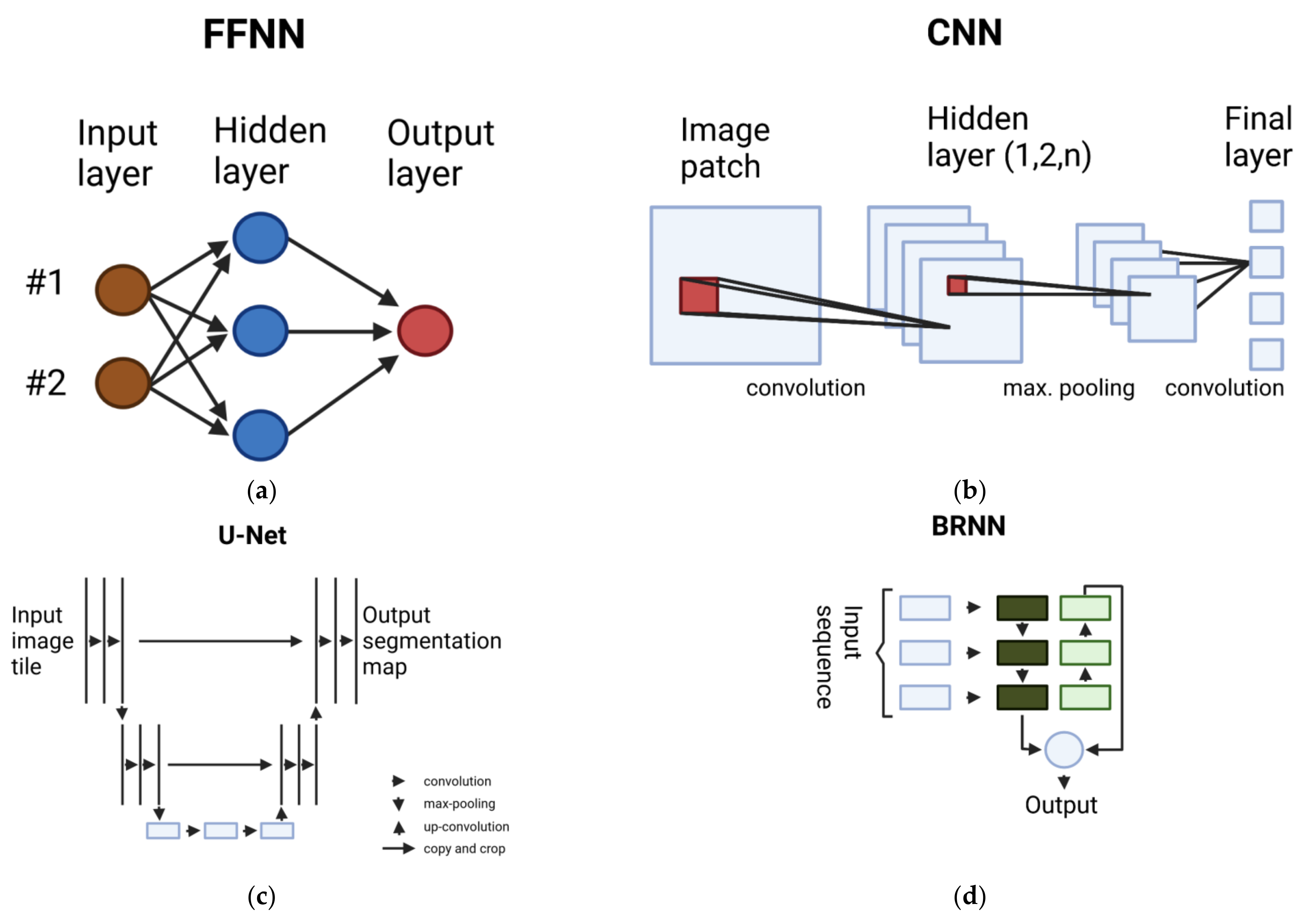
2.1. Overview of Deep Learning Methods
2.1.1. Multilayer Perceptron (MLP)
2.1.2. Convolutional Neural Network (CNN)
2.1.3. Fully Convolutional Network (FCN)
2.1.4. Recurrent Neural Network (RNN)
3. Machine Learning Applications in Cancer Diagnostics
4. A Practical Perspective on the Challenges and Limitations
4.1. Data-Driven Approaches and Human Interaction Complement Each Other
4.2. Dataset Shift and the Inhomogeneity of Patient Populations
4.3. The Clinical Applicability of Accuracy Metrics Is Limited
4.4. Prospective Evaluation and Peer-Reviewed Reporting
4.5. Safety and Protection from Adversarial Attacks
4.6. Ethical Implications for ML in Health Care
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Assessing National Capacity for the Prevention and Control of Noncommunicable Diseases: Report of the 2019 Global Survey. Available online: https://www.who.int/publications-detail-redirect/9789240002319 (accessed on 22 December 2021).
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Elmore, L.W.; Greer, S.F.; Daniels, E.C.; Saxe, C.C.; Melner, M.H.; Krawiec, G.M.; Cance, W.G.; Phelps, W.C. Blueprint for cancer research: Critical gaps and opportunities. CA Cancer J. Clin. 2021, 71, 107–139. [Google Scholar] [CrossRef] [PubMed]
- Kocarnik, J.M.; Compton, K.; Dean, F.; Fu, W.; Gaw, B.; Harvey, J.; Henrikson, H.; Lu, D.; Pennini, A.; Xu, R.; et al. The global burden of 29 cancer groups from 2010 to 2019: A systematic analysis for the Global Burden of Disease study 2019. J. Clin. Oncol. 2021, 39, 10577. [Google Scholar] [CrossRef]
- Yabroff, K.R.; Gansler, T.; Wender, R.C.; Cullen, K.J.; Brawley, O.W. Minimizing the burden of cancer in the United States: Goals for a high-performing health care system. CA Cancer J. Clin. 2019, 69, 166–183. [Google Scholar] [CrossRef] [Green Version]
- Cao, B.; Bray, F.; Ilbawi, A.; Soerjomataram, I. Effect on longevity of one-third reduction in premature mortality from non-communicable diseases by 2030: A global analysis of the Sustainable Development Goal health target. Lancet Glob. Health 2018, 6, e1288–e1296. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.; Eklund, M.; Sawaya, G.F.; Black, W.C.; Kramer, B.S.; Esserman, L.J. Population-based screening for cancer: Hope and hype. Nat. Rev. Clin. Oncol. 2016, 13, 550–565. [Google Scholar] [CrossRef]
- Weller, D.P.; Patnick, J.; McIntosh, H.M.; Dietrich, A.J. Uptake in cancer screening programmes. Lancet Oncol. 2009, 10, 693–699. [Google Scholar] [CrossRef]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and Overtreatment of Prostate Cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef] [Green Version]
- Lagies, S.; Schlimpert, M.; Neumann, S.; Waldin, A.; Kammerer, B.; Borner, C.; Peintner, L. Cells grown in three-dimensional spheroids mirror in vivo metabolic response of epithelial cells. Commun. Biol. 2020, 3, 246. [Google Scholar] [CrossRef]
- Poole, D.; Mackworth, A.; Goebel, R. Computational Intelligence: A Logical Approach. Available online: https://www.cs.ubc.ca/~poole/ci.html (accessed on 23 December 2021).
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Graves, A.; Antonoglou, I.; Wierstra, D.; Riedmiller, M. Playing Atari with Deep Reinforcement Learning. arXiv 2013, arXiv:1312.5602. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing System, Long Beach, CA, USA, 4 December 2017; pp. 6000–6010. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. arXiv 2020, arXiv:2005.14165. [Google Scholar]
- Han, L.; Kamdar, M.R. MRI to MGMT: Predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. In Biocomputing 2018; WORLD SCIENTIFIC: Singapore, 2017; pp. 331–342. [Google Scholar]
- U-Net: Convolutional Networks for Biomedical Image Segmentation. Available online: https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/ (accessed on 22 December 2021).
- Using Deep Learning Models/Convolutional Neural Networks. Available online: https://docs.ecognition.com/eCognition_documentation/User%20Guide%20Developer/8%20Classification%20-%20Deep%20Learning.htm (accessed on 22 December 2021).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Clarendon Press: Oxford, UK, 1995. [Google Scholar]
- Zhu, W.; Xie, L.; Han, J.; Guo, X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers 2020, 12, 603. [Google Scholar] [CrossRef] [Green Version]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Tran, D.; Bourdev, L.; Fergus, R.; Torresani, L.; Paluri, M. Learning Spatiotemporal Features with 3D Convolutional Networks. arXiv 2015, arXiv:1412.0767. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Graves, A.; Mohamed, A.-R.; Hinton, G. Speech Recognition with Deep Recurrent Neural Networks. arXiv 2013, arXiv:1303.5778. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Shi, B.; Bai, X.; Yao, C. An End-to-End Trainable Neural Network for Image-Based Sequence Recognition and Its Application to Scene Text Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2298–2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yang, D.M.; Rong, R.; Zhan, X.; Xiao, G. Pathology Image Analysis Using Segmentation Deep Learning Algorithms. Am. J. Pathol. 2019, 189, 1686–1698. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.; Li, C.; Wang, X.; Li, A.; Yuan, Y.; Feng, D.; Khadra, M.; Kim, J. Automatic prostate segmentation on MR images with deep network and graph model. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 635–638. [Google Scholar]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Swiderski, B.; Kurek, J.; Osowski, S.; Kruk, M.; Barhoumi, W. Deep learning and non-negative matrix factorization in recognition of mammograms. In Proceedings of the Eighth International Conference on Graphic and Image Processing (ICGIP 2016), Tokyo, Japan, 29–31 October 2016; pp. 53–59. [Google Scholar]
- Huynh, B.Q.; Li, H.; Giger, M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 2016, 3, 034501. [Google Scholar] [CrossRef]
- Ertosun, M.G.; Rubin, D.L. Probabilistic visual search for masses within mammography images using deep learning. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1310–1315. [Google Scholar]
- Kumar, V.; Webb, J.M.; Gregory, A.; Denis, M.; Meixner, D.D.; Bayat, M.; Whaley, D.H.; Fatemi, M.; Alizad, A. Automated and real-time segmentation of suspicious breast masses using convolutional neural network. PLoS ONE 2018, 13, e0195816. [Google Scholar] [CrossRef]
- Benning, L.; Peintner, A.; Finkenzeller, G.; Peintner, L. Automated spheroid generation, drug application and efficacy screening using a deep learning classification: A feasibility study. Sci. Rep. 2020, 10, 11071. [Google Scholar] [CrossRef]
- Yang, Z.; Ran, L.; Zhang, S.; Xia, Y.; Zhang, Y. EMS-Net: Ensemble of Multiscale Convolutional Neural Networks for Classification of Breast Cancer Histology Images. Neurocomputing 2019, 366, 46–53. [Google Scholar] [CrossRef]
- Vaka, A.R.; Soni, B.; Reddy, S.K. Breast cancer detection by leveraging Machine Learning. ICT Express 2020, 6, 320–324. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Wang, C.; Ecker, R.; Ellinge, I. Skin Lesion Classification Using Hybrid Deep Neural Networks. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1229–1233. [Google Scholar]
- Pomponiu, V.; Nejati, H.; Cheung, N.-M. Deepmole: Deep neural networks for skin mole lesion classification. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 2623–2627. [Google Scholar]
- Han, S.; Kang, H.-K.; Jeong, J.-Y.; Park, M.-H.; Kim, W.; Bang, W.-C.; Seong, Y.-K. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys. Med. Biol. 2017, 62, 7714–7728. [Google Scholar] [CrossRef]
- Byra, M.; Galperin, M.; Ojeda-Fournier, H.; Olson, L.; O’Boyle, M.; Comstock, C.; Andre, M. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med. Phys. 2019, 46, 746–755. [Google Scholar] [CrossRef]
- Hua, K.-L.; Hsu, C.-H.; Hidayati, S.C.; Cheng, W.-H.; Chen, Y.-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015, 8, 2015–2022. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Elazab, A.; Wu, J.; Hu, Q. Lung nodule classification using deep feature fusion in chest radiography. Comput. Med. Imaging Graph. 2017, 57, 10–18. [Google Scholar] [CrossRef]
- Hussein, S.; Gillies, R.; Cao, K.; Song, Q.; Bagci, U. TumorNet: Lung nodule characterization using multi-view Convolutional Neural Network with Gaussian Process. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 1007–1010. [Google Scholar]
- Jiao, W.; Atwal, G.; Polak, P.; Karlic, R.; Cuppen, E.; Danyi, A.; de Ridder, J.; van Herpen, C.; Lolkema, M.P.; Steeghs, N.; et al. A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat. Commun. 2020, 11, 728. [Google Scholar] [CrossRef]
- Gao, F.; Wang, W.; Tan, M.; Zhu, L.; Zhang, Y.; Fessler, E.; Vermeulen, L.; Wang, X. DeepCC: A novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xing, Y.; Sun, K.; Guo, Y. OmiEmbed: A Unified Multi-Task Deep Learning Framework for Multi-Omics Data. Cancers 2021, 13, 3047. [Google Scholar] [CrossRef]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLOS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef] [PubMed]
- Korfiatis, P.; Kline, T.L.; Lachance, D.H.; Parney, I.F.; Buckner, J.C.; Erickson, B.J. Residual Deep Convolutional Neural Network Predicts MGMT Methylation Status. J. Digit. Imaging 2017, 30, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Al-antari, M.A.; Han, S.-M.; Kim, T.-S. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput. Methods Programs Biomed. 2020, 196, 105584. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Yu, L.; Qin, J.; Heng, P.-A. Multilevel Contextual 3-D CNNs for False Positive Reduction in Pulmonary Nodule Detection. IEEE Trans. Biomed. Eng. 2017, 64, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Zhu, X.; Garmire, L.X. Cox-nnet: An artificial neural network method for prognosis prediction of high-throughput omics data. PLOS Comput. Biol. 2018, 14, e1006076. [Google Scholar] [CrossRef] [PubMed]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Asada, K.; Takasawa, K.; Shimoyama, R.; Sakai, A.; Bolatkan, A.; Shinkai, N.; Kobayashi, K.; Komatsu, M.; Kaneko, S.; et al. Predicting Deep Learning Based Multi-Omics Parallel Integration Survival Subtypes in Lung Cancer Using Reverse Phase Protein Array Data. Biomolecules 2020, 10, 1460. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Mitchel, J.; Chatlin, K.; Wang, M.D. Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Med. Inform. Decis. Mak. 2020, 20, 225. [Google Scholar] [CrossRef]
- Lai, Y.-H.; Chen, W.-N.; Hsu, T.-C.; Lin, C.; Tsao, Y.; Wu, S. Overall survival prediction of non-small cell lung cancer by integrating microarray and clinical data with deep learning. Sci. Rep. 2020, 10, 4679. [Google Scholar] [CrossRef]
- Courtiol, P.; Maussion, C.; Moarii, M.; Pronier, E.; Pilcer, S.; Sefta, M.; Manceron, P.; Toldo, S.; Zaslavskiy, M.; Le Stang, N.; et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 2019, 25, 1519–1525. [Google Scholar] [CrossRef]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Brit. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef]
- Heo, S.H.; Jeong, E.S.; Lee, K.S.; Seo, J.H.; Jeong, D.G.; Won, Y.S.; Kwon, H.J.; Kim, H.C.; Kim, D.Y.; Choi, Y.K. Canonical Wnt signaling pathway plays an essential role in N-methyl-N-nitrosurea induced gastric tumorigenesis of mice. J. Vet. Med. Sci. 2013, 75, 299–307. [Google Scholar] [CrossRef] [Green Version]
- DeepVariant: Highly Accurate Genomes With Deep Neural Networks. Google AI Blog. Available online: https://ai.googleblog.com/2017/12/deepvariant-highly-accurate-genomes.html (accessed on 1 December 2021).
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef]
- Singh, R.; Lanchantin, J.; Robins, G.; Qi, Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016, 32, i639–i648. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wu, M.; Liu, Q.; Lv, H.; Jiang, R. DeepHistone: A deep learning approach to predicting histone modifications. BMC Genom. 2019, 20, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.M.; Kerlikowske, K.; Tosteson, A.N.A.; Miglioretti, D.L.; Breast Cancer Surveillance Consortium. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef]
- Sayres, R.; Taly, A.; Rahimy, E.; Blumer, K.; Coz, D.; Hammel, N.; Krause, J.; Narayanaswamy, A.; Rastegar, Z.; Wu, D.; et al. Using a Deep Learning Algorithm and Integrated Gradients Explanation to Assist Grading for Diabetic Retinopathy. Ophthalmology 2019, 126, 552–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nestor, B.; McDermott, M.B.A.; Chauhan, G.; Naumann, T.; Hughes, M.C.; Goldenberg, A.; Ghassemi, M. Rethinking clinical prediction: Why machine learning must consider year of care and feature aggregation. arXiv 2018, arXiv:1811.12583. [Google Scholar]
- Marcus, G. Deep Learning: A Critical Appraisal. arXiv 2018, arXiv:1801.00631. [Google Scholar]
- Eykholt, K.; Evtimov, I.; Fernandes, E.; Li, B.; Rahmati, A.; Xiao, C.; Prakash, A.; Kohno, T.; Song, D. Robust Physical-World Attacks on Deep Learning Visual Classification. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1625–1634. [Google Scholar]
- Park, J.H.; Shin, J.; Fung, P. Reducing Gender Bias in Abusive Language Detection. In Proceedings of the EMNLP 2018, Brussels, Belgium, 31 October–4 November 2018; pp. 2799–2804. [Google Scholar]
- Shah, N.H.; Milstein, A.; Bagley, P.; Steven, C. Making Machine Learning Models Clinically Useful. JAMA 2019, 322, 1351–1352. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD). Circulation 2015, 131, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Keane, P.A.; Topol, E.J. With an eye to AI and autonomous diagnosis. NPJ Digit. Med. 2018, 1, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, G.S.; Moons, K.G.M. Reporting of artificial intelligence prediction models. Lancet 2019, 393, 1577–1579. [Google Scholar] [CrossRef]
- Komkov, S.; Petiushko, A. AdvHat: Real-world adversarial attack on ArcFace Face ID system. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 819–826. [Google Scholar] [CrossRef]
- Cisse, M.; Adi, Y.; Neverova, N.; Keshet, J. Houdini: Fooling Deep Structured Prediction Models. arXiv 2017, arXiv:1707.05373. [Google Scholar]
- Finlayson, S.G.; Bowers, J.D.; Ito, J.; Zittrain, J.L.; Beam, A.L.; Kohane, I.S. Adversarial attacks on medical machine learning. Science 2019, 363, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Office of the Commissioner. U.S. Food and Drug Administration. Available online: https://www.fda.gov/home (accessed on 23 November 2021).
- Harvey, H.B.; Gowda, V. How the FDA Regulates AI. Acad. Radiol. 2020, 27, 58–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hintze, M.; El Emam, K. Comparing the benefits of pseudonymisation and anonymisation under the GDPR. J. Data Prot. Priv. 2018, 2, 145–158. [Google Scholar]
- Ghassemi, M.; Oakden-Rayner, L.; Beam, A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 2021, 3, e745–e750. [Google Scholar] [CrossRef]
- Peccoud, J.; Gallegos, J.E.; Murch, R.; Buchholz, W.G.; Raman, S. Cyberbiosecurity: From Naive Trust to Risk Awareness. Trends Biotechnol. 2018, 36, 4–7. [Google Scholar] [CrossRef]
- Global Partnership Against the Spread of Weapons and Materials of Mass Destruction. Available online: https://www.gpwmd.com/ (accessed on 12 December 2021).
| Task | Type of Data | ML Method | Disease Spectrum | References |
|---|---|---|---|---|
| Segmentation | MRI images | 3D-CNN, CNN | Brain tumour, prostate cancer | [37,38] |
| Mammograms | CNN | Breast cancer | [39,40,41] | |
| Ultrasound images | U-Net (FCN) | Breast cancer | [42] | |
| Classification | Histological images | CNN, CRNN | Breast cancer, colorectal cancer | [43,44,45] |
| Dermoscopic segmentation | CNN | Skin lesions | [46,47] | |
| Ultrasound images | CNN | Breast cancer | [48,49] | |
| (Volumetric) CT scans, slides | CNN | Lung cancer | [50,51,52] | |
| OMICs, multi-OMICs | DNN | Various | [53,54,55] | |
| H&E images, slides | CNN | Colorectal cancer | [56] | |
| MRI images | CNN, CRNN | Brain tumour | [19,57] | |
| Mammograms | CNN | Breast cancer | [58] | |
| Detection | Mammograms | CNN | Breast cancer | [58] |
| CT scans | 3D-CNN | Lung cancer | [59] | |
| Prognostic | OMICs, multi-OMICs | DNN | Various | [55,60,61,62,63,64] |
| Histological images | CNN | Soft tissue cancers | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benning, L.; Peintner, A.; Peintner, L. Advances in and the Applicability of Machine Learning-Based Screening and Early Detection Approaches for Cancer: A Primer. Cancers 2022, 14, 623. https://doi.org/10.3390/cancers14030623
Benning L, Peintner A, Peintner L. Advances in and the Applicability of Machine Learning-Based Screening and Early Detection Approaches for Cancer: A Primer. Cancers. 2022; 14(3):623. https://doi.org/10.3390/cancers14030623
Chicago/Turabian StyleBenning, Leo, Andreas Peintner, and Lukas Peintner. 2022. "Advances in and the Applicability of Machine Learning-Based Screening and Early Detection Approaches for Cancer: A Primer" Cancers 14, no. 3: 623. https://doi.org/10.3390/cancers14030623
APA StyleBenning, L., Peintner, A., & Peintner, L. (2022). Advances in and the Applicability of Machine Learning-Based Screening and Early Detection Approaches for Cancer: A Primer. Cancers, 14(3), 623. https://doi.org/10.3390/cancers14030623






