Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Non-Lymphoma Cohort
2.2. Imaging Data
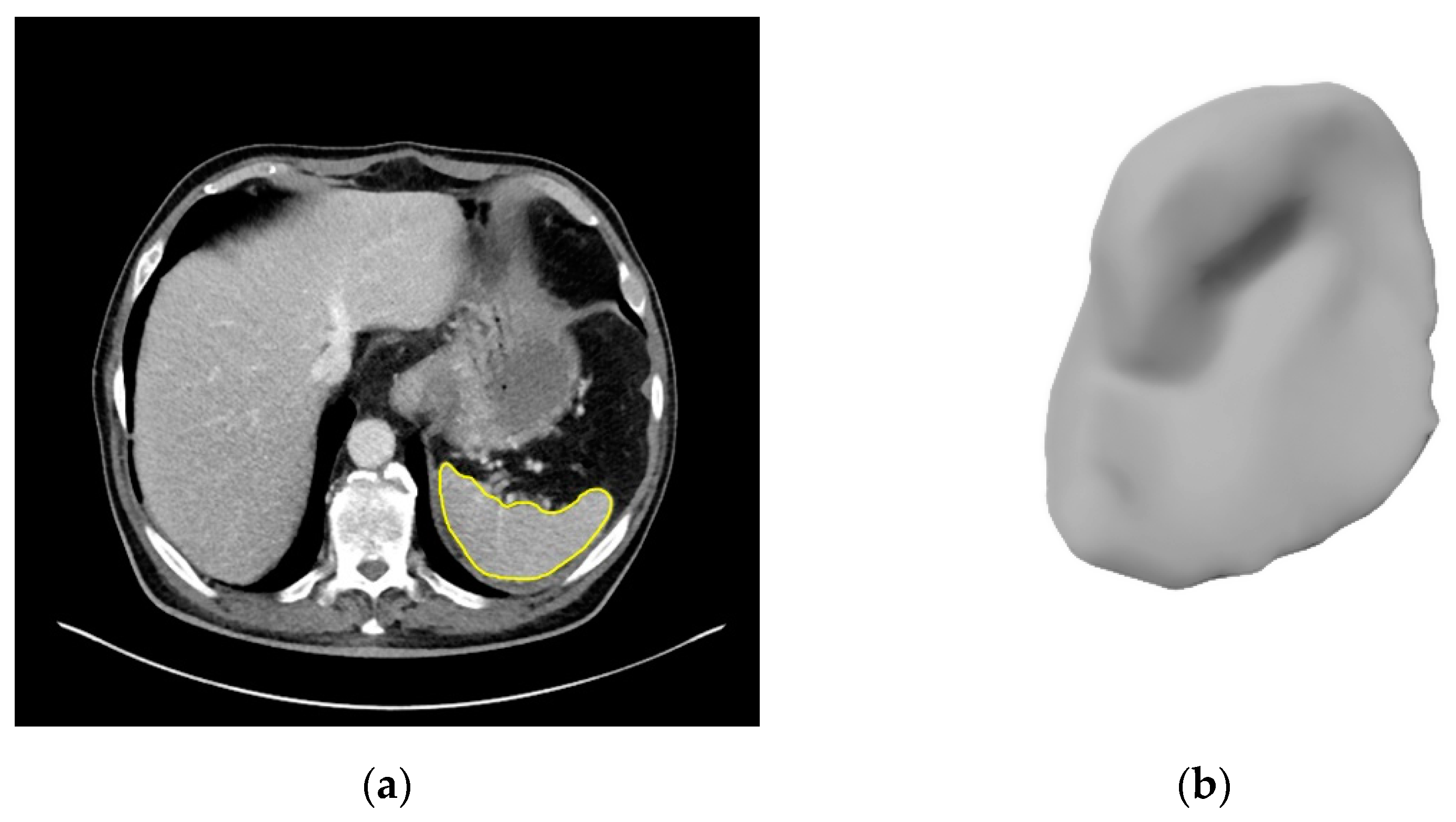
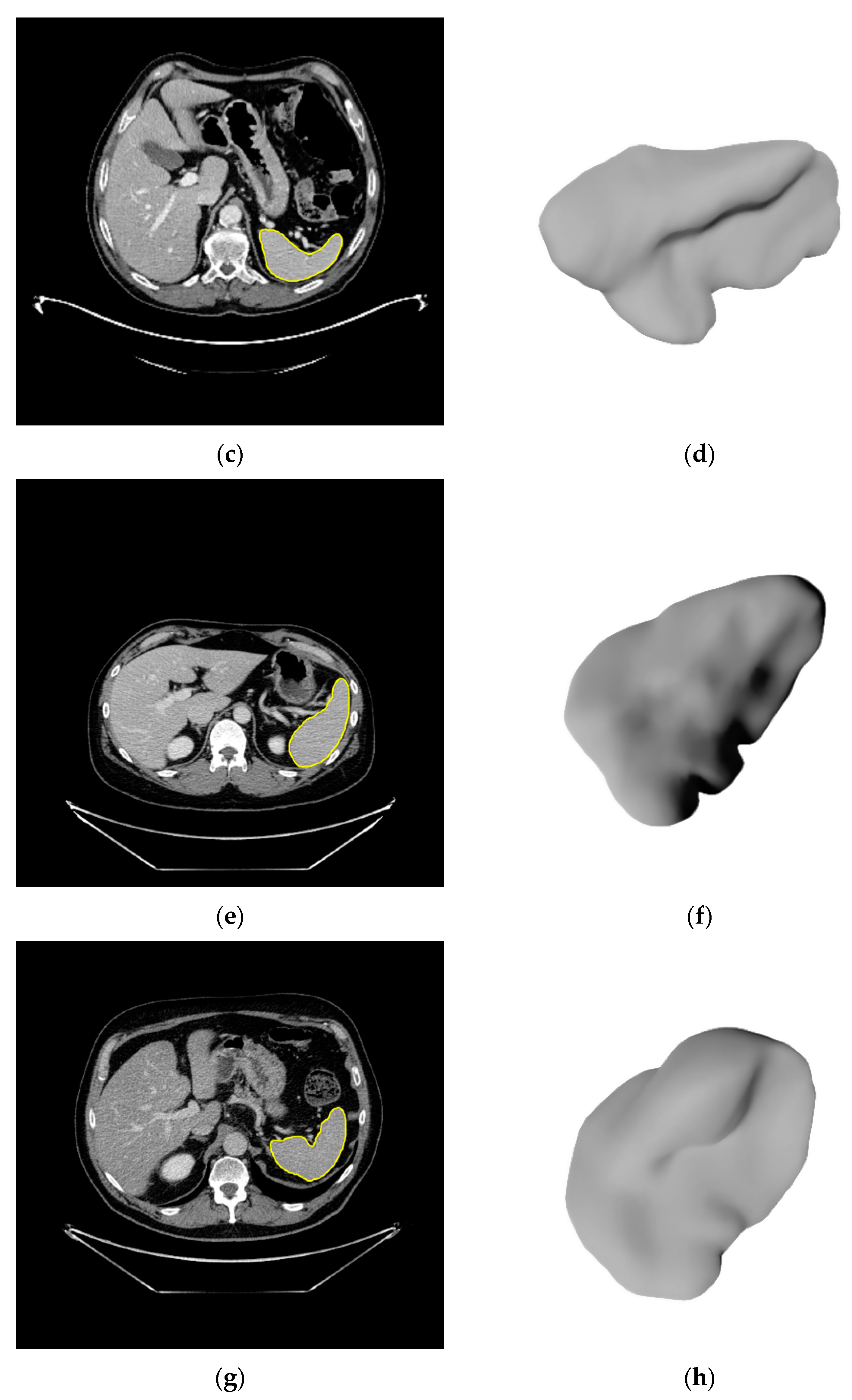
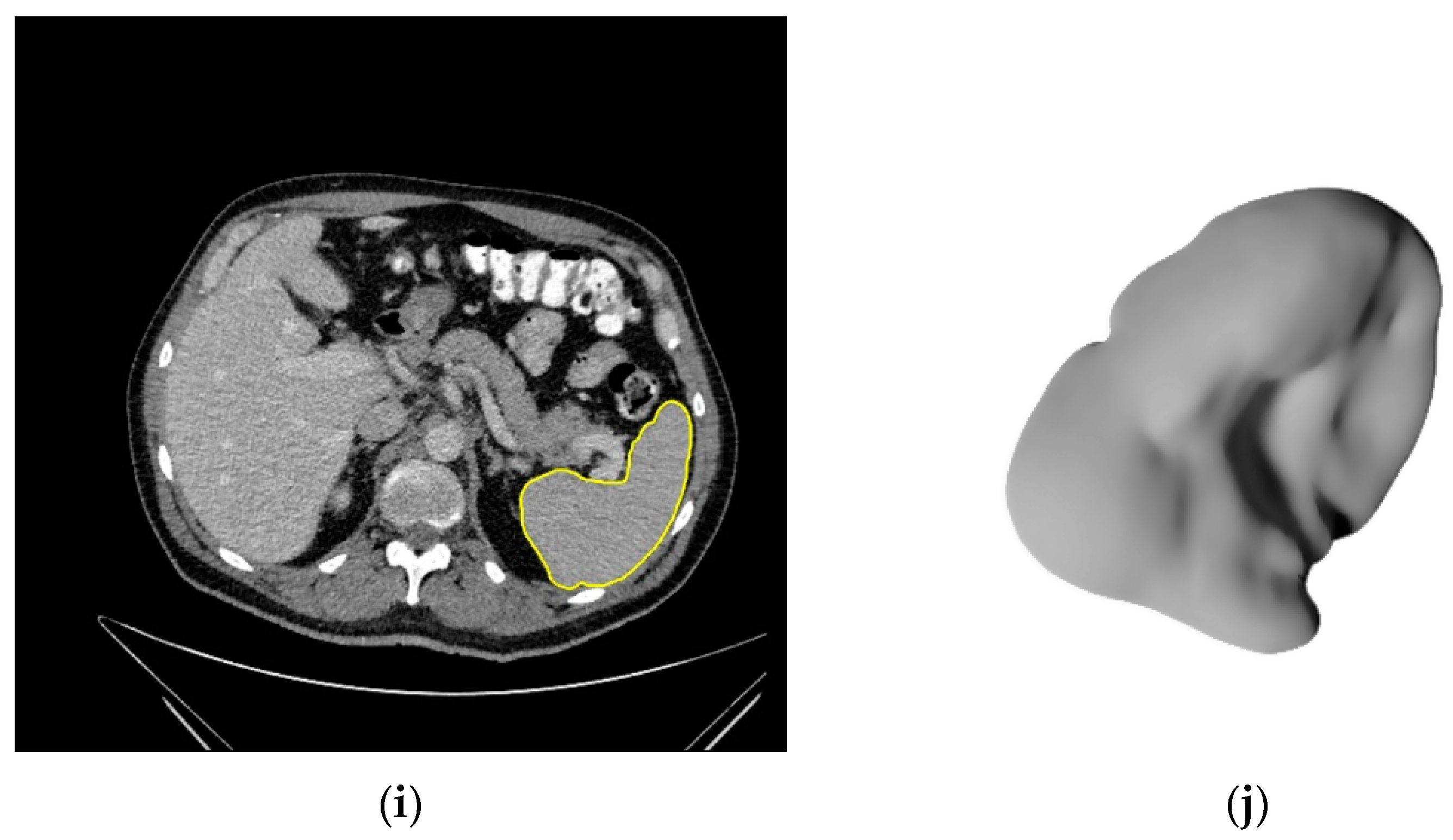
2.3. Image Segmentation
2.4. Statistical Analysis
2.5. Feature Extraction
2.6. Model Building and Analysis
3. Results
3.1. Demographic Data
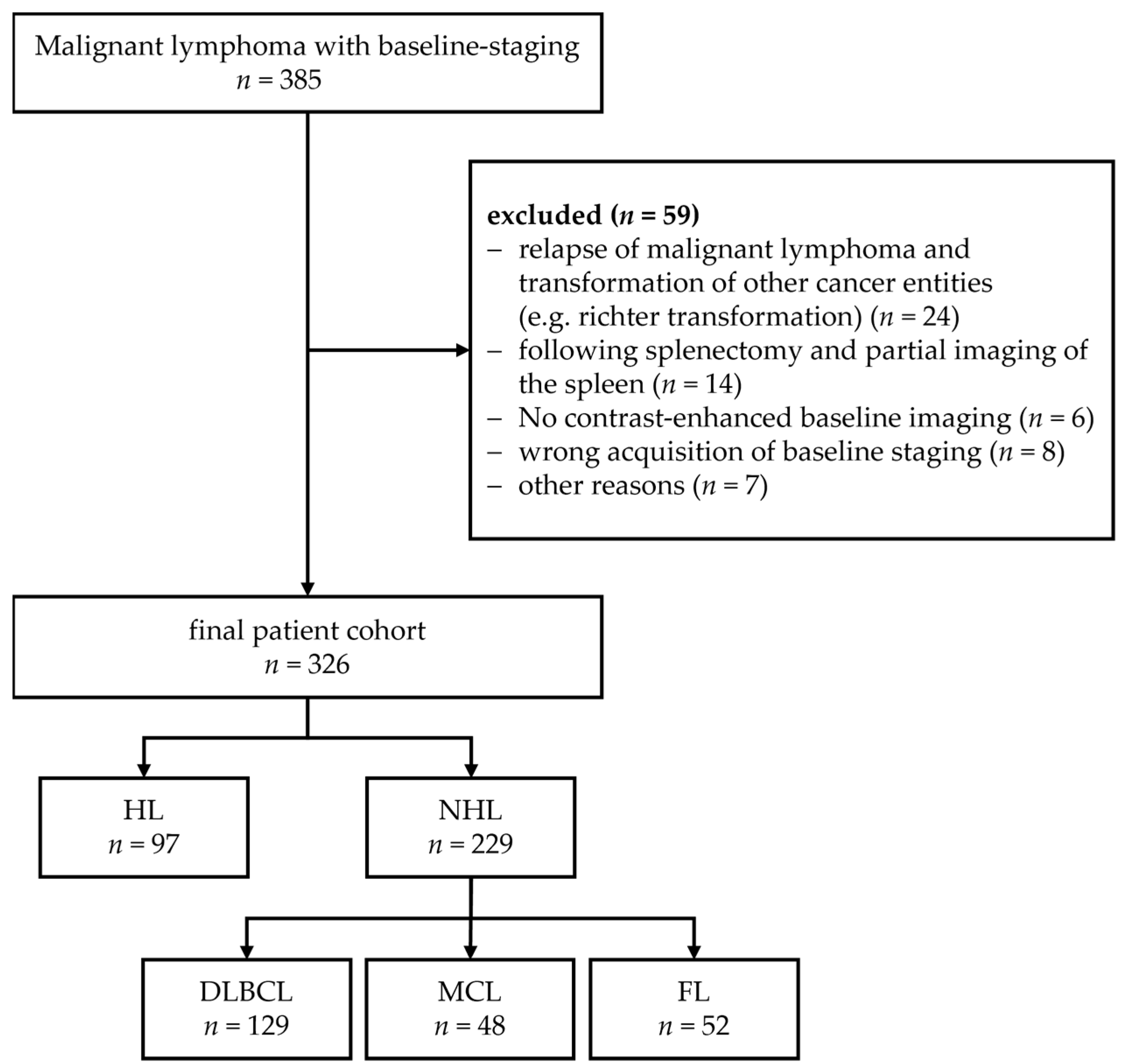
3.2. Segmentations of the Spleen
3.3. Classifying Malignant Lymphoma vs. Non-Lymphoma
3.4. Subtype Prediction in Malignant Lymphoma
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guermazi, A.; Brice, P.; de Kerviler, E.; Fermé, C.; Hennequin, C.; Meignin, V.; Frija, J. Extranodal Hodgkin Disease: Spectrum of Disease. Radiographics 2001, 21, 161–179. [Google Scholar] [CrossRef] [PubMed]
- De Jong, P.A.; Van Ufford, H.M.Q.; Baarslag, H.J.; De Haas, M.J.; Wittebol, S.H.; Quekel, L.G.; De Klerk, J.M. CT and 18F-FDG PET for noninvasive detection of splenic involvement in patients with malignant lymphoma. Am. J. Roentgenol. 2009, 192, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, K.; Sahdev, A.; Reznek, R.H. Lymphoma of the Spleen. Semin. Ultrasound, CT MRI 2007, 28, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Saboo, S.; Krajewski, K.M.; O’Regan, K.N.; Giardino, A.; Brown, J.R.; Ramaiya, N.; Jagannathan, J.P. Spleen in haematological malignancies: Spectrum of imaging findings. Br. J. Radiol. 2012, 85, 81–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Even-Sapir, E.; Lievshitz, G.; Perry, C.; Herishanu, Y.; Lerman, H.; Metser, U. Fluorine-18 Fluorodeoxyglucose PET/CT Patterns of Extranodal Involvement in Patients with Non-Hodgkin Lymphoma and Hodgkin’s Disease. Radiol. Clin. N. Am. 2007, 45, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Barrington, S.F.; Mikhaeel, N.G.; Kostakoglu, L.; Meignan, M.; Hutchings, M.; Müeller, S.P.; Schwartz, L.H.; Zucca, E.; Fisher, R.I.; Trotman, J.; et al. Role of imaging in the staging and response assessment of lymphoma: Consensus of the international conference on malignant lymphomas imaging working group. J. Clin. Oncol. 2014, 32, 3048–3058. [Google Scholar] [CrossRef] [PubMed]
- Cheson, B.D.; Fisher, R.I.; Barrington, S.F.; Cavalli, F.; Schwartz, L.H.; Zucca, E.; Lister, T.A. Recommendations for initial evaluation, staging, and response assessment of hodgkin and non-hodgkin lymphoma: The lugano classification. J. Clin. Oncol. 2014, 32, 3059–3067. [Google Scholar] [CrossRef]
- Cheson, B.D.; Pfistner, B.; Juweid, M.E.; Gascoyne, R.D.; Specht, L.; Horning, S.J.; Coiffier, B.; Fisher, R.I.; Hagenbeek, A.; Zucca, E.; et al. Revised response criteria for malignant lymphoma. J. Clin. Oncol. 2007, 25, 579–586. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4644. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, M.; Nakaura, T.; Namimoto, T.; Kitajima, M.; Uetani, H.; Tateishi, M.; Oda, S.; Utsunomiya, D.; Makino, K.; Nakamura, H.; et al. Machine learning based on multi-parametric magnetic resonance imaging to differentiate glioblastoma multiforme from primary cerebral nervous system lymphoma. Eur. J. Radiol. 2018, 108, 147–154. [Google Scholar] [CrossRef]
- Ba-Ssalamah, A.; Muin, D.; Schernthaner, R.; Kulinna-Cosentini, C.; Bastati, N.; Stift, J.; Gore, R.; Mayerhoefer, M.E. Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur. J. Radiol. 2013, 82, e537–e543. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, B.; Miles, K.A.; Babikir, S.; Shortman, R.; Afaq, A.; Ardeshna, K.M.; Groves, A.M.; Kayani, I. CT-based texture analysis potentially provides prognostic information complementary to interim fdg-pet for patients with hodgkin’s and aggressive non-hodgkin’s lymphomas. Eur. Radiol. 2017, 27, 1012–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebauer, L.; Moltz, J.; Mühlberg, A.; Holch, J.; Huber, T.; Enke, J.; Jäger, N.; Haas, M.; Kruger, S.; Boeck, S.; et al. Quantitative Imaging Biomarkers of the Whole Liver Tumor Burden Improve Survival Prediction in Metastatic Pancreatic Cancer. Cancers 2021, 13, 5732. [Google Scholar] [CrossRef]
- Mühlberg, A.; Holch, J.W.; Heinemann, V.; Huber, T.; Moltz, J.; Maurus, S.; Jäger, N.; Liu, L.; Froelich, M.F.; Katzmann, A.; et al. The relevance of CT-based geometric and radiomics analysis of whole liver tumor burden to predict survival of patients with metastatic colorectal cancer. Eur. Radiol. 2021, 31, 834–846. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Fang, M.; Huang, Y.; He, L.; Chen, X.; Liang, C.; Huang, X.; Cheng, Z.; Dong, D.; Liang, C.; et al. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. Eur. J. Radiol. 2017, 91, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, M.; He, D.; Wei, Y.; Yu, H.; Wang, Y.; Yuan, F.; Song, B. Two-dimensional Texture Analysis Based on CT Images to Differentiate Pancreatic Lymphoma and Pancreatic Adenocarcinoma: A Preliminary Study. Acad. Radiol. 2019, 26, e189–e195. [Google Scholar] [CrossRef]
- Reinert, C.P.; Federmann, B.; Hofmann, J.; Bösmüller, H.; Wirths, S.; Fritz, J.; Horger, M. Computed tomography textural analysis for the differentiation of chronic lymphocytic leukemia and diffuse large B cell lymphoma of Richter syndrome. Eur. Radiol. 2019, 29, 6911–6921. [Google Scholar] [CrossRef]
- Seidler, M.; Forghani, B.; Reinhold, C.; Pérez-Lara, A.; Romero-Sanchez, G.; Muthukrishnan, N.; Wichmann, J.L.; Melki, G.; Yu, E.; Forghani, R. Dual-Energy CT Texture Analysis With Machine Learning for the Evaluation and Characterization of Cervical Lymphadenopathy. Comput. Struct. Biotechnol. J. 2019, 17, 1009–1015. [Google Scholar] [CrossRef]
- Lippi, M.; Gianotti, S.; Fama, A.; Casali, M.; Barbolini, E.; Ferrari, A.; Fioroni, F.; Iori, M.; Luminari, S.; Menga, M.; et al. Texture analysis and multiple-instance learning for the classification of malignant lymphomas. Comput. Methods Programs Biomed. 2020, 185, 105153. [Google Scholar] [CrossRef]
- Wu, X.; Sikiö, M.; Pertovaara, H.; Järvenpää, R.; Eskola, H.; Dastidar, P.; Kellokumpu-Lehtinen, P.L. Differentiation of Diffuse Large B-cell Lymphoma From Follicular Lymphoma Using Texture Analysis on Conventional MR Images at 3.0 Tesla. Acad. Radiol. 2016, 23, 696–703. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.-P.; Cullen, N.; Sheline, Y.I.; Taylor, W.D.; Aselcioglu, I.; Cook, P.A.; Adams, P.; Cooper, C.; Fava, M.; McGrath, P.J.; et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 2018, 167, 104–120. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, T.A.; Crowther, D.; Sutcliffe, S.B.; Glatstein, E.; Canellos, G.P.; Young, R.C.; Rosenberg, S.A.; Coltman, C.A.; Tubiana, M. Report of a committee convened to discuss the evaluation and staging of patients with Hodgkin’s disease: Cotswolds meeting. J. Clin. Oncol. 1989, 7, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Park, J.E.; Lee, H.; Ham, S.; Kim, N.; Kim, H.S. Radiomic features and multilayer perceptron network classifier: A robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci. Rep. 2019, 9, 5746. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Cho, H.H.; Kim, S.T.; Park, H.; Nam, D.; Kong, D.S. Radiomics features to distinguish glioblastoma from primary central nervous system lymphoma on multi-parametric MRI. Neuroradiology 2018, 60, 1297–1305. [Google Scholar] [CrossRef]
- Campo, E.; Harris, N.L.; Jaffe, E.S.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Pileri, S.A., Jaffe, E.S., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 9789283244943.
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification oflymphoid neoplasms. Blood J. 2017, 127, 453–462. [Google Scholar] [CrossRef]
- Schürch, C.M.; Federmann, B.; Quintanilla-Martinez, L.; Fend, F. Tumor Heterogeneity in Lymphomas: A Different Breed. Pathobiology 2018, 85, 130–145. [Google Scholar] [CrossRef]
- Khan, S.; Naim, S.; Bilwani, R.; Salem, A.; Gorlin, D.; Muhammad, A.; Gul, M.; Imam, M.H.; Chaudhry, A. Radiogenomics and Its Role in Lymphoma. Curr. Hematol. Malig. Rep. 2020, 15, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Humpire-Mamani, G.E.; Bukala, J.; Scholten, E.T.; Prokop, M.; van Ginneken, B.; Jacobs, C. Fully Automatic Volume Measurement of the Spleen at CT Using Deep Learning. Radiol. Artif. Intell. 2020, 2, e190102. [Google Scholar] [CrossRef] [PubMed]
- Barat, M.; Hoeffel, C.; Aissaoui, M.; Dohan, A.; Oudjit, A.; Dautry, R.; Paisant, A.; Malgras, B.; Cottereau, A.-S.; Soyer, P. Focal splenic lesions: Imaging spectrum of diseases on CT, MRI and PET/CT. Diagn. Interv. Imaging 2021, 102, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Reinert, C.P.; Kloth, C.; Fritz, J.; Nikolaou, K.; Horger, M. Discriminatory CT-textural features in splenic infiltration of lymphoma versus splenomegaly in liver cirrhosis versus normal spleens in controls and evaluation of their role for longitudinal lymphoma monitoring. Eur. J. Radiol. 2018, 104, 129–135. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Image biomarker standardisation initiative. arXiv 2016, arXiv:1612.07003. [Google Scholar]
- Bezerra, A.S.; D’Ippolito, G.; Faintuch, S.; Szejnfeld, J.; Ahmed, M. Determination of splenomegaly by CT: Is there a place for a single measurement? Am. J. Roentgenol. 2005, 184, 1510–1513. [Google Scholar] [CrossRef]
- Abi Saad, G.S.; Hussein, M.; El-Saghir, N.S.; Termos, S.; Sharara, A.I.; Shamseddine, A. Isolated splenic metastasis from colorectal cancer. Int. J. Clin. Oncol. 2011, 16, 306–313. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Chu-Shern, J.L.; Loi, H.Y.; Khor, L.K.; Sinha, A.K.; Quek, S.T.; Tham, I.W.K.; Townsend, D. Impact of Image Reconstruction Settings on Texture Features in 18F-FDG PET. J. Nucl. Med. 2015, 56, 1667–1673. [Google Scholar] [CrossRef] [Green Version]
- Bae, K.T. Intravenous contrast medium administration and scan timing at CT: Considerations and approaches. Radiology 2010, 256, 32–61. [Google Scholar] [CrossRef]

| Characteristics | HL | DLBCL | MCL | FL | Non-Lymphoma |
|---|---|---|---|---|---|
| Cases (percentage of cohort) | 97 (29.8%) | 129 (39.6%) | 48 (14.7%) | 52 (16%) | 56 |
| Age | |||||
| Median (years) | 34.00 | 67.0 | 63.5 | 65.0 | 61.5 |
| Lower/upper quartile (years) | 25.0/45.0 | 50.0/74.5 | 51.0/68.8 | 53.3/71.0 | 51.3/67.0 |
| Gender | |||||
| Male | 49 (50.5%) | 70 (54.3%) | 34 (70.8%) | 34 (65.4%) | 20 (35.7%) |
| Female | 48 (49.5%) | 59 (45.7%) | 14 (29.2%) | 18 (34.6%) | 36 (64.3%) |
| Stage (Ann Arbor) | |||||
| III/IV—“advanced disease” | 38 (39.2%) | 49 (38.0%) | 40 (83.3%) | 39 (75%) | - |
| IV | 24 (24.7%) | 31 (24.0%) | 32 (66.7%) | 21 (40.4%) | - |
| Craniocaudal diameter | |||||
| Median (mm) | 101.60 | 93.00 | 131.50 | 101.20 | 89.50 |
| Lower/upper quartile (mm) | 90.00/116.25 | 77.15/110.70 | 100.00/189.00 | 91.88/119.80 | 76.50/101.75 |
| Minimum–maximum (mm) | 60.00–163.20 | 55.00–235.00 | 49.60–310.00 | 60.00–248.00 | 47.00–127.00 |
| Splenic involvement | 12 (12.4%) | 10 (7.8%) | 15 (31.6%) | 4 (7.7%) | - |
| Features | AUC [CI] All Patients | AUC [CI] All Patients +ComBat | AUC [CI] Patients without Splenic Involvement | Most Important Feature |
|---|---|---|---|---|
| All | 0.86 * [0.80, 0.90] | 0.85 * [0.80, 0.90] | 0.85 * [0.79, 0.89] | log-sigma-3-0-mm-3D_glszm_GrayLevelNonUniformity |
| Original | 0.85 * [0.78, 0.90] | 0.81 * [0.74, 0.86] | 0.83 * [0.78, 0.88] | original_shape_Sphericity |
| Shape | 0.77 * [0.70, 0.83] | 0.77 * [0.70, 0.83] | 0.75 * [0.69, 0.80] | original_shape_Sphericity |
| Volume | 0.67 * [0.60, 0.76] | 0.67 * [0.60, 0.76] | 0.65 * [0.58, 0.72] | - |
| CCD | 0.68 * [0.61, 0.75] | 0.68 * [0.61, 0.75] | 0.67 * [0.59, 0.76] | - |
| Features | AUC [CI] All Patients | AUC [CI] All Patients +ComBat | AUC [CI] Patients without Splenic Involvement | Most Important Feature |
|---|---|---|---|---|
| All | 0.75 * [0.69, 0.81] | 0.75 * [0.69, 0.80] | 0.73 * [0.65, 0.78] | log-sigma-5-0-mm-3D_firstorder_90Percentile |
| Original | 0.65 * [0.58, 0.71] | 0.65 * [0.58, 0.71] | 0.63 * [0.57, 0.69] | original_shape_Maximum2DDiameterRow |
| Shape | 0.61 * [0.54, 0.66] | 0.61 * [0.54, 0.66] | 0.63 * [0.56, 0.69] | original_shape_Sphericity |
| Volume | 0.56 * [0.51, 0.61] | 0.56 * [0.51, 0.61] | 0.57 * [0.51, 0.63] | - |
| CCD | 0.53 [0.46, 0.58] | 0.53 [0.46, 0.58] | 0.56 * [0.51, 0.62] | - |
| Features | AUC [CI] All Patients | AUC [CI] All Patients +ComBat | AUC [CI] Patients without Splenic Involvement | Most Important Feature |
|---|---|---|---|---|
| All | 0.65 * [0.56, 0.71] | 0.65 * [0.56, 0.71] | 0.64 * [0.58, 0.70] | log-sigma-2-0-mm-3D_glrlm_RunEntropy |
| Original | 0.63 * [0.55, 0.70] | 0.63 * [0.55, 0.70] | 0.66 * [0.60, 0.73] | original_shape_Maximum2DDiameterColumn |
| Shape | 0.62 * [0.55, 0.68] | 0.62 * [0.55, 0.68] | 0.63 * [0.56, 0.69] | original_shape_Maximum2DDiameterColumn |
| Volume | 0.52 [0.46, 0.59] | 0.52 [0.46, 0.59] | 0.53 [0.46, 0.61] | - |
| CCD | 0.60 * [0.52, 0.66] | 0.60 * [0.52, 0.66] | 0.57 * [0.50, 0.63] | - |
| Features | AUC [CI] All Patients | AUC [CI] All Patients +ComBat | AUC [CI] Patients without Splenic Involvement | Most Important Feature |
|---|---|---|---|---|
| All | 0.67 * [0.55, 0.79] | 0.67 * [0.55, 0.79] | 0.65 * [0.53, 0.76] | log-sigma-5-0-mm-3D_glszm_SizeZoneNonUniformity |
| Original | 0.64 * [0.54, 0.76] | 0.65 * [0.54, 0.76] | 0.64 * [0.52, 0.75] | original_shape_SurfaceVolumeRatio |
| Shape | 0.71 * [0.60, 0.80] | 0.71 * [0.60, 0.80] | 0.69 * [0.60, 0.80] | original_shape_Flatness |
| Volume | 0.59 [0.49, 0.70] | 0.59 [0.49, 0.70] | 0.58 [0.47, 0.70] | - |
| CCD | 0.59 [0.46, 0.69] | 0.59 [0.46, 0.69] | 0.71 * [0.56, 0.83] | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enke, J.S.; Moltz, J.H.; D'Anastasi, M.; Kunz, W.G.; Schmidt, C.; Maurus, S.; Mühlberg, A.; Katzmann, A.; Sühling, M.; Hahn, H.; et al. Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation. Cancers 2022, 14, 713. https://doi.org/10.3390/cancers14030713
Enke JS, Moltz JH, D'Anastasi M, Kunz WG, Schmidt C, Maurus S, Mühlberg A, Katzmann A, Sühling M, Hahn H, et al. Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation. Cancers. 2022; 14(3):713. https://doi.org/10.3390/cancers14030713
Chicago/Turabian StyleEnke, Johanna S., Jan H. Moltz, Melvin D'Anastasi, Wolfgang G. Kunz, Christian Schmidt, Stefan Maurus, Alexander Mühlberg, Alexander Katzmann, Michael Sühling, Horst Hahn, and et al. 2022. "Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation" Cancers 14, no. 3: 713. https://doi.org/10.3390/cancers14030713
APA StyleEnke, J. S., Moltz, J. H., D'Anastasi, M., Kunz, W. G., Schmidt, C., Maurus, S., Mühlberg, A., Katzmann, A., Sühling, M., Hahn, H., Nörenberg, D., & Huber, T. (2022). Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation. Cancers, 14(3), 713. https://doi.org/10.3390/cancers14030713







