Blood Circulating Non-Coding RNAs for the Clinical Management of Triple-Negative Breast Cancer
Abstract
:Simple Summary
Abstract
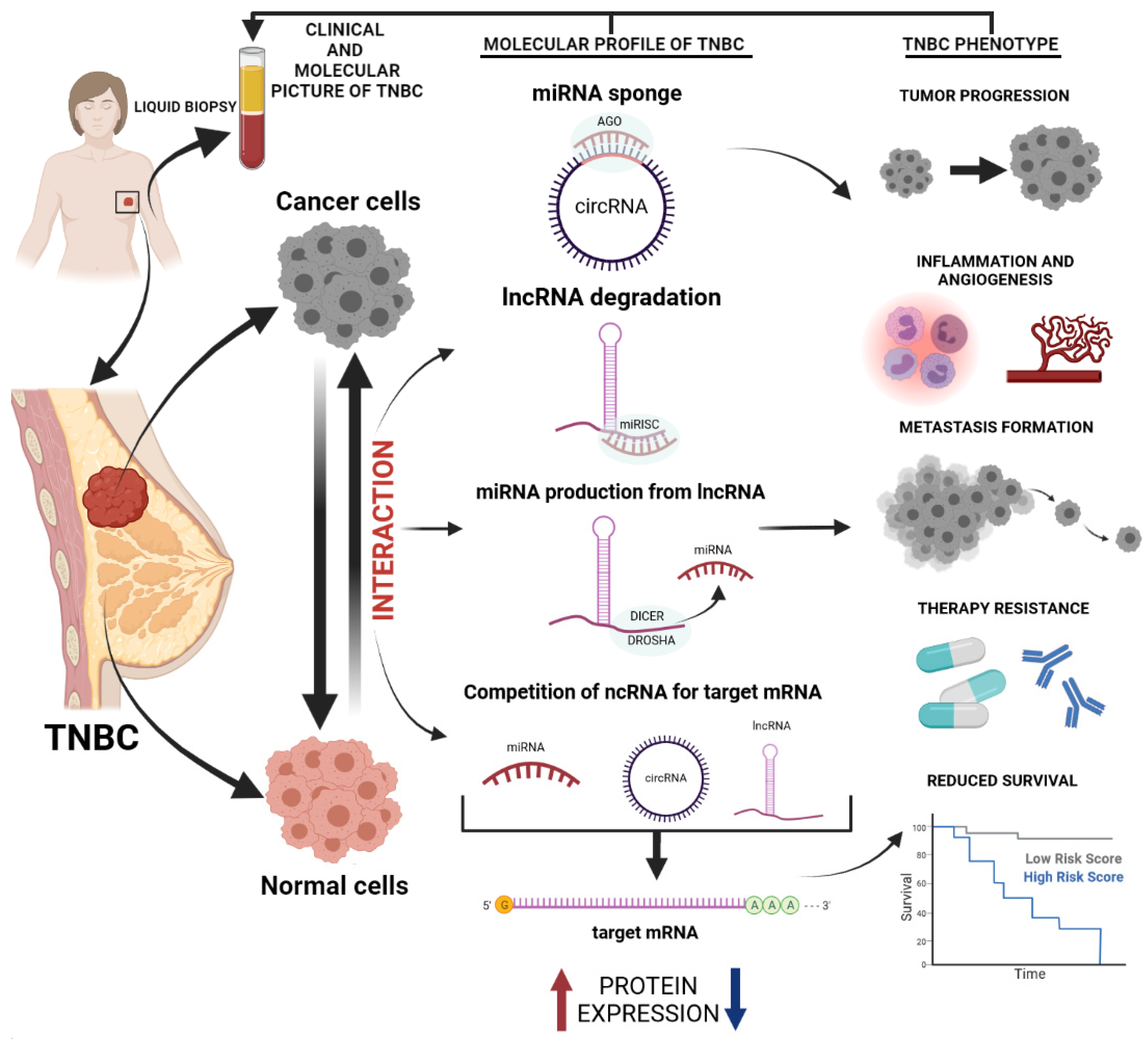
1. Introduction
1.1. Triple-Negative Breast Cancer
1.2. ncRNAs
2. Circulating ncRNAs for TNBC Detection
2.1. miRNAs
2.2. lncRNA
2.3. circRNAs
3. Circulating ncRNAs for TNBC Prediction and Prognosis
3.1. miRNAs
3.2. lncRNAs
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Levi, F.; Bosetti, C.; Lucchini, F.; Negri, E.; La Vecchia, C. Monitoring the decrease in breast cancer mortality in Europe. Eur. J. Cancer Prev. 2005, 14, 497–502. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2021; American Cancer Society: Atlanta, GA, USA, 2021. [Google Scholar]
- Serpico, D.; Molino, L.; Di Cosimo, S. microRNAs in breast cancer development and treatment. Cancer Treat. Rev. 2014, 40, 595–604. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2018; National Cancer Institute: Bethesda, MD, USA. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 20 January 2022).
- Griffiths, C.L.; Olin, J.L. Triple Negative Breast Cancer: A Brief Review of its Characteristics and Treatment Options. J. Pharm. Pr. 2012, 25, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2015, 293, 247–269. [Google Scholar] [CrossRef]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; Van Reesema, L.L.S.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers 2020, 12, 2392. [Google Scholar] [CrossRef] [PubMed]
- Mehanna, J.; Haddad, F.; Eid, R.; Lambertini, M.; Kourie, H.R. Triple-negative breast cancer: Current perspective on the evolving therapeutic lands-cape. Int. J. Womens Health 2019, 11, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Shimelis, H.; LaDuca, H.; Hu, C.; Hart, S.N.; Na, J.; Thomas, A.; Akinhanmi, M.; Moore, R.M.; Brauch, H.; Cox, A.; et al. Triple-Negative Breast Cancer Risk Genes Identified by Multigene Hereditary Cancer Panel Testing. JNCI J. Natl. Cancer Inst. 2018, 110, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nat. Cell Biol. 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Carmona, F.J.; Gomez, A.; Ferreira, H.; Bell, J.; Sayols, S.; Ward, K.; Stefansson, O.A.; Moran, S.; Sandoval, J.; et al. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis 2012, 34, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2021, 19, 91–113. [Google Scholar] [CrossRef]
- Davey, M.; Davies, M.; Lowery, A.; Miller, N.; Kerin, M. The Role of MicroRNA as Clinical Biomarkers for Breast Cancer Surgery and Treatment. Int. J. Mol. Sci. 2021, 22, 8290. [Google Scholar] [CrossRef]
- Győrffy, B.; Hatzis, C.; Sanft, T.; Hofstatter, E.; Aktas, B.; Pusztai, L. Multigene prognostic tests in breast cancer: Past, present, future. Breast Cancer Res. 2015, 17, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Cheang, M.; van de Rijn, M.; Nielsen, T. Gene expression profiling of breast cancer. Annu. Rev. Pathol. 2008, 3, 67–97. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Liu, Y.-R.; Jiang, Y.-Z.; Xu, X.-E.; Yu, K.-D.; Jin, X.; Hu, X.; Zuo, W.-J.; Hao, S.; Wu, J.; Liu, G.-Y.; et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res. 2016, 18, 33. [Google Scholar] [CrossRef] [Green Version]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.; Sun, T.; Li, H.; Hu, Z. Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2018, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- O´Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Piasecka, J.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in regulation of triple-negative breast cancer pro-gression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411. [Google Scholar] [CrossRef] [Green Version]
- Zubor, P.; Kubatka, P.; Danková, Z.; Gondova, A.; Kajo, K.; Hatok, J.; Samec, M.; Jagelkova, M.; Krivus, S.; Holubekova, V.; et al. miRNA in a multiomic context for diagnosis, treatment monitoring and personalized management of metastatic breast cancer. Future Oncol. 2018, 14, 1847–1867. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar]
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.-A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 2020, 10, 2423. [Google Scholar] [CrossRef]
- Zhao, X.; Cai, Y.; Xu, J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 2019, 20, 3926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.-Y.; Kuo, H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Zhang, S.; Deng, Y.; Wang, M.; Deng, X.; Yang, S.; Wu, Y.; Dai, Z. Regulatory mechanisms, functions, and clinical significance of CircRNAs in triple-negative breast cancer. J. Hematol. Oncol. 2021, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Li, P.; Zhang, Z.; Wu, M. Insights Into Exosomal Non-Coding RNAs Sorting Mechanism and Clinical Application. Front. Oncol. 2021, 11, 664904. [Google Scholar] [CrossRef]
- Li, G.; Sun, Y. Liquid Biopsy: Advances, Limitations and Clinical Applications. JSM Biotechnol. Bioeng. 2017, 4, 1078. [Google Scholar]
- Tay, T.K.Y.; Tan, P.H. Liquid Biopsy in Breast Cancer: A Focused Review. Arch. Pathol. Lab. Med. 2020, 145, 678–686. [Google Scholar] [CrossRef]
- Shin, V.Y.; Siu, J.M.; Cheuk, W.; Ng, E.K.O.; Kwong, A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef]
- Ye, T.; Liang, Y.; Zhang, D.; Zhang, X. MicroRNA-16-1-3p Represses Breast Tumor Growth and Metastasis by Inhibiting PGK1-Mediated Warburg Effect. Front. Cell Dev. Biol. 2020, 8, 1457. [Google Scholar] [CrossRef]
- Thakur, S.; Grover, R.K.; Gupta, S.; Yadav, A.K.; Das, B.C. Identification of Specific miRNA Signature in Paired Sera and Tissue Samples of Indian Women with Triple Negative Breast Cancer. PLoS ONE 2016, 11, e0158946. [Google Scholar] [CrossRef] [Green Version]
- Qattan, A.; Intabli, H.; Alkhayal, W.; Eltabache, C.; Tweigieri, T.; Bin Amer, S. Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pan, M.; You, C.; Zhao, F.; Wu, D.; Guo, M.; Xu, H.; Shi, F.; Zheng, D.; Dou, J. MiR-7 reduces the BCSC subset by inhibiting XIST to modulate the miR-92b/Slug/ESA axis and inhibit tumor growth. Breast Cancer Res. 2020, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pan, M.; Wang, J.; You, C.; Zhao, F.; Zheng, D.; Guo, M.; Xu, H.; Wu, D.; Wang, L.; et al. miR-7 Reduces Breast Cancer Stem Cell Metastasis via Inhibiting RELA to Decrease ESAM Expression. Mol. Ther. Oncolytics 2020, 18, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Li, X.; Li, J.; Kong, X.; Zhang, J.; Chen, L.; Huang, Y.; Fang, L. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int. J. Oncol. 2013, 43, 1212–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Yadav, V.; Kumar, S.; Saini, N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci. Rep. 2015, 5, 17454. [Google Scholar] [CrossRef]
- Li, H.Y.; Liang, J.L.; Kuo, Y.L.; Lee, H.-H.; Calkins, M.J.; Chang, H.-T.; Lin, F.-C.; Chen, Y.-C.; Hsu, T.-I.; Hsiao, M.; et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diag-nostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017, 19, 133. [Google Scholar] [CrossRef]
- Lin, B.; Liu, C.; Shi, E.; Jin, Q.; Zhao, W.; Wang, J.; Ji, R. MiR-105-3p acts as an oncogene to promote the proliferation and metastasis of breast cancer cells by targeting GOLIM4. BMC Cancer 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Kahraman, M.; Röske, A.; Laufer, T.; Fehlmann, T.; Backes, C.; Kern, F.; Kohlhaas, J.; Schrörs, H.; Saiz, A.; Zabler, C.; et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Hong, Z.; Hong, C.; Ma, B.; Wang, Q.; Zhang, X.; Li, L.; Wang, C.; Chen, D. MicroRNA-126-3p inhibits the proliferation, migration, invasion, and angiogenesis of tri-ple-negative breast cancer cells by targeting RGS3. Technol. Cancer Res. Treat. 2020, 19, 1533033820965574. [Google Scholar]
- Wang, C.-Z.; Yuan, P.; Li, Y. MiR-126 regulated breast cancer cell invasion by targeting ADAM9. Int. J. Clin. Exp. Pathol. 2015, 8, 6547–6553. [Google Scholar]
- Kia, V.; Beigli, M.S.; Hosseini, V.; Koochaki, A.; Paryan, M.; Mohammadi-Yeganeh, S. Is miR-144 an effective inhibitor of PTEN mRNA: A controversy in breast cancer. Vitro Cell. Dev. Biol. Anim. 2018, 54, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-T.; Jia, L.-T.; Liu, N.-N.; Zhu, X.-S.; Liu, Q.-Q.; Wang, X.-L.; Yu, F.; Liu, Y.-L.; Yang, A.-G.; Gao, C.-F. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget 2015, 6, 30818–30830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitobe, Y.; Ikeda, K.; Suzuki, T.; Takagi, K.; Kawabata, H.; Horie-Inoue, K.; Inoue, S. MiR-301a-3p Suppresses Estrogen Signaling by Directly Inhibiting ESR1 in ERα Positive Breast Cancer. Mol. Cell Biol. 2019, 39, e00261-19. [Google Scholar] [PubMed]
- Milevskiy, M.J.; Sandhu, G.K.; Wronski, A.; Korbie, D.; Brewster, B.L.; Shewan, A.; Edwards, S.L.; French, J.D.; Brown, M.A. MiR-29b-1-5p is altered in BRCA1 mutant tumours and is a biomarker in basal-like breast cancer. Oncotarget 2018, 9, 33577–33588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Raduly, L.; Morar-Bolba, G.; Cojocneanu, R.; Jurj, A.; Pop, L.-A.; Pileczki, V.; Ciocan, C.; Moldovan, A.; Irimie, A.; et al. Aberrant miRNAs expressed in HER-2 negative breast cancers patient. J. Exp. Clin. Cancer Res. 2018, 37, 257. [Google Scholar] [CrossRef] [Green Version]
- Akhavantabasi, S.; Sapmaz, A.; Tuna, S.; Erson-Bensan, A.E. miR-125b Targets ARID3B in Breast Cancer Cells. Cell Struct. Funct. 2012, 37, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Hulin, J.-A.; Tommasi, S.; Elliot, D.; Hu, D.G.; Lewis, B.C.; Mangoni, A.A. MiR-193b regulates breast cancer cell migration and vasculogenic mimicry by targeting dimethylarginine dimethylaminohydrolase 1. Sci. Rep. 2017, 7, 13996. [Google Scholar] [CrossRef]
- Patel, Y.; Shah, N.; Lee, J.S.; Markoutsa, E.; Jie, C.; Liu, S.; Botbyl, R.; Reisman, D.; Xu, P.; Chen, H. A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK sig-naling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget 2016, 7, 18295–18308. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, J.; Shahid, S.; Shahzadi, S.; Akhtar, M.W.; Sadaf, S. Identification of Circulating miRNAs as Non-Invasive Biomarkers of Triple Negative Breast Cancer in the Population of Pakistan. Pak. J. Zool. 2019, 51, 1113. [Google Scholar] [CrossRef]
- Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010, 12, 210. [Google Scholar] [CrossRef] [Green Version]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.; Callen, D. The Oncogenic Role of miR-155 in Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1236–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.; Zhong, M.; Pei, W.; Tian, B.; Cai, Y. miR-376c-3p modulates the properties of breast cancer stem cells by targeting RAB2A. Exp. Ther. Med. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Adam-Artigues, A.; Garrido-Cano, I.; Simón, S.; Ortega, B.; Moragón, S.; Lameirinhas, A.; Constâncio, V.; Salta, S.; Burgués, O.; Bermejo, B.; et al. Circulating miR-30b-5p levels in plasma as a novel potential biomarker for early detection of breast cancer. ESMO Open 2021, 6, 100039. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xing, L.-Q.; Liu, Y.-J. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine 2017, 96, e6222. [Google Scholar] [CrossRef]
- Du, Q.; Yang, Y.; Kong, X.; Lan, F.; Sun, J.; Zhu, H.; Ni, Y.; Pan, A. Circulating lncRNAs acting as diagnosis fingerprints for predicting triple negative breast cancer. Int. J. Clin. Exp. Med. 2018, 11, 8139–8145. [Google Scholar]
- Xu, S.T.; Xu, J.H.; Zheng, Z.R.; Zhao, Q.Q.; Zeng, X.S.; Cheng, S.X.; Liang, T.H.; Hu, Q.F. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in tri-ple-negative breast cancer. Biomed. Pharmacother. 2017, 96, 14–21. [Google Scholar] [CrossRef]
- Beckedorff, F.; Ayupe, A.C.; Crocci-Souza, R.; Amaral, M.S.; Nakaya, H.I.; Soltys, D.T.; Menck, C.F.M.; Reis, E.M.; Verjovski-Almeida, S. The Intronic Long Noncoding RNA ANRASSF1 Recruits PRC2 to the RASSF1A Promoter, Reducing the Expression of RASSF1A and Increasing Cell Proliferation. PLoS Genet. 2013, 9, e1003705. [Google Scholar] [CrossRef]
- Lan, F.; Zhang, X.; Li, H.; Yue, X.; Sun, Q. Serum exosomal lncRNA XIST is a potential non-invasive biomarker to diagnose recurrence of triple-negative breast cancer. J. Cell. Mol. Med. 2021, 25, 7602–7607. [Google Scholar] [CrossRef]
- Li, X.; Hou, L.; Yin, L.; Zhao, S. LncRNA XIST interacts with miR-454 to inhibit cells proliferation, epithelial mesenchymal transition and induces apoptosis in triple-negative breast cancer. J. Biosci. 2020, 45, 1–11. [Google Scholar] [CrossRef]
- Zhang, M.L.; Liu, W.W.; Li, W.D. Imbalance of Molecular Module of TINCR-miR-761 Promotes the Metastatic Potential of Early Triple Negative Breast Cancer and Partially Offsets the Anti-Tumor Activity of Luteolin. Cancer Manag. Res. 2021, 13, 1877–1886. [Google Scholar] [CrossRef]
- Lou, N.; Liu, G.; Pan, Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Futur. Oncol. 2020, 16, 2981–2995. [Google Scholar] [CrossRef]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long Noncoding RNAs as Biomarkers in Cancer. Dis. Markers 2017, 2017, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ma, F.; Wu, L.; Zhang, X.; Tian, J.; Li, J.; Cao, J.; Ma, Y.; Zhang, L.; Wang, L. Identification of Hsa_circ_0104824 as a Potential Biomarkers for Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820960745. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Wang, D.D.; Zhong, S.L.; Chen, W.Q.; Wang, F.L.; Zhang, J.; Xu, W.X.; Xu, D.; Zhang, Q.; Li, J.; et al. Tumor-derived exosomal circPSMA1 facilitates the tumorigenesis, metastasis, and migration in triple-negative breast cancer (TNBC) through miR-637/Akt1/β-catenin (cyclin D1) axis. Cell Death Dis. 2021, 12, 420. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modu-lating NFIB expression and translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef] [PubMed]
- Sahlberg, K.K.; Bottai, G.; Naume, B.; Burwinkel, B.; Calin, G.; Børresen-Dale, A.-L.; Santarpia, L. A Serum MicroRNA Signature Predicts Tumor Relapse and Survival in Triple-Negative Breast Cancer Patients. Clin. Cancer Res. 2014, 21, 1207–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, D.; Clayton, W.M.; Yoshimatsu, T.; Chen, J.; Olopade, O.I. Identification of a circulating MicroRNA signature to distinguish recurrence in breast cancer patients. Oncotarget 2016, 7, 55231–55248. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Chen, X.; Zhu, D.; Luo, Z.; Yang, Z. Low Expression of Circulating MicroRNA-34c is Associated with Poor Prognosis in Tri-ple-Negative Breast Cancer. Yonsei Med. J. 2017, 58, 697–702. [Google Scholar] [CrossRef]
- Shao, B.; Wang, X.; Zhang, L.; Li, D.; Liu, X.; Song, G.; Cao, H.; Zhu, J.; Li, H. Plasma microRNAs Predict Chemoresistance in Patients With Metastatic Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819828709. [Google Scholar] [CrossRef]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Wang, X.; Li, S.; Xiao, H.; Deng, X. Serum lncRNA TINCR Serve as a Novel Biomarker for Predicting the Prognosis in Tri-ple-Negative Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820965574. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, S.; Zhang, Z.; Li, J. Long non-coding RNA BRE-AS1 inhibits the proliferation, migration, and invasion of cancer cells in triple-negative breast cancer and predicts patients’ survival by downregulating miR-21. BMC Cancer 2021, 21, 745. [Google Scholar] [CrossRef] [PubMed]
- Na-Er, A.; Xu, Y.-Y.; Liu, Y.-H.; Gan, Y.-J. Upregulation of serum exosomal SUMO1P3 predicts unfavorable prognosis in triple negative breast cancer. Vitro Cell. Dev. Biol. Anim. 2021, 25, 154–160. [Google Scholar]
- Liu, Y.; Cai, Q.; Bao, P.-P.; Su, Y.; Cai, H.; Wu, J.; Ye, F.; Guo, X.; Zheng, W.; Zheng, Y.; et al. Tumor tissue microRNA expression in association with triple-negative breast cancer outcomes. Breast Cancer Res. Treat. 2015, 152, 183–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, F.; Nakata, A.; Gotoh, N.; Fujita, A. Large miRNA survival analysis reveals a prognostic four-biomarker signature for triple negative breast cancer. Genet. Mol. Biol. 2020, 43. [Google Scholar] [CrossRef]
- Wu, X.; Ding, M.; Lin, J. Three-microRNA expression signature predicts survival in triple-negative breast cancer. Oncol. Lett. 2020, 19, 301–308. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Tan, C.; He, Y.; Zhang, G.; Xu, Y.; Tang, J. Functional miRNAs in breast cancer drug resistance. OncoTargets Ther. 2018, 11, 1529–1541. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, M.; Li, Y.; Ye, S.; Ma, J.; Lu, L.; Lv, W.; Chang, G.; Li, X.; Li, Q.; Wang, S.; et al. MicroRNA Profiling Implies New Markers of Chemoresistance of Triple-Negative Breast Cancer. PLoS ONE 2014, 9, e96228. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Dashti, S.; Taheri, M.; Omrani, M.D. TINCR: An lncRNA with dual functions in the carcinogenesis process. Non-Coding RNA Res. 2020, 5, 109–115. [Google Scholar] [CrossRef]
- Xu, S.; Kong, D.; Chen, Q.; Ping, Y.; Pang, D. Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer 2017, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Liu, J.; You, Z.; Yin, Y.; Liu, L.; Kang, Y.; Li, S.; Ning, S.; Li, H.; Gong, Y.; et al. LncRNA TINCR favors tumorigenesis via STAT3–TINCR–EGFR-feedback loop by recruiting DNMT1 and acting as a competing endogenous RNA in human breast cancer. Cell Death Dis. 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, Y.; Hu, X.; Zhao, L.; Xia, W. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer 2018, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Di, S.; Zhuo, S.; Zhou, L.; Bai, R.; Ma, T.; Zou, Z.; Chen, C.; Sun, M.; Tang, J.; et al. The long noncoding RNA TINCR promotes breast cancer cell proliferation and migration by regulating OAS1. Cell Death Discov. 2021, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hu, J.; Zou, K.; Ye, M.; Chen, Y.; Wu, C.; Chen, X.; Han, M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer 2019, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhu, X.; Zhao, Q.; Huang, Q. miR-589-3p sponged by the lncRNA TINCR inhibits the proliferation, migration and invasion and promotes the apoptosis of breast cancer cells by suppressing the Akt pathway via IGF1R. Int. J. Mol. Med. 2020, 46, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Shadbad, M.A.; Safaei, S.; Brunetti, O.; Derakhshani, A.; Lotfinejad, P.; Mokhtarzadeh, A.; Hemmat, N.; Racanelli, V.; Solimando, A.G.; Argentiero, A.; et al. A Systematic Review on the Therapeutic Potentiality of PD-L1-Inhibiting MicroRNAs for Triple-Negative Breast Cancer: Toward Single-Cell Sequen-cing-Guided Biomimetic Delivery. Genes 2021, 12, 1206. [Google Scholar] [CrossRef]
- Selem, N.; Nafae, H.; Youness, R.A.; Gad, M.Z. Immunoregulatory loop between let-7a and CCAT1 lncRNA coordinated by c-Myc underlies the PD-1/PD-L1 immunoresistance in triple negative breast cancer patients. Ann. Oncol. 2021, 32 (Suppl. 6), S1355. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, N.; Song, P.; Fu, Y.; Ren, Y.; Li, Z.; Wang, J. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif. 2020, 53, e12855. [Google Scholar] [CrossRef]
- Loi, S.; Schmid, P.; Cortes, J.; Cescon, D.W.; Winer, E.P.; Toppmeyer, D.; Rugo, H.S.; De Laurentiis, M.; Nanda, R.; Iwata, H.; et al. RNA molecular signatures as predictive bio-markers of response to monotherapy pembrolizumab in patients with metastatic triple-negative breast cancer: KEYNOTE-086. Cancer Res. 2019, 79 (Suppl. 1). [Google Scholar] [CrossRef]
- Perdas, E.; Stawski, R.; Kaczka, K.; Zubrzycka, M. Analysis of Let-7 Family miRNA in Plasma as Potential Predictive Biomarkers of Diagnosis for Papillary Thyroid Cancer. Diagnostics 2020, 10, 130. [Google Scholar] [CrossRef] [Green Version]
- Spagnuolo, M.; Costantini, M.; Ferriero, M.; Varmi, M.; Sperduti, I.; Regazzo, G.; Cicchillitti, L.; Mendez, A.B.D.; Cigliana, G.; Pompeo, V.; et al. Urinary expression of let-7c cluster as non-invasive tool to assess the risk of disease progression in patients with high grade non-muscle invasive bladder Cancer: A pilot study. J. Exp. Clin. Cancer Res. 2020, 39, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, H.; Wang, Y.; Su, G.; Zhao, S. Decreased plasma let-7c and miR-152 as noninvasive biomarker for non-small-cell lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 9291–9298. [Google Scholar] [PubMed]
- Ali, S.; Almhanna, K.; Chen, W.; A Philip, P.; Sarkar, F.H. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am. J. Transl. Res. 2010, 3, 28–47. [Google Scholar]
- Liu, W.-J.; Xu, Q.; Sun, L.-P.; Dong, Q.-G.; He, C.-Y.; Yuan, Y. Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease. Tumor Biol. 2014, 36, 3337–3343. [Google Scholar] [CrossRef] [PubMed]
- Langhe, R.; Norris, L.; Abu Saadeh, F.; Blackshields, G.; Varley, R.; Harrison, A.; Gleeson, N.; Spillane, C.; Martin, C.; O’Donnell, D.M.; et al. A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. 2015, 356, 628–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; A Joosse, S.; Müller, V.; Trillsch, F.; Milde-Langosch, K.; Mahner, S.; Geffken, M.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br. J. Cancer 2015, 113, 1358–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, K.; Gu, W.; Gu, C.; Zhang, J.; Qwang, W.; Ren, G.; Tian, J. Relationship between miR-7 expression and treatment outcomes with gefitinib in non-small cell lung cancer. Oncol. Lett. 2016, 12, 4613–4617. [Google Scholar] [CrossRef]
- Roth, C.; Kasimir-Bauer, S.; Pantel, K.; Schwarzenbach, H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 2011, 5, 281–291. [Google Scholar] [CrossRef]
- Xu, H.; Yao, Y.; Meng, F.; Qian, X.; Jiang, X.; Li, X.; Gao, Z.; Gao, L. Predictive Value of Serum miR-10b, miR-29c, and miR-205 as Promising Biomarkers in Esopha-geal Squamous Cell Carcinoma Screening. Medicine (Baltimore) 2015, 94, e1558. [Google Scholar] [CrossRef]
- Yoon, E.L.; Yeon, J.E.; Ko, E.; Lee, H.J.; Je, J.H.; Yoo, Y.J.; Kang, S.H.; Suh, S.J.; Kim, J.H.; Seo, Y.S.; et al. An Explorative Analysis for the Role of Serum miR-10b-3p Levels in Predicting Response to Sorafenib in Patients with Advanced Hepatocellular Carcinoma. J. Korean Med. Sci. 2017, 32, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Tolle, A.; Buckendahl, L.; Jung, K. Plasma miR-15b-5p and miR-590-5p for distinguishing patients with bladder cancer from healthy individuals. Oncol. Rep. 2019, 42, 1609–1620. [Google Scholar] [PubMed]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non–Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.M.; Yao, T.-J.; Wang, W.; Wong, K.-F.; Lee, N.P.; Fan, S.T.; Poon, R.T.P.; Gao, C.; Luk, J.M. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open 2012, 2, e000825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sromek, M.; Glogowski, M.; Chechlinska, M.; Kulinczak, M.; Szafron, L.; Zakrzewska, K.; Owczarek, J.; Wisniewski, P.; Wlodarczyk, R.; Talarek, L.; et al. Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell. Oncol. 2017, 40, 529–536. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Y.; Zhang, C.; Zhi, X.; Fu, H.; Ma, Y.; Chen, Y.; Pan, F.; Wang, K.; Ni, J.; et al. Circulating MiR-16-5p and MiR-19b-3p as Two Novel Potential Biomarkers to Indicate Pro-gression of Gastric Cancer. Theranostics 2015, 5, 733–745. [Google Scholar] [CrossRef] [Green Version]
- El-Abd, N.E.; Fawzy, N.A.; El-Sheikh, S.M.; Soliman, M.E. Circulating miRNA-122, miRNA-199a, and miRNA-16 as Biomarkers for Early Detection of Hepatocellular Carcinoma in Egyptian Patients with Chronic Hepatitis C Virus Infection. Theranostics 2015, 19, 213–220. [Google Scholar] [CrossRef]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Xu, W.; Wang, M.; Gu, H.; Wang, S.; Qian, H.; Zhu, W.; Zhang, L.; Zhao, C.; Tao, Y. Circulating miR-17-5p and miR-20a: Molecular markers for gastric cancer. Mol. Med. Rep. 2012, 5, 1514–1520. [Google Scholar] [CrossRef]
- Wei, J.; Gao, W.; Zhu, C.j. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non–small cell lung cancer. Chin. J. Cancer 2011, 30, 407–414. [Google Scholar] [CrossRef] [Green Version]
- Qu, K.; Zhang, X.; Lin, T. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: Evidence from com-prehensive miRNA expression profiling analysis and clinical validation. Sci. Rep. 2017, 7, 1692. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Zhang, X.; Min, M.; Zou, L. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prog-nosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 44893–44909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekouian, R.; Emami, S.S.; Akbari, A.; Faraji, A.; Abbasi, V.; Agah, S. Evaluation of circulating miR-21 and miR-222 as diagnostic biomarkers for gastric cancer. J. Cancer Res. Ther. 2018, 15, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Tusong, H.; Maolakuerban, N.; Guan, J. Functional analysis of serum microRNAs miR-21 and miR-106a in renal cell carcinoma. Cancer Biomark. 2017, 18, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Kartika, A.I.; Chasanah, S.N.; Fitriawan, A.S.; Tanjung, D.S.; Trirahmanto, A.; Pradjatmo, H.; Aryandono, T.; Haryana, S.M. MicroRNA-21 as a biomarker for ovarian cancer detection. Indones. J. Biotechnol. 2018, 23, 35–39. [Google Scholar] [CrossRef]
- Khan, I.A.; Rashid, S.; Singh, N. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adeno-carcinoma. Sci. Rep. 2021, 11, 2824. [Google Scholar] [CrossRef]
- Hojbjerg, J.A.; Ebert, E.B.F.; Clement, M.S.; Winther-Larsen, A.; Meldgaard, P.; Sorensen, B. Circulating miR-30b and miR-30c predict erlotinib response in EGFR-mutated non-small cell lung cancer patients. Lung Cancer 2019, 135, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xu, S.; Liu, X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol. Lett. 2019, 17, 5601–5607. [Google Scholar] [CrossRef] [Green Version]
- Zedan, A.H.; Hansen, T.; Assenholt, J.; Pleckaitis, M.; Madsen, J.S.; Osther, P.J.S. microRNA expression in tumour tissue and plasma in patients with newly diagnosed metastatic prostate cancer. Tumor Biol. 2018, 40, 1010428318775864. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Zeng, Y.; Luo, Y.; Guo, S.; Bao, L.; Zhang, Q. Urine miR-93-5p is a promising biomarker for early detection of HBV-related hepatocellular carcinoma. Eur. J. Surg. Oncol. (EJSO) 2021, 48, 95–102. [Google Scholar] [CrossRef]
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Miyamae, M.; Okajima, W.; Ohashi, T.; Kiuchi, J.; Nishibeppu, K.; Kosuga, T.; Konishi, H.; et al. Low plasma levels of miR-101 are associated with tumor progression in gastric cancer. Oncotarget 2017, 8, 106538–106550. [Google Scholar] [CrossRef] [Green Version]
- Moshiri, F.; Salvi, A.; Gramantieri, L.; Sangiovanni, A.; Guerriero, P.; De Petro, G.; Bassi, C.; Lupini, L.; Sattari, A.; Cheung, D.; et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018, 9, 15350–15364. [Google Scholar] [CrossRef]
- He, D.; Yue, Z.; Li, G.; Chen, L.; Feng, H.; Sun, J. Low Serum Levels of miR-101 Are Associated with Poor Prognosis of Colorectal Cancer Patients After Curative Resection. Med. Sci. Monit. 2018, 24, 7475–7481. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Chang, M.; Song, X. Plasma miR-1247-5p, miR-301b-3p and miR-105-5p as potential biomarkers for early diag-nosis of non-small cell lung cancer. Thorac. Cancer 2021, 12, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Cui, E.; Li, H.; Hua, F. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients re-ceiving cisplatin-based chemotherapy. Acta Pharmacol. Sin. 2013, 34, 309–313. [Google Scholar]
- Liu, W.; Hu, J.; Zhou, K.; Chen, F.; Wang, Z.; Liao, B.; Dai, Z.; Cao, Y.; Fan, J.; Zhou, J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 3843–3851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuberi, M.; Khan, I.; Mir, R.; Gandhi, G.; Ray, P.C.; Saxena, A. Utility of Serum miR-125b as a Diagnostic and Prognostic Indicator and Its Alliance with a Panel of Tumor Suppressor Genes in Epithelial Ovarian Cancer. PLoS ONE 2016, 11, e0153902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, Q.; Lin, Y.; Jiang, F.; Lee, C.-J.; Zhan, M.; Fang, H.; Wang, Y.; Jiang, F. A plasma miRNA signature for lung cancer early detection. Oncotarget 2017, 8, 111902–111911. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.; Carlsen, A.L.; Heegaard, N.H.H.; Sørensen, F.B.; Jakobsen, A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br. J. Cancer 2015, 112, 624–629. [Google Scholar] [CrossRef]
- Tan, Y.; Lin, J.-J.; Yang, X.; Gou, D.-M.; Fu, L.; Li, F.-R.; Yu, X.-F. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019, 60, 67–76. [Google Scholar] [CrossRef]
- Liu, S.; Suo, J.; Wang, C.; Sun, X.; Wang, D.; He, L.; Zhang, Y.; Li, W. Prognostic significance of low miR-144 expression in gastric cancer. Cancer Biomark. 2017, 20, 547–552. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Song, S.-Y.; Lim, K.-H.; Park, C.-W. 490P Serum microRNAs as potential biomarkers for lung cancer. Ann. Oncol. 2015, 26, ix148–ix150. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Yang, F.; Qin, Z. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systemat-ic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, S.; Liu, H.; Gao, B. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol. Lett. 2019, 18, 3381–3387. [Google Scholar]
- Saeidi, N.; Saeidi, G.; Kheirandish, K. Evaluation of Circulating miRNA146a, miRNA155 and miRNA373 as Potential Biomarkers in Ovarian Cancer Detection. J. Mol. Genet. Med. 2018, 12, 100358. [Google Scholar]
- Chan, C.M.; Lai, K.K.Y.; Ng, E.K.O.; Na Kiang, M.; Kwok, T.W.H.; Wang, H.K.; Chan, K.W.; Law, T.T.; Tong, D.K.; Chan, K.T.; et al. Serum microRNA-193b as a promising biomarker for prediction of chemoradiation sensitivity in esophageal squamous cell carcinoma patients. Oncol. Lett. 2017, 15, 3273–3280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadal, E.; Truini, A.; Nakata, A.; Lin, J.; Reddy, R.M.; Chang, A.; Ramnath, N.; Gotoh, N.; Beer, D.G.; Chen, G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci. Rep. 2015, 5, 12464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zhao, J.; Zhang, R. Prognostic significance of serum miR-193b in colorectal cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 9509–9514. [Google Scholar]
- Su, K.; Zhang, T.; Wang, Y.; Hao, G. RETRACTED ARTICLE: Diagnostic and prognostic value of plasma microRNA-195 in patients with non-small cell lung cancer. World J. Surg. Oncol. 2016, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, A. Clinical significance of miR-195 in hepatocellular carcinoma and its biological function in tumor progression. OncoTargets Ther. 2019, 12, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Sui, D. Expression of serum microRNA-195 in patients with esophageal cancer and its clinical significance. Chin. J. Postgrad. Med. 2021, 36, 58–62. [Google Scholar]
- Nonaka, R.; Nishimura, J.; Kagawa, Y.; Osawa, H.; Hasegawa, J.; Murata, K.; Okamura, S.; Ota, H.; Uemura, M.; Hata, T.; et al. Circulating miR-199a-3p as a novel serum biomarker for colorectal cancer. Oncol. Rep. 2014, 32, 2354–2358. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, J.; Luo, Z.; Feng, C.; Hu, P.; He, X.-F.; Li, Y. Serum microRNA-199a/b-3p as a predictive biomarker for treatment response in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. OncoTargets Ther. 2016, 9, 2667–2674. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Li, J.F.; Cai, Q.; Qiu, Q.Q.; Yan, M.; Liu, B.Y.; Zhu, Z.G. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J. Surg. Oncol. 2013, 108, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Mir, R.; Das, J. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 2015, 17, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Fei, X.; Wang, X. Circulating miRNAs as Biomarkers for Prostate Cancer Diagnosis in Subjects with Benign Prostat-ic Hyperplasia. J. Immuno. Res. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Yuan, Z.; Baker, K.; Redman, M. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recur-rence in colorectal cancer. Br. J. Cancer 2017, 117, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhi, X.; Zhang, Y.; An, G.; Feng, G. Role of plasma MicroRNAs in the early diagnosis of non-small-cell lung cancers: A case-control study. J. Thorac. Dis. 2016, 8, 1645–1652. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Li, Y.; Hu, B.; He, X.; Huang, J.; Zhao, Y.; Fu, S.; Lu, L. Serum MicroRNA-210 as a Predictive Biomarker for Treatment Response and Prognosis in Patients with Hepatocellular Carcinoma undergoing Transarterial Chemoembolization. J. Vasc. Interv. Radiol. 2014, 25, 1279–1287.e1. [Google Scholar] [CrossRef]
- Fathy, M.; Hany, N.; Bahgat, A.; Youssef, O.; Fayyad, A.; Kotb, A.; Al-Khatib, S. Circulating miR-210 and miR-23b in bladder Cancer. Urol. Sci. 2021, 32, 64. [Google Scholar] [CrossRef]
- Daoud, A.Z.; Mulholland, E.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, T.; Komatsu, S.; Ichikawa, D.; Morimura, R.; Tsujiura, M.; Konishi, H.; Takeshita, H.; Nagata, H.; Arita, T.; Hirajima, S.; et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br. J. Cancer 2013, 108, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Pu, X.-X.; Huang, G.-L.; Guo, H.-Q.; Guo, C.-C.; Li, H.; Ye, S.; Ling, S.; Jiang, L.; Tian, Y.; Lin, T.-Y. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 2010, 25, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Li, Y.; Xu, Y.; Zhu, L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J. Int. Med. Res. 2013, 41, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Karimi, N.; Feizi, M.A.H.; Safaralizadeh, R.; Hashemzadeh, S.; Baradaran, B.; Shokouhi, B.; Teimourian, S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J. Chin. Med. Assoc. 2019, 82, 215–220. [Google Scholar] [CrossRef]
- Dias, F.; Teixeira, A.; Nogueira, I. Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers 2020, 12, 1450. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.S.; Chen, C.Y.; Chen, W.T. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS ONE 2017, 12, e0177346. [Google Scholar]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M.; et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 2016, 37, 941–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, H.; Guan, J.; Yuan, Z.-C.; Lin, X.; Wu, Z.-J.; Liu, B.; He, J.-L. Expression and predictive value of miR-489 and miR-21 in melanoma metastasis. World J. Clin. Cases 2019, 7, 2930–2941. [Google Scholar] [CrossRef]
- Kurt, B.; Tuncer, S.; Odemis, D. The Aberrant Expression Levels of miR-423-5p and miR-664b-5p in Peripheral Blood of Patients with Familial and Sporadic Ovarian Carcinoma. 2021. Available online: https://assets.researchsquare.com/files/rs-84141/v1/80f2b33a-71ed-411e-a4b7-099113665f78.pdf?c=1631856701 (accessed on 20 January 2022).
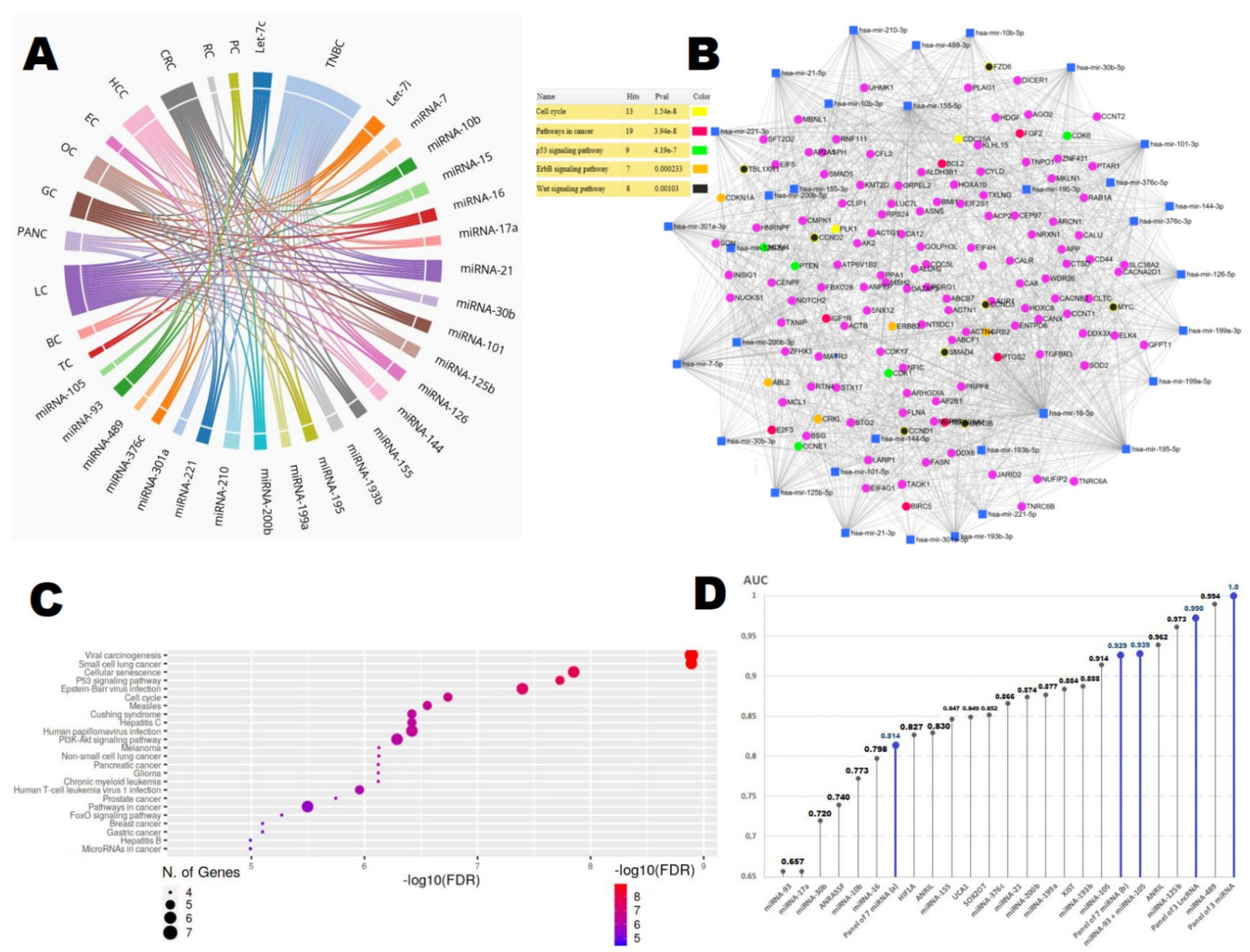

| Sample Size (*-Plasma; #-Serum) | ncRNA Expression in TNBC (Method of Detection) | Validated Targets | Biological and/or Clinical Significance of ncRNA for TNBC | Diagnostic Accuracy (AUC) | Study |
|---|---|---|---|---|---|
| miRNA | |||||
| 67 TNBC 90 HC * | ↓:miRNA-16, 21, 199a-5p | miRNA-16: AKT3, PGK1 miRNA-199a-5p: GRP78 [41] | Warburg effect mediation, cyclin E regulation, endothelial cell migration miRNA-199a-5p is associated with tumor stage | miRNA-16: 0.798 miRNA-21: 0.874 miRNA-199a-5p: 0.884 | Shin 2015 [40] |
| 23 TNBC 85 HC # | ↑: miRNA-21, 221,210 | miRNA-21: PDCD4, PTEN miRNA-221: p27Kip1, ERα miRNA-210: RAD52, HIF-1α | Oncogenic, DNA repair, cell migration, translation inhibitors, cell proliferation Correlation with tumor grade, Ki67 expression, clinical stage, lymph node status, BMI | Combination of 3 miRNA: 1.00 | Thakur 2016 [42] |
| 36 TNBC 34 HC * | ↑: miRNA-Let-7c-5p, Let-7i-5p, 7, 15, 195-5p, 489-3p ↓: miRNA-199a-3p | miRNA-7: lncRNA-XIST, RELA miRNA-15: CCNE1 miRNA-195: FASN, HMGCR, ACACA, CYP27B1 [44,45,46,47] | Cancer growth Metastasis formation Cell migration Apoptosis | Combination of 7 miRNA: 0.929 | Qattan 2017 [43] |
| 74 TNBC 12 HC * | ↑: miRNA-93-3p, 105 | miRNA-93-3p: SFRP1 miRNA-105: GOLIM4 [49] | Promotes stemness, chemoresistance, and metastasis in TNBC Correlation with distant metastases | miRNA-93-3p: 0.657 miRNA-105: 0.928 Panel of 2: 0.939 | Li 2017 [48] |
| 31 TNBC 34 HC | ↑: miRNA-126-5p, 126-3p, 144-5p, 144-3p, 301a-3p, 101-3p ↓: miRNA-664b-5p | miRNA-101: CXCR7 miRNA-126: ADAM9, RGS3 miRNA-144: PTEN miRNA-301a: ESR1 miRNA-664b: BRIP1 [51,52,53,54,55,56] | Oncogenic or tumor-suppressive regulators Cell proliferation, migration, and tumor growth Estrogen signaling pathway | Combination of 7 miRNA: 0.814 | Kahraman 2018 [50] |
| 24 TNBC 28 HC * | ↑: miRNA-125b, 193b, 200b, 489 | miRNA-125b: ARID3B miRNA-193b: DDAH1 miRNA-200b: VEGF-A, RARA miRNA-489: SHP2, HER2 [58,59,60] | Tumor invasion and metastasis, cell migration, angiogenesis MAPK signaling | miRNA-125b: 0.973 miRNA-193b: 0.914 miRNA-200b: 0.877 miRNA-489: 0.994 | Braicu 2018 [57] |
| 37 TNBC 34 HC * | ↑: miRNA-10b, 17a, 155, 376c | miRNA-10b: HOXD4, KLF4 miRNA-17a: TIMP2, TIMP3 miRNA-155: SOCS1, Smad2, FGF, E2F miRNA-376c: RAB2A [62,63,64] | DNA repair, cell cycle procession Metastasis formation, tumor aggressiveness Correlation with tumor stage, size, lymph node status and metastasis | miRNA-10b: 0.773 miRNA-17a: 0.657 miRNA-155: 0.847 miRNA-376c: 0.866 | Shaheen 2019 [61] |
| 13 TNBC 83 HC * | ↑miRNA-30b-5p | miRNA-30b-5p: CDH11, ITGA5, ITGB3 | Enrichment in Wnt and p53 signaling Apoptosis Correlation with lymph node status and distant metastases | 0.720 | Adam-Artigues 2021 [65] |
| lncRNA | |||||
| 25 TNBC 40 HC # | ↑: ANRIL, HIF1A-AS2, UCA1 | UCA1-miRNA-143 | Invasiveness of tumor cells Activation of Wnt/β-catenin signaling Tumor progression and metastasis Correlation with lymph node status and tumor size | lncRNA-ANRIL: 0.830 lncRNA-HIF1A-AS2: 0.827 lncRNA-UCA1: 0.849 | Liu 2017 [66] |
| 100 TNBC 50 HC * | ↑: ANRIL, SOX2OT, ANRASSF1 | ANRIL-miRNA-199a ANRASSF1-RASSF1A [68,69] | Tumor growth and proliferation Promotion of carcinogenesis | lncRNA-ANRIL: 0.962 lncRNA-SOX2OT: 0.852 lncRNA-ANRASSF1: 0.740 Combination of 3: 0.990 | Du 2018 [67] |
| 91 TNBC 50 HC # | ↑XIST | XIST-miRNA-7 XIST-miRNA-454 [71] | Tumor aggressiveness and proliferation, metastasis formation Correlation with tumor stage | 0.888 | Lan 2021 [70] |
| 50 TNBC 40 BC # | ↑TINCR | TINCR-miRNA-761, 125b, 503 | Tumor progression, cell growth and proliferation, apoptosis regulation | TINCR allow to distinguish TNBC from BC with AUC of 0.868 | Zhang 2021 [72] |
| circRNA | |||||
| 83 BC (TNBC) 49 HC * | ↓circ0104824 | Interaction with miRNA-140, 197, 599, 677 and 1278 | Cell cycle and cell proliferation Tumor stage, grading and metastasis Correlation with tumor size, estrogen, and progesterone receptor status | AUC for total BC: 0.849 Significant difference in expression between TNBC and non-TNBC and controls | Li 2020 [75] |
| 20 TNBC 20 HC # | ↑circPSMA1 | PSMA1-miRNA-637 | Facilitates the tumorigenesis, metastasis, cell migration through miR-637/Akt1/β-catenin axis and immunosuppression | AUC not assessed Significant difference in expression between TNBC and controls | Yang 2021 [76] |
| 24 TNBC 68 HC * | ↑circHIF1A | circHIF1A-miRNA-149-5p Interaction with NFIB | Promotion of cell proliferation and metastasis | 0.897 | Chen 2021 [77] |
| ncRNA | Role | Study Findings | Study |
|---|---|---|---|
| Unfavorable: ↑ miRNA-18b, ↑ miRNA-103, ↑ miRNA-107, ↑ miRNA-652 (all ↑ considered as a high risk score signature) | Prognosis/OS |
| Sahlberg 2015 [78] |
| Tumor relapse/RFS |
| ||
| Unfavorable: ↑ miRNA-21, ↑ miRNA-194, ↑ miRNA-205, ↑ miRNA-375 ↓ miRNA-376c, ↓ miRNA-382, ↓ miRNA-411 | Tumor relapse |
| Huo 2016 [79] |
| Unfavorable: ↓ miRNA-34a, 34c | Prognosis/OS |
| Zeng 2017 [80] |
| Unfavorable: ↓ miRNA-29c | Prognosis/OS |
| Braicu 2018 [57] |
| Unfavorable: ↑ miRNA-200a ↑ miRNA-210 | Chemoresistance |
| Shao 2019 [81] |
| Unfavorable: ↓miRNA-4448, miRNA-2392, miRNA-2467, miRNA-4800 | Response to chemotherapy |
| Sueta 2021 [82] |
| Unfavorable: ↑ lncRNA-TINCR | Tumor relapse/RFS |
| Wang 2020 [83] |
| Prognosis/OS |
| ||
| Unfavorable: ↑ miRNA-21 ↓ lncRNA-BRE-AS1 | Prognosis/OS |
| Gao 2021 [84] |
| Unfavorable: ↑ lncRNA-XIST | Tumor relapse |
| Lan 2021 [70] |
| Prognosis/OS |
| ||
| Unfavorable: ↑ lncRNA-SUMO1P3 | Prognosis/OS |
| Na-Er 2021 [85] |
| Response to chemotherapy |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powrózek, T.; Ochieng Otieno, M. Blood Circulating Non-Coding RNAs for the Clinical Management of Triple-Negative Breast Cancer. Cancers 2022, 14, 803. https://doi.org/10.3390/cancers14030803
Powrózek T, Ochieng Otieno M. Blood Circulating Non-Coding RNAs for the Clinical Management of Triple-Negative Breast Cancer. Cancers. 2022; 14(3):803. https://doi.org/10.3390/cancers14030803
Chicago/Turabian StylePowrózek, Tomasz, and Michael Ochieng Otieno. 2022. "Blood Circulating Non-Coding RNAs for the Clinical Management of Triple-Negative Breast Cancer" Cancers 14, no. 3: 803. https://doi.org/10.3390/cancers14030803





