Telomerase in Cancer: Function, Regulation, and Clinical Translation
Abstract
Simple Summary
Abstract
1. Introduction
2. Regulation of Telomerase Component Expression and Function
2.1. Transcriptional Regulation of TERT
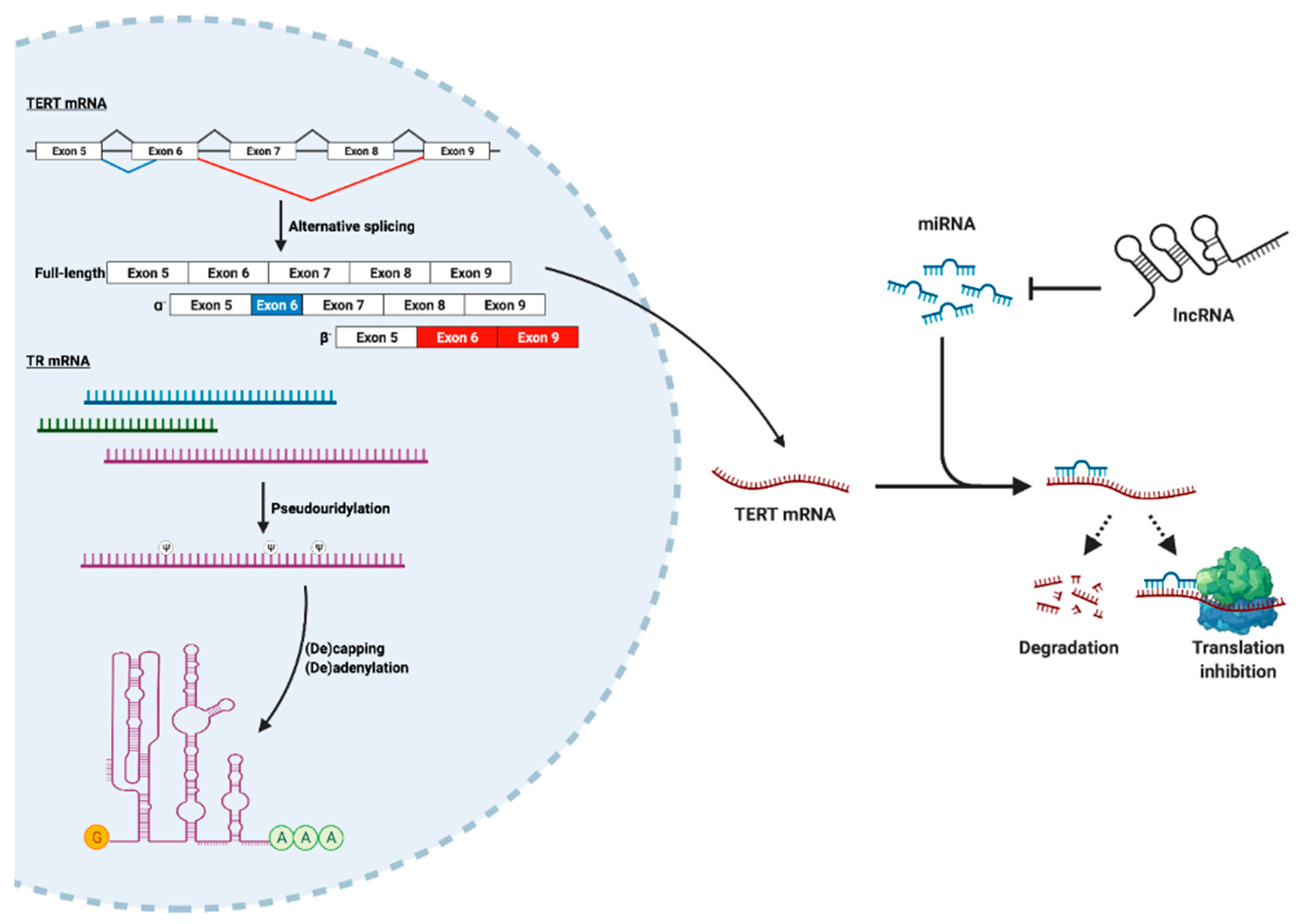
2.2. Post-Transcriptional Regulation of TERT and TR
2.3. Regulation of Other Telomerase Components
3. Extratelomeric Functions of Telomerase: Implications for Cancer Initiation and Progression
3.1. TERT as a Transcription Factor and Signaling Effector
3.2. TERT as a Regulator of Cellular Energetics
3.3. Telomerase, DNA Damage, and Genome Stability
3.4. Extratelomeric Functions of TR and Other Telomerase Components
4. Clinical Applications of Telomeres and Telomerase in Oncology
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALT | Alternative Lengthening of Telomeres |
| CAR | chimeric antigen receptor |
| CB | Cajal body |
| CST | CTC1-STN1-TEN1 |
| CTCF | CCCTC-binding factor |
| DC | dendritic cell |
| DDR | DNA damage response |
| DSBs | DNA double-strand breaks |
| EGF | epidermal growth factor |
| HAT | histone acetyltransferase |
| HDAC | histone deacetylase |
| hTERP | human telomerase RNA protein |
| miRNA | microRNA |
| ncRNA | noncoding RNA |
| PARN | poly(A) ribonuclease |
| PARP | poly(ADP-ribose) polymerase |
| POT1 | protection of telomeres 1 |
| RAP1 | repressor/activator protein 1 |
| RNP | ribonucleoprotein |
| RT | reverse transcriptase |
| Sp | specificity protein |
| TERRA | telomeric repeat containing RNA |
| TERT | protein component of telomerase |
| TF | transcription factor |
| TGF-β | transforming growth factor-β |
| TIN2 | TRF1-interacting nuclear protein 2 |
| TMM | telomere maintenance mechanism |
| TR or TERC | RNA component of telomerase |
| TRF1 | telomeric repeat-binding factor 1 |
| TRF2 | telomeric repeat-binding factor 1 |
| VEGF | vascular endothelial growth factor |
References
- Herbert, B.; Pitts, A.E.; Baker, S.I.; Hamilton, S.E.; Wright, W.E.; Shay, J.W.; Corey, D.R. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. USA 1999, 96, 14276–14281. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Hastie, N.D.; Dempster, M.; Dunlop, M.G.; Thompson, A.M.; Green, D.K.; Allshire, R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990, 346, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. Targeting the limitless replicative potential of cancer: The telomerase/telomere pathway. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 4960–4963. [Google Scholar] [CrossRef]
- Masutomi, K.; Possemato, R.; Wong, J.M.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227. [Google Scholar] [CrossRef]
- Singhapol, C.; Pal, D.; Czapiewski, R.; Porika, M.; Nelson, G.; Saretzki, G.C. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PLoS ONE 2013, 8, e52989. [Google Scholar] [CrossRef]
- Choi, J.; Southworth, L.K.; Sarin, K.Y.; Venteicher, A.S.; Ma, W.; Chang, W.; Cheung, P.; Jun, S.; Artandi, M.K.; Shah, N.; et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008, 4, e10. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Nehyba, J.; Bose, H.R., Jr. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol. Cell. Biol. 2012, 32, 4283–4296. [Google Scholar] [CrossRef]
- Stampfer, M.R.; Garbe, J.; Levine, G.; Lichtsteiner, S.; Vasserot, A.P.; Yaswen, P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor beta growth inhibition in p16INK4A(-) human mammary epithelial cells. Proc. Natl. Acad. Sci. USA 2001, 98, 4498–4503. [Google Scholar] [CrossRef]
- Cao, Y.; Li, H.; Deb, S.; Liu, J.P. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 2002, 21, 3130–3138. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zheng, D.; Wang, M.; Cong, Y.S. Telomerase reverse transcriptase activates the expression of vascular endothelial growth factor independent of telomerase activity. Biochem. Biophys. Res. Commun. 2009, 386, 739–743. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Banik, N.L.; Ray, S.K. Combination of hTERT knockdown and IFN-gamma treatment inhibited angiogenesis and tumor progression in glioblastoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 7186–7195. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Saginc, G.; Leow, S.C.; Khattar, E.; Shin, E.M.; Yan, T.D.; Wong, M.; Zhang, Z.; Li, G.; Sung, W.K.; et al. Telomerase directly regulates NF-kappaB-dependent transcription. Nat. Cell Biol. 2012, 14, 1270–1281. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Q.; Li, K.; Chen, L.; Li, W.; Hou, M.; Liu, T.; Yang, J.; Lindvall, C.; Bjorkholm, M.; et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene 2013, 32, 4203–4213. [Google Scholar] [CrossRef]
- Canela, A.; Vera, E.; Klatt, P.; Blasco, M.A. High-throughput telomere length quantification by FISH and its application to human population studies. Proc. Natl. Acad. Sci. USA 2007, 104, 5300–5305. [Google Scholar] [CrossRef]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef]
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef]
- Watson, J.D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Soudet, J.; Jolivet, P.; Teixeira, M.T. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol. Cell 2014, 53, 954–964. [Google Scholar] [CrossRef]
- McClintock, B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics 1941, 26, 234–282. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. How telomeres solve the end-protection problem. Science 2009, 326, 948–952. [Google Scholar] [CrossRef]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef]
- Doksani, Y.; Wu, J.Y.; de Lange, T.; Zhuang, X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013, 155, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.; Shastrula, P.K.; Kossenkov, A.V.; Hills, R.; Baird, D.M.; Showe, L.C.; Doukov, T.; Janicki, S.; Skordalakes, E. Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat. Commun. 2017, 8, 14928. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zheng, Y.L.; Harris, C.C. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 2005, 25, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Denchi, E.L.; de Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef]
- Palm, W.; Hockemeyer, D.; Kibe, T.; de Lange, T. Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 2009, 29, 471–482. [Google Scholar] [CrossRef]
- Ye, J.Z.; Donigian, J.R.; van Overbeek, M.; Loayza, D.; Luo, Y.; Krutchinsky, A.N.; Chait, B.T.; de Lange, T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 2004, 279, 47264–47271. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Y.; van Overbeek, M.; Donigian, J.R.; Baciu, P.; de Lange, T.; Lei, M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 2008, 319, 1092–1096. [Google Scholar] [CrossRef]
- Shibuya, H.; Ishiguro, K.; Watanabe, Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 2014, 16, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Miralles Fuste, J.; Simavorian, T.; Bartocci, C.; Tsai, J.; Karlseder, J.; Lazzerini Denchi, E. TZAP: A telomere-associated protein involved in telomere length control. Science 2017, 355, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; van Overbeek, M.; Rooney, S.; de Lange, T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell 2010, 39, 606–617. [Google Scholar] [CrossRef]
- Lenain, C.; Bauwens, S.; Amiard, S.; Brunori, M.; Giraud-Panis, M.J.; Gilson, E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006, 16, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Hu, C.; Broton, C.; Chen, Y.; Lei, M.; Chang, S. NBS1 Phosphorylation Status Dictates Repair Choice of Dysfunctional Telomeres. Mol. Cell 2017, 65, 801–817 e804. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Yin, J.; Horvath, K.; Sarkar, J.; Chen, Y.; Wu, J.; Wan, K.; Lu, J.; Gu, P.; Yu, E.Y.; et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013, 4, 861–869. [Google Scholar] [CrossRef]
- Lim, C.J.; Cech, T.R. Shaping human telomeres: From shelterin and CST complexes to telomeric chromatin organization. Nat Rev. Mol. Cell Biol. 2021, 22, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.; Ding, H.; Boulton, S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; de Lange, T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000, 10, 1299–1302. [Google Scholar] [CrossRef]
- Cook, B.D.; Dynek, J.N.; Chang, W.; Shostak, G.; Smith, S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 2002, 22, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S.; Garcia-Cao, M.; Fraga, M.F.; Schotta, G.; Peters, A.H.; Cotter, S.E.; Eguia, R.; Dean, D.C.; Esteller, M.; Jenuwein, T.; et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005, 7, 420–428. [Google Scholar] [CrossRef]
- Garcia-Cao, M.; O’Sullivan, R.; Peters, A.H.; Jenuwein, T.; Blasco, M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004, 36, 94–99. [Google Scholar] [CrossRef]
- Dejardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic Loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef]
- Udugama, M.; FT, M.C.; Chan, F.L.; Tang, M.C.; Pickett, H.A.; JD, R.M.; Mayne, L.; Collas, P.; Mann, J.R.; Wong, L.H. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015, 43, 10227–10237. [Google Scholar] [CrossRef]
- Jones, B.; Su, H.; Bhat, A.; Lei, H.; Bajko, J.; Hevi, S.; Baltus, G.A.; Kadam, S.; Zhai, H.; Valdez, R.; et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008, 4, e1000190. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, J.; Zhang, Q.D.; Lv, D.K.; Wu, N.F.; Zhou, J.Q. Rad6-Bre1-mediated H2B ubiquitination regulates telomere replication by promoting telomere-end resection. Nucleic Acids Res. 2017, 45, 3308–3322. [Google Scholar] [CrossRef]
- Ghanim, G.E.; Fountain, A.J.; van Roon, A.M.; Rangan, R.; Das, R.; Collins, K.; Nguyen, T.H.D. Structure of human telomerase holoenzyme with bound telomeric DNA. Nature 2021, 593, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- Liu, L.; Bailey, S.M.; Okuka, M.; Munoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere lengthening early in development. Nat. Cell Biol. 2007, 9, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Allen, N.D.; Baird, D.M. Telomere length maintenance in stem cell populations. Biochim. Biophys. Acta 2009, 1792, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Capper, R.; Britt-Compton, B.; Tankimanova, M.; Rowson, J.; Letsolo, B.; Man, S.; Haughton, M.; Baird, D.M. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007, 21, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Counter, C.M.; Avilion, A.A.; LeFeuvre, C.E.; Stewart, N.G.; Greider, C.W.; Harley, C.B.; Bacchetti, S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992, 11, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, R.C.; Chang, S.; Maser, R.S.; Mohan, R.; Artandi, S.E.; Chin, L.; DePinho, R.A. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell 2002, 2, 149–155. [Google Scholar] [CrossRef]
- Murnane, J.P. Telomere dysfunction and chromosome instability. Mutat. Res. 2012, 730, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Artandi, S.E.; Shen, Q.; Tam, A.; Lee, S.L.; Gottlieb, G.J.; Greider, C.W.; DePinho, R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 1999, 97, 527–538. [Google Scholar] [CrossRef]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef]
- Saretzki, G. Extra-telomeric functions of human telomerase: Cancer, mitochondria and oxidative stress. Curr. Pharm. Des. 2014, 20, 6386–6403. [Google Scholar] [CrossRef]
- Podlevsky, J.D.; Chen, J.J. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat. Res. 2012, 730, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.S.; Wright, W.E.; Shay, J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002, 66, 407–425, table of contents. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mukherjee, A.K.; Roy, S.S.; Bagri, S.; Lier, S.; Verma, M.; Sengupta, A.; Kumar, M.; Nesse, G.; Pandey, D.P.; et al. Human telomerase is directly regulated by non-telomeric TRF2-G-quadruplex interaction. Cell Rep. 2021, 35, 109154. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.S.; Aroh, C.; Vadgama, J.V. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PLoS ONE 2013, 8, e83971. [Google Scholar] [CrossRef]
- Khattar, E.; Tergaonkar, V. Transcriptional Regulation of Telomerase Reverse Transcriptase (TERT) by MYC. Front Cell Dev. Biol. 2017, 5, 1. [Google Scholar] [CrossRef]
- Yin, L.; Hubbard, A.K.; Giardina, C. NF-kappa B regulates transcription of the mouse telomerase catalytic subunit. J. Biol. Chem. 2000, 275, 36671–36675. [Google Scholar] [CrossRef]
- Avin, B.A.; Umbricht, C.B.; Zeiger, M.A. Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (Review). Int. J. Oncol. 2016, 49, 2199–2205. [Google Scholar] [CrossRef]
- Poos, A.M.; Kordass, T.; Kolte, A.; Ast, V.; Oswald, M.; Rippe, K.; Konig, R. Modelling TERT regulation across 19 different cancer types based on the MIPRIP 2.0 gene regulatory network approach. BMC Bioinform. 2019, 20, 737. [Google Scholar] [CrossRef]
- Wu, K.J.; Grandori, C.; Amacker, M.; Simon-Vermot, N.; Polack, A.; Lingner, J.; Dalla-Favera, R. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999, 21, 220–224. [Google Scholar] [CrossRef]
- Xu, D.; Popov, N.; Hou, M.; Wang, Q.; Bjorkholm, M.; Gruber, A.; Menkel, A.R.; Henriksson, M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3826–3831. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Q.L.; Sun, W.; Chandrasekharan, P.; Cheng, H.S.; Ying, Z.; Lakshmanan, M.; Raju, A.; Tenen, D.G.; Cheng, S.Y.; et al. Non-canonical NF-kappaB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 2015, 17, 1327–1338. [Google Scholar] [CrossRef]
- Konnikova, L.; Simeone, M.C.; Kruger, M.M.; Kotecki, M.; Cochran, B.H. Signal transducer and activator of transcription 3 (STAT3) regulates human telomerase reverse transcriptase (hTERT) expression in human cancer and primary cells. Cancer Res. 2005, 65, 6516–6520. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Broome, M.A.; Sinibaldi, D.; Wharton, W.; Pledger, W.J.; Sedivy, J.M.; Irby, R.; Yeatman, T.; Courtneidge, S.A.; Jove, R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 7319–7324. [Google Scholar] [CrossRef]
- Kidder, B.L.; Yang, J.; Palmer, S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE 2008, 3, e3932. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Takakura, M.; Taira, T.; Kanaya, T.; Itoh, H.; Yutsudo, M.; Ariga, H.; Inoue, M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 2000, 28, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, Y.; Yang, H.; Cheng, J.Q.; Kruk, P.A. Pyk2/ERK 1/2 mediate Sp1- and c-Myc-dependent induction of telomerase activity by epidermal growth factor. Growth Factors 2008, 26, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pore, N.; Liu, S.; Shu, H.K.; Li, B.; Haas-Kogan, D.; Stokoe, D.; Milanini-Mongiat, J.; Pages, G.; O’Rourke, D.M.; Bernhard, E.; et al. Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol. Biol. Cell 2004, 15, 4841–4853. [Google Scholar] [CrossRef]
- Baudino, T.A.; McKay, C.; Pendeville-Samain, H.; Nilsson, J.A.; Maclean, K.H.; White, E.L.; Davis, A.C.; Ihle, J.N.; Cleveland, J.L. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002, 16, 2530–2543. [Google Scholar] [CrossRef]
- Jungert, K.; Buck, A.; von Wichert, G.; Adler, G.; Konig, A.; Buchholz, M.; Gress, T.M.; Ellenrieder, V. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 2007, 67, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; D’Cruz, C.M.; Chodosh, L.A.; Cole, M.D. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol. Cell. Biol. 2007, 27, 5135–5146. [Google Scholar] [CrossRef] [PubMed]
- Rennoll, S.; Yochum, G. Regulation of MYC gene expression by aberrant Wnt/beta-catenin signaling in colorectal cancer. World J. Biol. Chem. 2015, 6, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.; Sharma, A.; Pradhan, S.J.; Galande, S. Regulation of Transcription Factor SP1 by the beta-Catenin Destruction Complex Modulates Wnt Response. Mol. Cell. Biol. 2018, 38, e00188-18. [Google Scholar] [CrossRef] [PubMed]
- Lorbeer, F.K.; Hockemeyer, D. TERT promoter mutations and telomeres during tumorigenesis. Curr. Opin. Genet. Dev. 2020, 60, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Hafezi, F.; Perez Bercoff, D. The Solo Play of TERT Promoter Mutations. Cells 2020, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, T.; Zhu, G.; Xing, M. Regulation of mutant TERT by BRAF V600E/MAP kinase pathway through FOS/GABP in human cancer. Nat. Commun. 2018, 9, 579. [Google Scholar] [CrossRef]
- Song, Y.S.; Yoo, S.K.; Kim, H.H.; Jung, G.; Oh, A.R.; Cha, J.Y.; Kim, S.J.; Cho, S.W.; Lee, K.E.; Seo, J.S.; et al. Interaction of BRAF-induced ETS factors with mutant TERT promoter in papillary thyroid cancer. Endocr. Relat. Cancer 2019, 26, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Song, Y.H.; Yim, J.; Kim, T.K. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene 2000, 19, 1485–1490. [Google Scholar] [CrossRef]
- Kanaya, T.; Kyo, S.; Hamada, K.; Takakura, M.; Kitagawa, Y.; Harada, H.; Inoue, M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 1239–1247. [Google Scholar]
- Xu, D.; Wang, Q.; Gruber, A.; Bjorkholm, M.; Chen, Z.; Zaid, A.; Selivanova, G.; Peterson, C.; Wiman, K.G.; Pisa, P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene 2000, 19, 5123–5133. [Google Scholar] [CrossRef] [PubMed]
- Renaud, S.; Loukinov, D.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005, 33, 6850–6860. [Google Scholar] [CrossRef]
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007, 35, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef]
- Liu, C.; Fang, X.; Ge, Z.; Jalink, M.; Kyo, S.; Bjorkholm, M.; Gruber, A.; Sjoberg, J.; Xu, D. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007, 67, 2626–2631. [Google Scholar] [CrossRef]
- Thakur, R.K.; Yadav, V.K.; Kumar, P.; Chowdhury, S. Mechanisms of non-metastatic 2 (NME2)-mediated control of metastasis across tumor types. Naunyn. Schmiedebergs Arch. Pharm. 2011, 384, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Singh, A.; Hussain, T.; Srivastava, V.; Sengupta, S.; Kar, A.; Dhapola, P.; Dhople, V.; Ummanni, R.; Chowdhury, S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J. Biol. Chem. 2017, 292, 15205–15215. [Google Scholar] [CrossRef] [PubMed]
- Kar, A.; Saha, D.; Purohit, G.; Singh, A.; Kumar, P.; Yadav, V.K.; Kumar, P.; Thakur, R.K.; Chowdhury, S. Metastases suppressor NME2 associates with telomere ends and telomerase and reduces telomerase activity within cells. Nucleic Acids Res. 2012, 40, 2554–2565. [Google Scholar] [CrossRef]
- Guilleret, I.; Yan, P.; Grange, F.; Braunschweig, R.; Bosman, F.T.; Benhattar, J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer 2002, 101, 335–341. [Google Scholar] [CrossRef]
- Saeboe-Larssen, S.; Fossberg, E.; Gaudernack, G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): Analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol. Biol. 2006, 7, 26. [Google Scholar] [CrossRef]
- Yi, X.; White, D.M.; Aisner, D.L.; Baur, J.A.; Wright, W.E.; Shay, J.W. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2000, 2, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Listerman, I.; Sun, J.; Gazzaniga, F.S.; Lukas, J.L.; Blackburn, E.H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013, 73, 2817–2828. [Google Scholar] [CrossRef]
- Colgin, L.M.; Wilkinson, C.; Englezou, A.; Kilian, A.; Robinson, M.O.; Reddel, R.R. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2000, 2, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, A.; Kalthoff, H.; Schuermann, M.; Schafer, B.; Boukamp, P. Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J. Cell Sci. 2002, 115, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Wang, R.; Guo, J.; Liu, X.; Wang, F.; Qi, Y.; Wan, H.; Liu, M.; Li, X.; Tang, H. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci. Rep. 2015, 5, 15793. [Google Scholar] [CrossRef]
- Ohira, T.; Naohiro, S.; Nakayama, Y.; Osaki, M.; Okada, F.; Oshimura, M.; Kugoh, H. miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci. Rep. 2015, 5, 8201. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.H.; Tang, B.; Zeng, S.; Hu, C.J.; Xie, R.; Wu, Y.Y.; Wang, S.M.; He, F.T.; Yang, S.M. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene 2016, 35, 3524–3534. [Google Scholar] [CrossRef]
- Zhang, X.L.; Xu, L.L.; Wang, F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 2017, 41, 1056–1064. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef]
- Fernandes, R.V.; Feretzaki, M.; Lingner, J. The makings of TERRA R-loops at chromosome ends. Cell Cycle 2021, 20, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; Lopez-Silanes, I.; Megias, D.; Fraga, M.F.; Castells-Garcia, A.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Chow, T.T.; Shi, X.; Wei, J.H.; Guan, J.; Stadler, G.; Huang, B.; Blackburn, E.H. Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection. Nat. Commun. 2018, 9, 3583. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Yi, X.; Tesmer, V.M.; Savre-Train, I.; Shay, J.W.; Wright, W.E. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999, 19, 3989–3997. [Google Scholar] [CrossRef]
- Goldfarb, K.C.; Cech, T.R. 3’ terminal diversity of MRP RNA and other human noncoding RNAs revealed by deep sequencing. BMC Mol. Biol. 2013, 14, 23. [Google Scholar] [CrossRef]
- Tseng, C.K.; Wang, H.F.; Schroeder, M.R.; Baumann, P. The H/ACA complex disrupts triplex in hTR precursor to permit processing by RRP6 and PARN. Nat. Commun. 2018, 9, 5430. [Google Scholar] [CrossRef]
- Kim, N.K.; Theimer, C.A.; Mitchell, J.R.; Collins, K.; Feigon, J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 2010, 38, 6746–6756. [Google Scholar] [CrossRef]
- Tseng, C.K.; Wang, H.F.; Burns, A.M.; Schroeder, M.R.; Gaspari, M.; Baumann, P. Human Telomerase RNA Processing and Quality Control. Cell Rep. 2015, 13, 2232–2243. [Google Scholar] [CrossRef]
- Moon, D.H.; Segal, M.; Boyraz, B.; Guinan, E.; Hofmann, I.; Cahan, P.; Tai, A.K.; Agarwal, S. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet 2015, 47, 1482–1488. [Google Scholar] [CrossRef]
- Roake, C.M.; Chen, L.; Chakravarthy, A.L.; Ferrell, J.E., Jr.; Raffa, G.D.; Artandi, S.E. Disruption of Telomerase RNA Maturation Kinetics Precipitates Disease. Mol. Cell 2019, 74, 688–700 e683. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Schmidt, J.C.; Goldfarb, K.C.; Cech, T.R.; Parker, R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol. 2016, 23, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Kannan, R.; Blanchette, M.; Baumann, P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 2012, 484, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Roake, C.M.; Galati, A.; Bavasso, F.; Micheli, E.; Saggio, I.; Schoeftner, S.; Cacchione, S.; Gatti, M.; Artandi, S.E.; et al. Loss of Human TGS1 Hypermethylase Promotes Increased Telomerase RNA and Telomere Elongation. Cell Rep. 2020, 30, 1358–1372 e1355. [Google Scholar] [CrossRef]
- Savelyev, N.V.; Shepelev, N.M.; Lavrik, O.I.; Rubtsova, M.P.; Dontsova, O.A. PARP1 Regulates the Biogenesis and Activity of Telomerase Complex Through Modification of H/ACA-Proteins. Front. Cell Dev. Biol. 2021, 9, 621134. [Google Scholar] [CrossRef]
- Beneke, S.; Cohausz, O.; Malanga, M.; Boukamp, P.; Althaus, F.; Burkle, A. Rapid regulation of telomere length is mediated by poly(ADP-ribose) polymerase-1. Nucleic Acids Res. 2008, 36, 6309–6317. [Google Scholar] [CrossRef]
- Hamma, T.; Ferre-D’Amare, A.R. The box H/ACA ribonucleoprotein complex: Interplay of RNA and protein structures in post-transcriptional RNA modification. J. Biol. Chem. 2010, 285, 805–809. [Google Scholar] [CrossRef]
- Stanek, D.; Pridalova-Hnilicova, J.; Novotny, I.; Huranova, M.; Blazikova, M.; Wen, X.; Sapra, A.K.; Neugebauer, K.M. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol. Biol. Cell 2008, 19, 2534–2543. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Henriksson, S.; Weibrecht, I.; Smith, S.; Soderberg, O.; Stromblad, S.; Wiman, K.G.; Farnebo, M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010, 8, e1000521. [Google Scholar] [CrossRef]
- Wang, Q.; Sawyer, I.A.; Sung, M.H.; Sturgill, D.; Shevtsov, S.P.; Pegoraro, G.; Hakim, O.; Baek, S.; Hager, G.L.; Dundr, M. Cajal bodies are linked to genome conformation. Nat. Commun. 2016, 7, 10966. [Google Scholar] [CrossRef]
- Gu, P.; Jia, S.; Takasugi, T.; Tesmer, V.M.; Nandakumar, J.; Chen, Y.; Chang, S. Distinct functions of POT1 proteins contribute to the regulation of telomerase recruitment to telomeres. Nat. Commun. 2021, 12, 5514. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Bell, C.F.; Weidenfeld, I.; Zaug, A.J.; Leinwand, L.A.; Cech, T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 2012, 492, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.K.; Tran, D.C.; Qu, R.W.; Stohr, B.A.; Segal, D.J.; Xu, L. The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres. PLoS Genet. 2015, 11, e1005410. [Google Scholar] [CrossRef]
- Greider, C.W. Regulating telomere length from the inside out: The replication fork model. Genes Dev. 2016, 30, 1483–1491. [Google Scholar] [CrossRef]
- Xu, M.; Axhemi, A.; Malgowska, M.; Chen, Y.; Leonard, D.; Srinivasan, S.; Jankowsky, E.; Taylor, D.J. Active and Passive Destabilization of G-Quadruplex DNA by the Telomere POT1-TPP1 Complex. J. Mol. Biol. 2021, 433, 166846. [Google Scholar] [CrossRef]
- Walker, J.R.; Zhu, X.D. Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012, 133, 421–434. [Google Scholar] [CrossRef]
- Smith, L.L.; Coller, H.A.; Roberts, J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003, 5, 474–479. [Google Scholar] [CrossRef]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.P.; Song, Z.; et al. hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef]
- Zaccagnini, G.; Gaetano, C.; Della Pietra, L.; Nanni, S.; Grasselli, A.; Mangoni, A.; Benvenuto, R.; Fabrizi, M.; Truffa, S.; Germani, A.; et al. Telomerase mediates vascular endothelial growth factor-dependent responsiveness in a rat model of hind limb ischemia. J. Biol. Chem. 2005, 280, 14790–14798. [Google Scholar] [CrossRef]
- Coleman, C.; Levine, D.; Kishore, R.; Qin, G.; Thorne, T.; Lambers, E.; Sasi, S.P.; Yaar, M.; Gilchrest, B.A.; Goukassian, D.A. Inhibition of melanoma angiogenesis by telomere homolog oligonucleotides. J. Oncol. 2010, 2010, 928628. [Google Scholar] [CrossRef] [PubMed]
- Pallini, R.; Sorrentino, A.; Pierconti, F.; Maggiano, N.; Faggi, R.; Montano, N.; Maira, G.; Larocca, L.M.; Levi, A.; Falchetti, M.L. Telomerase inhibition by stable RNA interference impairs tumor growth and angiogenesis in glioblastoma xenografts. Int. J. Cancer 2006, 118, 2158–2167. [Google Scholar] [CrossRef]
- Zhang, X.; Gaspard, J.P.; Chung, D.C. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001, 61, 6050–6054. [Google Scholar] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeyer, K.; Raggioli, A.; Rudloff, S.; Anton, R.; Hierholzer, A.; Del Valle, I.; Hein, K.; Vogt, R.; Kemler, R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012, 336, 1549–1554. [Google Scholar] [CrossRef]
- Ding, D.; Xi, P.; Zhou, J.; Wang, M.; Cong, Y.S. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-kappaB-dependent transcription. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 4375–4383. [Google Scholar] [CrossRef]
- Huber, M.A.; Azoitei, N.; Baumann, B.; Grunert, S.; Sommer, A.; Pehamberger, H.; Kraut, N.; Beug, H.; Wirth, T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Investig. 2004, 114, 569–581. [Google Scholar] [CrossRef]
- Qin, Y.; Tang, B.; Hu, C.J.; Xiao, Y.F.; Xie, R.; Yong, X.; Wu, Y.Y.; Dong, H.; Yang, S.M. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget 2016, 7, 351–361. [Google Scholar] [CrossRef]
- Akiyama, M.; Hideshima, T.; Hayashi, T.; Tai, Y.T.; Mitsiades, C.S.; Mitsiades, N.; Chauhan, D.; Richardson, P.; Munshi, N.C.; Anderson, K.C. Cytokines modulate telomerase activity in a human multiple myeloma cell line. Cancer Res. 2002, 62, 3876–3882. [Google Scholar]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Haendeler, J.; Drose, S.; Buchner, N.; Jakob, S.; Altschmied, J.; Goy, C.; Spyridopoulos, I.; Zeiher, A.M.; Brandt, U.; Dimmeler, S. Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arter. Thromb. Vasc. Biol. 2009, 29, 929–935. [Google Scholar] [CrossRef]
- Ahmed, S.; Passos, J.F.; Birket, M.J.; Beckmann, T.; Brings, S.; Peters, H.; Birch-Machin, M.A.; von Zglinicki, T.; Saretzki, G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 2008, 121, 1046–1053. [Google Scholar] [CrossRef]
- Sahin, E.; Colla, S.; Liesa, M.; Moslehi, J.; Muller, F.L.; Guo, M.; Cooper, M.; Kotton, D.; Fabian, A.J.; Walkey, C.; et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 2011, 470, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Ale-Agha, N.; Jakobs, P.; Goy, C.; Zurek, M.; Rosen, J.; Dyballa-Rukes, N.; Metzger, S.; Greulich, J.; von Ameln, F.; Eckermann, O.; et al. Mitochondrial Telomerase Reverse Transcriptase Protects From Myocardial Ischemia/Reperfusion Injury by Improving Complex I Composition and Function. Circulation 2021, 144, 1876–1890. [Google Scholar] [CrossRef]
- Martens, A.; Schmid, B.; Akintola, O.; Saretzki, G. Telomerase Does Not Improve DNA Repair in Mitochondria upon Stress but Increases MnSOD Protein under Serum-Free Conditions. Int. J. Mol. Sci. 2019, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Indran, I.R.; Hande, M.P.; Pervaiz, S. hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 2011, 71, 266–276. [Google Scholar] [CrossRef]
- Nishikawa, M. Reactive oxygen species in tumor metastasis. Cancer Lett. 2008, 266, 53–59. [Google Scholar] [CrossRef]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef]
- Ahmad, F.; Patrick, S.; Sheikh, T.; Sharma, V.; Pathak, P.; Malgulwar, P.B.; Kumar, A.; Joshi, S.D.; Sarkar, C.; Sen, E. Telomerase reverse transcriptase (TERT)—enhancer of zeste homolog 2 (EZH2) network regulates lipid metabolism and DNA damage responses in glioblastoma. J. Neurochem. 2017, 143, 671–683. [Google Scholar] [CrossRef]
- Shaheen, F.; Grammatopoulos, D.K.; Muller, J.; Zammit, V.A.; Lehnert, H. Extra-nuclear telomerase reverse transcriptase (TERT) regulates glucose transport in skeletal muscle cells. Biochim. Biophys. Acta 2014, 1842, 1762–1769. [Google Scholar] [CrossRef]
- Viswanath, P.; Batsios, G.; Ayyappan, V.; Taglang, C.; Gillespie, A.M.; Larson, P.E.Z.; Luchman, H.A.; Costello, J.F.; Pieper, R.O.; Ronen, S.M. Metabolic imaging detects elevated glucose flux through the pentose phosphate pathway associated with TERT expression in low-grade gliomas. Neuro-Oncology 2021, 23, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Ozsvari, B.; Bonuccelli, G.; Smith, D.L.; Pestell, R.G.; Martinez-Outschoorn, U.E.; Clarke, R.B.; Sotgia, F.; Lisanti, M.P. Dissecting tumor metabolic heterogeneity: Telomerase and large cell size metabolically define a sub-population of stem-like, mitochondrial-rich, cancer cells. Oncotarget 2015, 6, 21892–21905. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Ducray, C.; Pommier, J.P.; Martins, L.; Boussin, F.D.; Sabatier, L. Telomere dynamics, end-to-end fusions and telomerase activation during the human fibroblast immortalization process. Oncogene 1999, 18, 4211–4223. [Google Scholar] [CrossRef] [PubMed]
- Suram, A.; Kaplunov, J.; Patel, P.L.; Ruan, H.; Cerutti, A.; Boccardi, V.; Fumagalli, M.; Di Micco, R.; Mirani, N.; Gurung, R.L.; et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012, 31, 2839–2851. [Google Scholar] [CrossRef]
- Meena, J.K.; Cerutti, A.; Beichler, C.; Morita, Y.; Bruhn, C.; Kumar, M.; Kraus, J.M.; Speicher, M.R.; Wang, Z.Q.; Kestler, H.A.; et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 2015, 34, 1371–1384. [Google Scholar] [CrossRef]
- Matmati, S.; Lambert, S.; Geli, V.; Coulon, S. Telomerase Repairs Collapsed Replication Forks at Telomeres. Cell Rep. 2020, 30, 3312–3322 e3313. [Google Scholar] [CrossRef]
- Blasco, M.A.; Rizen, M.; Greider, C.W.; Hanahan, D. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nat. Genet. 1996, 12, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Y.; Ge, Y.; Liu, J.; Zhao, Y. TERC promotes cellular inflammatory response independent of telomerase. Nucleic Acids Res. 2019, 47, 8084–8095. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, P.; Zheng, Q.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Tong, T.; et al. Mitochondrial Trafficking and Processing of Telomerase RNA TERC. Cell Rep. 2018, 24, 2589–2595. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.; Naraykina, Y.; Vasilkova, D.; Meerson, M.; Zvereva, M.; Prassolov, V.; Lazarev, V.; Manuvera, V.; Kovalchuk, S.; Anikanov, N.; et al. Protein encoded in human telomerase RNA is involved in cell protective pathways. Nucleic Acids Res. 2018, 46, 8966–8977. [Google Scholar] [CrossRef]
- Gazzaniga, F.S.; Blackburn, E.H. An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity. Blood 2014, 124, 3675–3684. [Google Scholar] [CrossRef]
- Kroupa, M.; Rachakonda, S.K.; Liska, V.; Srinivas, N.; Urbanova, M.; Jiraskova, K.; Schneiderova, M.; Vycital, O.; Vymetalkova, V.; Vodickova, L.; et al. Relationship of telomere length in colorectal cancer patients with cancer phenotype and patient prognosis. Br. J. Cancer 2019, 121, 344–350. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, X.; Li, L.; Zhou, Y.; Wang, C.; Hou, S. The Association between Telomere Length and Cancer Prognosis: Evidence from a Meta-Analysis. PLoS ONE 2015, 10, e0133174. [Google Scholar] [CrossRef]
- Ennour-Idrissi, K.; Maunsell, E.; Diorio, C. Telomere Length and Breast Cancer Prognosis: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2017, 26, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.K.; Kim, H.; Park, Y.N.; Yoo, J.E.; Choi, J.; Kim, K.S.; Lee, J.J.; Park, C. High telomerase activity and long telomeres in advanced hepatocellular carcinomas with poor prognosis. Lab. Investig. 2008, 88, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Gallicchio, L.; Gadalla, S.M.; Murphy, J.D.; Simonds, N.I. The Effect of Cancer Treatments on Telomere Length: A Systematic Review of the Literature. J. Natl. Cancer Inst. 2018, 110, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Subhawong, A.P.; Heaphy, C.M.; Argani, P.; Konishi, Y.; Kouprina, N.; Nassar, H.; Vang, R.; Meeker, A.K. The alternative lengthening of telomeres phenotype in breast carcinoma is associated with HER-2 overexpression. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2009, 22, 1423–1431. [Google Scholar] [CrossRef]
- Sampl, S.; Pramhas, S.; Stern, C.; Preusser, M.; Marosi, C.; Holzmann, K. Expression of telomeres in astrocytoma WHO grade 2 to 4: TERRA level correlates with telomere length, telomerase activity, and advanced clinical grade. Transl. Oncol. 2012, 5, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Vitelli, V.; Falvo, P.; Nergadze, S.G.; Santagostino, M.; Khoriauli, L.; Pellanda, P.; Bertino, G.; Occhini, A.; Benazzo, M.; Morbini, P.; et al. Telomeric Repeat-Containing RNAs (TERRA) Decrease in Squamous Cell Carcinoma of the Head and Neck Is Associated with Worsened Clinical Outcome. Int. J. Mol. Sci. 2018, 19, 274. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhai, Y.; Ji, X.; Wang, Y.; Zhao, J.; Xing, J.; An, J.; Ren, T. Noncoding telomeric repeat-containing RNA inhibits the progression of hepatocellular carcinoma by regulating telomerase-mediated telomere length. Cancer Sci. 2020, 111, 2789–2802. [Google Scholar] [CrossRef]
- Leao, R.; Apolonio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef]
- Bell, R.J.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015, 348, 1036–1039. [Google Scholar] [CrossRef]
- Robinson, N.J.; Schiemann, W.P. Means to the ends: The role of telomeres and telomere processing machinery in metastasis. Biochim. Biophys. Acta 2016, 1866, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Hochreiter, A.E.; Xiao, H.; Goldblatt, E.M.; Gryaznov, S.M.; Miller, K.D.; Badve, S.; Sledge, G.W.; Herbert, B.S. Telomerase template antagonist GRN163L disrupts telomere maintenance, tumor growth, and metastasis of breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 3184–3192. [Google Scholar] [CrossRef]
- Dikmen, Z.G.; Gellert, G.C.; Jackson, S.; Gryaznov, S.; Tressler, R.; Dogan, P.; Wright, W.E.; Shay, J.W. In vivo inhibition of lung cancer by GRN163L: A novel human telomerase inhibitor. Cancer Res. 2005, 65, 7866–7873. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, W.; Huang, W.; Plucinsky, B.; Garcia-Vazquez, N.; Robinson, N.J.; Schiemann, W.P.; Berdis, A.J.; Skordalakes, E.; Taylor, D.J. A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity. PLoS Biol. 2019, 17, e3000204. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M.; et al. A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Queisser, A.; Heeg, S.; Thaler, M.; von Werder, A.; Opitz, O.G. Inhibition of telomerase induces alternative lengthening of telomeres during human esophageal carcinogenesis. Cancer Genet. 2013, 206, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hwang, S.S.; Liesa, M.; Gan, B.; Sahin, E.; Jaskelioff, M.; Ding, Z.; Ying, H.; Boutin, A.T.; Zhang, H.; et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 2012, 148, 651–663. [Google Scholar] [CrossRef]
- Gan, Y.; Mo, Y.; Johnston, J.; Lu, J.; Wientjes, M.G.; Au, J.L. Telomere maintenance in telomerase-positive human ovarian SKOV-3 cells cannot be retarded by complete inhibition of telomerase. FEBS Lett. 2002, 527, 10–14. [Google Scholar] [CrossRef]
- Robinson, N.J.; Morrison-Smith, C.D.; Gooding, A.J.; Schiemann, B.J.; Jackson, M.W.; Taylor, D.J.; Schiemann, W.P. SLX4IP and telomere dynamics dictate breast cancer metastasis and therapeutic responsiveness. Life Sci. Alliance 2020, 3, e201900427. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.K.; Bryant, J.E.; Fordyce, C.A.; Gilliland, F.D.; Joste, N.E.; Moyzis, R.K. Reduced telomere DNA content is correlated with genomic instability and metastasis in invasive human breast carcinoma. Breast Cancer Res. Treat. 1999, 54, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Seimiya, H.; Muramatsu, Y.; Ohishi, T.; Tsuruo, T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell 2005, 7, 25–37. [Google Scholar] [CrossRef]
- Tian, X.H.; Hou, W.J.; Fang, Y.; Fan, J.; Tong, H.; Bai, S.L.; Chen, Q.; Xu, H.; Li, Y. XAV939, a tankyrase 1 inhibitior, promotes cell apoptosis in neuroblastoma cell lines by inhibiting Wnt/beta-catenin signaling pathway. J. Exp. Clin. Cancer Res. 2013, 32, 100. [Google Scholar] [CrossRef] [PubMed]
- Waaler, J.; Machon, O.; Tumova, L.; Dinh, H.; Korinek, V.; Wilson, S.R.; Paulsen, J.E.; Pedersen, N.M.; Eide, T.J.; Machonova, O.; et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012, 72, 2822–2832. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Hernandez-Sanchez, W.; Xu, M.; Whited, T.L.; Baus, D.; Zhang, J.; Berdis, A.J.; Taylor, D.J. Administration of a Nucleoside Analog Promotes Cancer Cell Death in a Telomerase-Dependent Manner. Cell Rep. 2018, 23, 3031–3041. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rosenberg, J.E.; Donjacour, A.A.; Botchkina, I.L.; Hom, Y.K.; Cunha, G.R.; Blackburn, E.H. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004, 64, 4833–4840. [Google Scholar] [CrossRef] [PubMed]
- Cerone, M.A.; Londono-Vallejo, J.A.; Autexier, C. Mutated telomeres sensitize tumor cells to anticancer drugs independently of telomere shortening and mechanisms of telomere maintenance. Oncogene 2006, 25, 7411–7420. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; De Palma, R.; Filaci, G. Anti-cancer Immunotherapies Targeting Telomerase. Cancers 2020, 12, 2260. [Google Scholar] [CrossRef]
- Nava-Parada, P.; Emens, L.A. GV-1001, an injectable telomerase peptide vaccine for the treatment of solid cancers. Curr. Opin. Mol. 2007, 9, 490–497. [Google Scholar]
- Fenoglio, D.; Parodi, A.; Lavieri, R.; Kalli, F.; Ferrera, F.; Tagliamacco, A.; Guastalla, A.; Lamperti, M.G.; Giacomini, M.; Filaci, G. Immunogenicity of GX301 cancer vaccine: Four (telomerase peptides) are better than one. Hum. Vaccines Immunother. 2015, 11, 838–850. [Google Scholar] [CrossRef]
- Vetsika, E.K.; Konsolakis, G.; Aggouraki, D.; Kotsakis, A.; Papadimitraki, E.; Christou, S.; Menez-Jamet, J.; Kosmatopoulos, K.; Georgoulias, V.; Mavroudis, D. Immunological responses in cancer patients after vaccination with the therapeutic telomerase-specific vaccine Vx-001. Cancer Immunol. Immunother. 2012, 61, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Patente, T.A.; Pinho, M.P.; Oliveira, A.A.; Evangelista, G.C.M.; Bergami-Santos, P.C.; Barbuto, J.A.M. Human Dendritic Cells: Their Heterogeneity and Clinical Application Potential in Cancer Immunotherapy. Front. Immunol. 2018, 9, 3176. [Google Scholar] [CrossRef]
- Nair, S.K.; Heiser, A.; Boczkowski, D.; Majumdar, A.; Naoe, M.; Lebkowski, J.S.; Vieweg, J.; Gilboa, E. Induction of cytotoxic T cell responses and tumor immunity against unrelated tumors using telomerase reverse transcriptase RNA transfected dendritic cells. Nat. Med. 2000, 6, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Dannull, J.; Yang, B.K.; Dahm, P.; Coleman, D.; Yancey, D.; Sichi, S.; Niedzwiecki, D.; Boczkowski, D.; Gilboa, E.; et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J. Immunol. 2005, 174, 3798–3807. [Google Scholar] [CrossRef] [PubMed]
- Berntsen, A.; Trepiakas, R.; Wenandy, L.; Geertsen, P.F.; thor Straten, P.; Andersen, M.H.; Pedersen, A.E.; Claesson, M.H.; Lorentzen, T.; Johansen, J.S.; et al. Therapeutic dendritic cell vaccination of patients with metastatic renal cell carcinoma: A clinical phase 1/2 trial. J. Immunother. 2008, 31, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Vik-Mo, E.O.; Nyakas, M.; Mikkelsen, B.V.; Moe, M.C.; Due-Tonnesen, P.; Suso, E.M.; Saeboe-Larssen, S.; Sandberg, C.; Brinchmann, J.E.; Helseth, E.; et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013, 62, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Khoury, H.J.; Collins, R.H., Jr.; Blum, W.; Stiff, P.S.; Elias, L.; Lebkowski, J.S.; Reddy, A.; Nishimoto, K.P.; Sen, D.; Wirth, E.D., 3rd; et al. Immune responses and long-term disease recurrence status after telomerase-based dendritic cell immunotherapy in patients with acute myeloid leukemia. Cancer 2017, 123, 3061–3072. [Google Scholar] [CrossRef] [PubMed]
- Lilleby, W.; Gaudernack, G.; Brunsvig, P.F.; Vlatkovic, L.; Schulz, M.; Mills, K.; Hole, K.H.; Inderberg, E.M. Phase I/IIa clinical trial of a novel hTERT peptide vaccine in men with metastatic hormone-naive prostate cancer. Cancer Immunol. Immunother. 2017, 66, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Middleton, G.; Silcocks, P.; Cox, T.; Valle, J.; Wadsley, J.; Propper, D.; Coxon, F.; Ross, P.; Madhusudan, S.; Roques, T.; et al. Gemcitabine and capecitabine with or without telomerase peptide vaccine GV1001 in patients with locally advanced or metastatic pancreatic cancer (TeloVac): An open-label, randomised, phase 3 trial. Lancet. Oncol. 2014, 15, 829–840. [Google Scholar] [CrossRef]
- Thalmensi, J.; Pliquet, E.; Liard, C.; Escande, M.; Bestetti, T.; Julithe, M.; Kostrzak, A.; Pailhes-Jimenez, A.S.; Bourges, E.; Loustau, M.; et al. Anticancer DNA vaccine based on human telomerase reverse transcriptase generates a strong and specific T cell immune response. Oncoimmunology 2016, 5, e1083670. [Google Scholar] [CrossRef]
- Teixeira, L.; Medioni, J.; Garibal, J.; Adotevi, O.; Doucet, L.; Durey, M.D.; Ghrieb, Z.; Kiladjian, J.J.; Brizard, M.; Laheurte, C.; et al. A First-in-Human Phase I Study of INVAC-1, an Optimized Human Telomerase DNA Vaccine in Patients with Advanced Solid Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 588–597. [Google Scholar] [CrossRef]
- Sandri, S.; Bobisse, S.; Moxley, K.; Lamolinara, A.; De Sanctis, F.; Boschi, F.; Sbarbati, A.; Fracasso, G.; Ferrarini, G.; Hendriks, R.W.; et al. Feasibility of Telomerase-Specific Adoptive T-cell Therapy for B-cell Chronic Lymphocytic Leukemia and Solid Malignancies. Cancer Res. 2016, 76, 2540–2551. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, K.; Hirai, Y.; Kyoizumi, S.; Akiyama, M.; Hiyama, E.; Piatyszek, M.A.; Shay, J.W.; Ishioka, S.; Yamakido, M. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 1995, 155, 3711–3715. [Google Scholar] [PubMed]
- Hiyama, E.; Tatsumoto, N.; Kodama, T.; Hiyama, K.; Shay, J.; Yokoyama, T. Telomerase activity in human intestine. Int. J. Oncol. 1996, 9, 453–458. [Google Scholar] [CrossRef]
- Pech, M.F.; Garbuzov, A.; Hasegawa, K.; Sukhwani, M.; Zhang, R.J.; Benayoun, B.A.; Brockman, S.A.; Lin, S.; Brunet, A.; Orwig, K.E.; et al. High telomerase is a hallmark of undifferentiated spermatogonia and is required for maintenance of male germline stem cells. Genes Dev. 2015, 29, 2420–2434. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Taylor, D.J.; Schiemann, W.P. Stem cells, immortality, and the evolution of metastatic properties in breast cancer: Telomere maintenance mechanisms and metastatic evolution. J. Cancer Metastasis Treat. 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Sotillo-Pineiro, E.; Sierrasesumaga, L.; Patinno-Garcia, A. Telomerase activity and telomere length in primary and metastatic tumors from pediatric bone cancer patients. Pediatr. Res. 2004, 55, 231–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suva, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef]
- Robinson, N.J.; Miyagi, M.; Scarborough, J.A.; Scott, J.G.; Taylor, D.J.; Schiemann, W.P. SLX4IP promotes RAP1 SUMOylation by PIAS1 to coordinate telomere maintenance through NF-kappaB and Notch signaling. Sci. Signal 2021, 14, eabe9613. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, N.J.; Schiemann, W.P. Telomerase in Cancer: Function, Regulation, and Clinical Translation. Cancers 2022, 14, 808. https://doi.org/10.3390/cancers14030808
Robinson NJ, Schiemann WP. Telomerase in Cancer: Function, Regulation, and Clinical Translation. Cancers. 2022; 14(3):808. https://doi.org/10.3390/cancers14030808
Chicago/Turabian StyleRobinson, Nathaniel J., and William P. Schiemann. 2022. "Telomerase in Cancer: Function, Regulation, and Clinical Translation" Cancers 14, no. 3: 808. https://doi.org/10.3390/cancers14030808
APA StyleRobinson, N. J., & Schiemann, W. P. (2022). Telomerase in Cancer: Function, Regulation, and Clinical Translation. Cancers, 14(3), 808. https://doi.org/10.3390/cancers14030808





