Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation: Biopsy Performance, Clinical and Histopathological Examination
2.2. Histological and Clinical Diagnosis
2.3. Raman Spectroscopy (RS): Instrumentation and Spectra Acquisition
2.4. Data Analysis
3. Results and Discussions
3.1. Raman Spectra of MM, DN and CN Skin Lesions Show Similar Spectral Features but Different Band Intensities
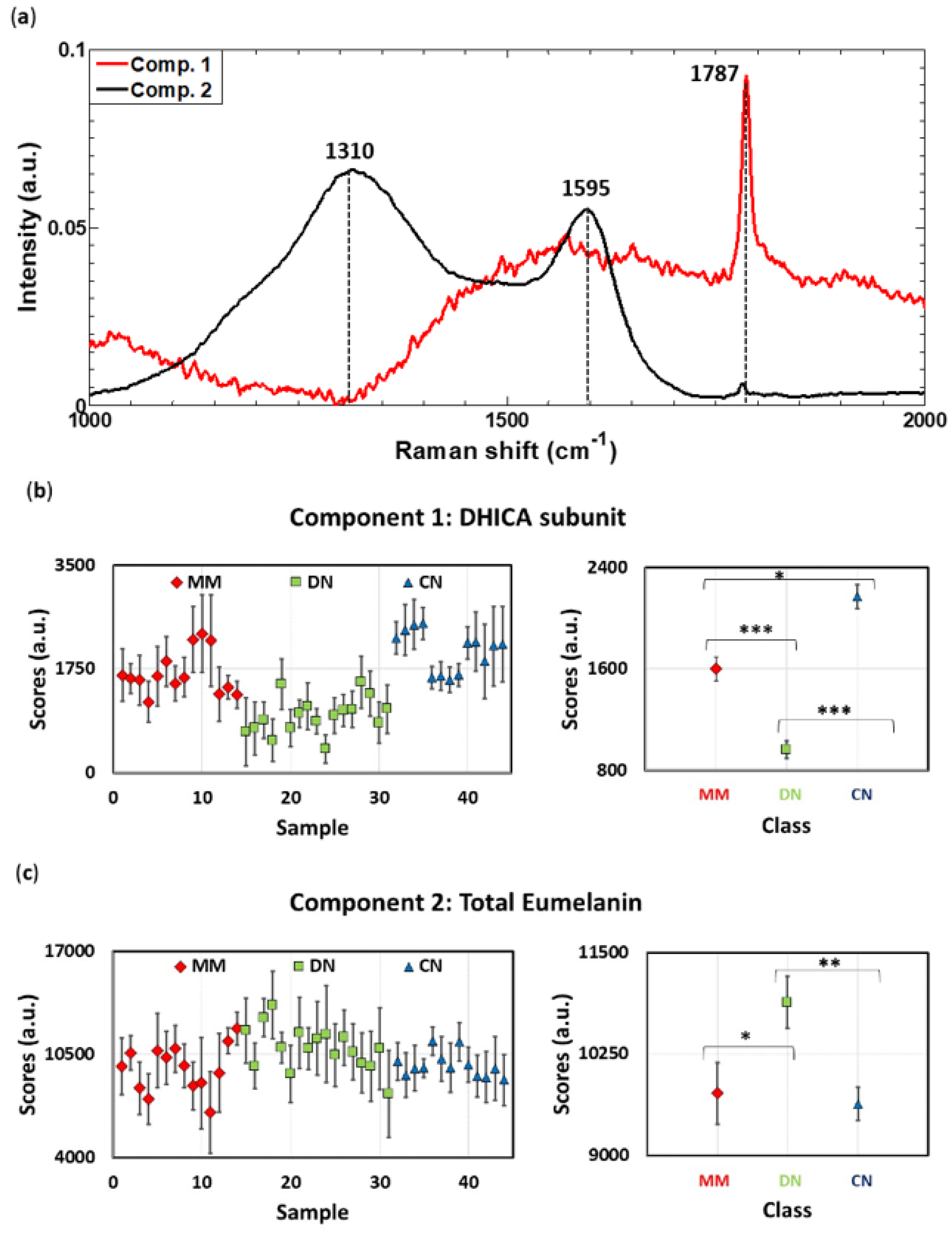
3.2. Raman Spectra Decomposition by Means of MCR-ALS Algorithm Enables Whole and Subunit Eumelanin Quantification in Skin Lesions
3.3. Raman Spectroscopy Imaging Coupled with MCR-ALS Algorithm Enables Whole and Subunit Eumelanin Quantification and Localization in Skin Lesions
3.4. Raman Spectroscopy Coupled with PLS-DA Efficiently Classifies MM, DN and CN Skin Lesions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fink, C.; Haenssle, H.A. Non-invasive tools for the diagnosis of cutaneous melanoma. Skin. Res. Technol. 2017, 23, 261–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Panzella, L.; Micillo, R.; Bentley, W.E.; Napolitano, A.; Payne, G.F. Reverse Engineering Applied to Red Human Hair Pheomelanin Reveals Redox-Buffering as a Pro-Oxidant Mechanism. Sci. Rep. 2015, 5, 18447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, K.; Grossman, D. The dysplastic nevus: From historical perspective to management in the modern era: Part I. Historical, histologic, and clinical aspects. J. Am. Acad. Dermatol. 2012, 67, 1.e1–1.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elder, D.E. Dysplastic naevi: An update. Histopathology 2010, 56, 112–120. [Google Scholar] [CrossRef]
- Zhang, G.; Li, G. Novel multiple markers to distinguish melanoma from dysplastic nevi. PLoS ONE 2012, 7, e45037. [Google Scholar] [CrossRef] [Green Version]
- Newton Bishop, J.A. Lentigos, Melanocytic Naevi and Melanoma. In Rook’s Textbook of Dermatology, 8th ed.; Burns, T., Breathnach, S., Cox, N., Griffiths, C., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 54.1–54.57. [Google Scholar]
- Santa Cruz, D.J. Tumors of the skin. In Diagnostic Histopathology of Tumors, 3th ed.; Fletcher, C.D.M., Ed.; Churchill Livingstone-Elsevier: London, UK, 2007; pp. 1423–1526. [Google Scholar]
- Arumi-Uria, M.; McNutt, N.S.; Finnerty, B. Grading of atypia in nevi: Correlation with melanoma risk. Mod. Pathol. 2003, 16, 764–771. [Google Scholar] [CrossRef] [Green Version]
- Berwick, M.; Buller, D.B.; Cust, A.; Gallagher, R.; Lee, T.K.; Meyskens, F.; Pandey, S.; Thomas, N.E.; Veierod, M.B.; Ward, S. Melanoma; Springer International Publishing: Cham, Switzerland, 2016; Volume 167. [Google Scholar]
- Saida, T. Histogenesis of cutaneous malignant melanoma: The vast majority do not develop from melanocytic nevus but arise de novo as melanoma in situ. J. Dermatol. 2019, 46, 80–94. [Google Scholar] [CrossRef]
- Strazzula, L.; Vedak, P.; Hoang, M.P.; Sober, A.; Tsao, H.; Kroshinsky, D. The utility of re-excising mildly and moderately dysplastic nevi: A retrospective analysis. J. Am. Acad. Dermatol. 2014, 71, 1071–1076. [Google Scholar] [CrossRef]
- Kim, C.C.; Swetter, S.M.; Curiel-Lewandrowski, C.; Grichnik, J.M.; Grossman, D.; Halpern, A.C.; Kirkwood, J.M.; Leachman, S.A.; Marghoob, A.A.; Ming, M.E.; et al. Addressing the knowledge gap in clinical recommendations for management and complete excision of clinically atypical nevi/dysplastic nevi: Pigmented Lesion Subcommittee consensus statement. JAMA Dermatol. 2015, 151, 212–218. [Google Scholar] [CrossRef]
- Lieber, C.A.; Majumder, S.K.; Billheimer, D.; Ellis, D.L.; Mahadevan-Jansen, A. Raman microspectroscopy for skin cancer detection in vitro. J. Biomed. Opt. 2008, 13, 024013. [Google Scholar] [CrossRef]
- Menzies, S.W.; Bischof, L.; Talbot, H.; Gutenev, A.; Avramidis, M.; Wong, L.; Lo, S.K.; Mackellar, G.; Skladnev, V.; McCarthy, W.; et al. The performance of SolarScan: An automated dermoscopy image analysis instrument for the diagnosis of primary melanoma. Arch. Dermatol. 2005, 141, 1388–1396, Correction in Arch Dermatol. 2006, 142, 558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, C.A.; Majumder, S.K.; Ellis, D.L.; Billheimer, D.D.; Mahadevan-Jansen, A. In vivo nonmelanoma skin cancer diagnosis using Raman microspectroscopy. Lasers Surg. Med. 2008, 40, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman Techniques: Fundamentals and Frontiers. Nanoscale Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marro, M.; Nieva, C.; de Juan, A. Unravelling the Metabolic Progression of Breast Cancer Cells to Bone Metastasis by Coupling Raman Spectroscopy and a Novel Use of Mcr-Als Algorithm. Anal. Chem. 2018, 90, 5594–5602. [Google Scholar] [CrossRef] [Green Version]
- Jermyn, M.; Desroches, J.; Aubertin, K.; St-Arnaud, K.; Madore, W.J.; De Montigny, E.; Guiot, M.C.; Trudel, D.; Wilson, B.C.; Petrecca, K.; et al. A review of Raman spectroscopy advances with an emphasis on clinical translation challenges in oncology. Phys. Med. Biol. 2016, 61, R370–R400. [Google Scholar] [CrossRef]
- Lui, H.; Zhao, J.; McLean, D.; Zeng, H. Real-time Raman spectroscopy for in vivo skin cancer diagnosis. Cancer Res. 2012, 72, 2491–2500. [Google Scholar] [CrossRef] [Green Version]
- Kourkoumelis, N.; Balatsoukas, I.; Moulia, V.; Elka, A.; Gaitanis, G.; Bassukas, I.D. Advances in the in Vivo Raman Spectroscopy of Malignant Skin Tumors Using Portable Instrumentation. Int. J. Mol. Sci. 2015, 16, 14554–14570. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zeng, H.; Kalia, S.; Lui, H. Incorporating patient demographics into Raman spectroscopy algorithm improves in vivo skin cancer diagnostic specificity. Transl. Biophotonics 2019, 1, e201900016. [Google Scholar] [CrossRef]
- Philipsen, P.A.; Knudsen, L.; Gniadecka, M.; Ravnbak, M.H.; Wulf, H.C. Diagnosis of malignant melanoma and basal cell carcinoma by in vivo NIR-FT Raman spectroscopy is independent of skin pigmentation. Photochem. Photobiol. Sci. 2012, 12, 770–776, Correction in Photochem. Photobiol. Sci. 2014, 13, 1360. [Google Scholar] [CrossRef]
- Lim, L.; Nichols, B.; Migden, M.R.; Rajaram, N.; Reichenberg, J.S.; Markey, M.K.; Ross, M.I.; Tunnell, J.W. Clinical study of noninvasive in vivo melanoma and nonmelanoma skin cancers using multimodal spectral diagnosis. J. Biomed. Opt. 2014, 19, 117003. [Google Scholar] [CrossRef] [Green Version]
- Santos, I.P.; Van Doorn, R.; Caspers, P.J.; Schut, T.C.B.; Barroso, E.; Nijsten, T.E.C.; Hegt, V.N.; Koljenović, S.; Puppels, G.J. Improving clinical diagnosis of early-stage cutaneous melanoma based on Raman spectroscopy. Br. J. Cancer 2018, 119, 1339–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Moy, A.J.; Nguyen, H.; Zhang, J.; Fox, M.C.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Raman active components of skin cancer. Biomed. Opt. Express 2017, 8, 2835–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Moy, A.J.; Nguyen, H.; Zhang, Y.; Zhang, J.; Fox, M.C.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Raman biophysical markers in skin cancer diagnosis. J. Biomed. Opt. 2018, 23, 057002. [Google Scholar] [CrossRef]
- Ito, S.; Nakanishi, Y.; Valenzuela, R.K.; Brilliant, M.H.; Kolbe, L.; Wakamatsu, K. Usefulness of alkaline hydrogen peroxide oxidation to analyze eumelanin and pheomelanin in various tissue samples: Application to chemical analysis of human hair melanins. Pigment Cell Melanoma Res. 2011, 24, 605–613. [Google Scholar] [CrossRef]
- Lerche, C.M.; Olsen, P.; Nissen, C.V.; Philipsen, P.A.; Wulf, H.C. A novel LC-MS/MS method to quantify eumelanin and pheomelanin and their relation to UVR sensitivity-A study on human skin biopsies. Pigment Cell Melanoma Res. 2019, 32, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Marro, M.; Rodríguez-Rivero, A.M.; Araujo-Andrade, C.; Fernández-Figueras, M.T.; Pérez-Roca, L.; Castellà, E.; Navinés, J.; Mariscal, A.; Julián, J.F.; Turon, P.; et al. Unravelling the Encapsulation of DNA and Other Biomolecules in HAp Microcalcifications of Human Breast Cancer Tissues by Raman Imaging. Cancers 2021, 13, 2658. [Google Scholar] [CrossRef] [PubMed]
- de Juan, A.; Tauler, R. Multivariate Curve Resolution: 50 years addressing the mixture analysis problem-A review. Anal. Chim. Acta 2021, 1145, 59–78. [Google Scholar] [CrossRef]
- Essendoubi, M.; Alsamad, F.; Noël, P.; Meunier, M.; Scandolera, A.; Sandré, J.; Manfait, M.; Gobinet, C.; Reynaud, R.; Piot, O. Combining Raman imaging and MCR-ALS analysis for monitoring retinol permeation in human skin. Skin. Res. Technol. 2021, 27, 1100–1109. [Google Scholar] [CrossRef]
- Annessi, G.; Bono, R.; Sampogna, F.; Faraggiana, T.; Abeni, D. Sensitivity, specificity, and diagnostic accuracy of three dermoscopic algorithmic methods in the diagnosis of doubtful melanocytic lesions: The importance of light brown structureless areas in differentiating atypical melanocytic nevi from thin melanomas. J. Am. Acad. Dermatol. 2007, 56, 759–767. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Teichmann, H.O.; Herrmann, T.R.; Bach, T. Technical aspects of lasers in urology. World J. Urol. 2007, 25, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Mikhailyuk, I.K.; Razzhivin, A.P. Background Subtraction in Experimental Data Arrays Illustrated by the Example of Raman Spectra and Fluorescent Gel Electrophoresis Patterns. Instrum. Exp. Tech. 2003, 46, 765–769. [Google Scholar] [CrossRef]
- Mian, S.A.; Colley, H.E.; Thornhill, M.; Rehman, I.U. Development of a Dewaxing Protocol for Tissue-Engineered Models of the Oral Mucosa Used for Raman Spectroscopic Analysis. Appl. Spectrosc. Rev. 2014, 49, 614–617. [Google Scholar] [CrossRef]
- Tuschel, D. Selecting an Excitation Wavelength for Raman Spectroscopy. Spectroscopy 2016, 31, 14–23. [Google Scholar]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Galván, I.; Jorge, A.; Solano, F.; Wakamatsu, K. Vibrational characterization of pheomelanin and trichochrome F by Raman spectroscopy. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2013, 110, 55–59. [Google Scholar] [CrossRef]
- Huang, Z.; Lui, H.; Chen, X.K.; Alajlan, A.; McLean, D.I.; Zeng, H. Raman spectroscopy of in vivo cutaneous melanin. J. Biomed. Opt. 2004, 9, 1198–1205. [Google Scholar] [CrossRef]
- Galván, I.; Araujo-Andrade, C.; Marro, M.; Loza-Alvarez, P.; Wakamatsu, K. Raman spectroscopy quantification of eumelanin subunits in natural unaltered pigments. Pigment. Cell Melanoma Res. 2018, 31, 673–682. [Google Scholar] [CrossRef]
- Elder, D.E. Precursors to melanoma and their mimics: Nevi of special sites. Mod. Pathol. 2006, 19 (Suppl. 2), S4–S20. [Google Scholar] [CrossRef] [Green Version]
- Van Nieuwpoort, A.F. Biochemical and Molecular Studies of Atypical Nevi. Ph.D. Thesis, Leiden University Medical Center (LUMC), Leiden University, Leiden, The Netherlands, 16 March 2011. Available online: https://hdl.handle.net/1887/16632 (accessed on 18 February 2022).
- Galván, I.; Solano, F. Melanin chemistry and the ecology of stress. Physiol. Biochem. Zool. 2015, 88, 352–355. [Google Scholar] [CrossRef] [Green Version]
- Chong, I.; Jun, C. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 78–103. [Google Scholar] [CrossRef]
- Vincensi, M.R.; d’Ischia, M.; Napolitano, A.; Procaccini, E.M.; Riccio, G.; Monfrecola, G.; Santoianni, P.; Prota, G. Phaeomelanin versus eumelanin as a chemical indicator of ultraviolet sensitivity in fair-skinned subjects at high risk for melanoma: A pilot study. Melanoma Res. 1998, 8, 53–58. [Google Scholar] [CrossRef] [PubMed]


| Model 1: DN vs. [MM and CN] | ||
| DN | MM and CN | |
| Sensitivity (%) | 94.1 | 100 |
| Specificity (%) | 100 | 94.1 |
| Model 2: DN vs. MM | ||
| DN | MM | |
| Sensitivity (%) | 82.4 | 92.9 |
| Specificity (%) | 92.9 | 82.4 |
| Model 3: DN vs. CN | ||
| DN | CN | |
| Sensitivity (%) | 100 | 100 |
| Specificity (%) | 100 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz, J.J.; Marro, M.; Galván, I.; Bernabeu-Wittel, J.; Conejo-Mir, J.; Zulueta-Dorado, T.; Guisado-Gil, A.B.; Loza-Álvarez, P. Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis. Cancers 2022, 14, 1056. https://doi.org/10.3390/cancers14041056
Ruiz JJ, Marro M, Galván I, Bernabeu-Wittel J, Conejo-Mir J, Zulueta-Dorado T, Guisado-Gil AB, Loza-Álvarez P. Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis. Cancers. 2022; 14(4):1056. https://doi.org/10.3390/cancers14041056
Chicago/Turabian StyleRuiz, José Javier, Monica Marro, Ismael Galván, José Bernabeu-Wittel, Julián Conejo-Mir, Teresa Zulueta-Dorado, Ana Belén Guisado-Gil, and Pablo Loza-Álvarez. 2022. "Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis" Cancers 14, no. 4: 1056. https://doi.org/10.3390/cancers14041056
APA StyleRuiz, J. J., Marro, M., Galván, I., Bernabeu-Wittel, J., Conejo-Mir, J., Zulueta-Dorado, T., Guisado-Gil, A. B., & Loza-Álvarez, P. (2022). Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis. Cancers, 14(4), 1056. https://doi.org/10.3390/cancers14041056







