The Roles of microRNAs in Cancer Multidrug Resistance
Abstract
:Simple Summary
Abstract
1. Introduction
2. Cell Resistance to Multiple Drugs
3. The Role of microRNAs in the Development of Neoplastic Cell Resistance to Chemotherapy
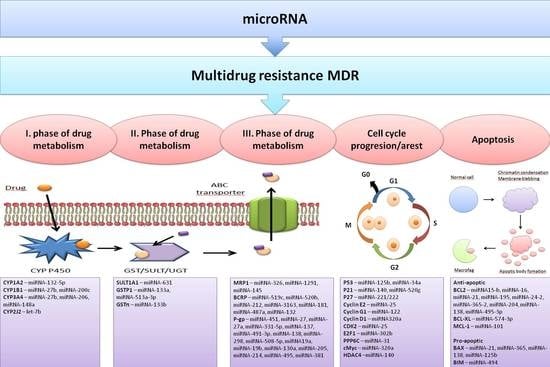
4. Effects of miRNAs on Cell Detoxification Pathways
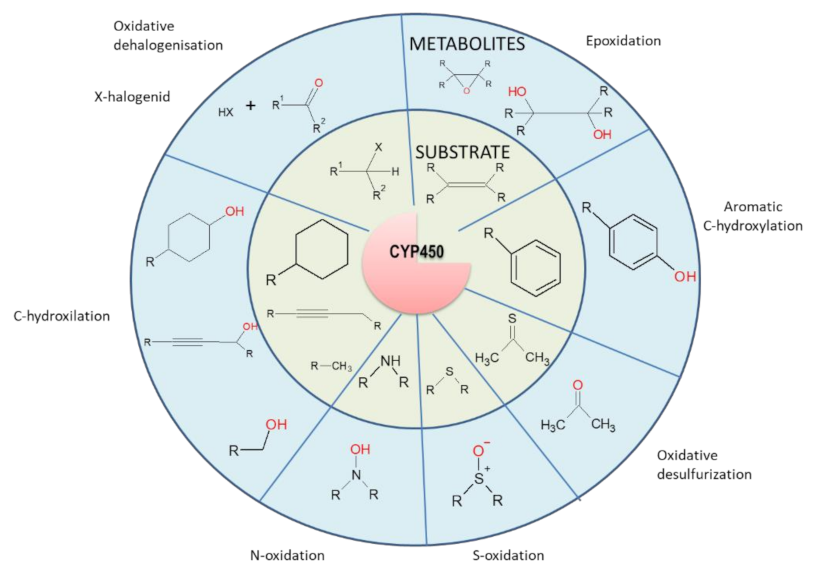
4.1. The First Phase of Drug Metabolism–Oxidation
4.2. The Second Phase of Drug Metabolism–Conjugation

4.3. The Third Phase of Drug Metabolism-Transporters
4.3.1. Permeability Glycoprotein P-gp/ABCB1/MDR1
4.3.2. Breast Cancer Resistance Protein BCRP/ABCG2
4.3.3. Multidrug Resistance-Associated Protein MRP1/ABCC1
4.4. Effects of miRNAs on Cell Cycle Progression and Apoptosis Induction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Choi, J.W.; Hua, T.N.M. Impact of lifestyle behaviors on cancer risk and prevention. J. Lifestyle Med. 2021, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J. The hidden dangers of fast and processed food. Am. J. Lifestyle Med. 2018, 12, 375–381. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Report 2014; International Agency for Research on Cancer: Lyon, France, 2014.
- Bray, F.; Jemal, A.; Grey, N.; Ferlay, J.; Forman, D. Global cancer transitions according to the human development index (2008–2030): A population-based study. Lancet Oncol. 2012, 13, 790–801. [Google Scholar] [CrossRef]
- United Nations Population Division. World Population Prospects: The 2008 Revision; United Nations: New York, NY, USA, 2007.
- Arruebo, M.; Vilaboa, N.; Sáez-Gutierrez, B.; Lambea, J.; Tres, A.; Valladares, M.; González-Fernández, Á. Assessment of the evolution of cancer treatment therapies. Cancers 2011, 3, 3279–3330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, S.D.; Konig, H. Treatment of acute myeloid leukemia in the era of genomics-achievements and persisting challenges. Front. Genet. 2020, 11, 480. [Google Scholar] [CrossRef]
- O’Donnell, M.R.; Tallman, M.S.; Abboud, C.N.; Altman, J.K.; Appelbaum, F.R.; Arber, D.A.; Bhatt, V.; Bixby, D.; Blum, W.; Coutre, S.E.; et al. Acute myeloid leukemia, version 3.2017, nccn clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 926–957. [Google Scholar] [CrossRef]
- Mueller, B.U.; Seipel, K.; Bacher, U.; Pabst, T. Autologous transplantation for older adults with AML. Cancers 2018, 10, 340. [Google Scholar] [CrossRef] [Green Version]
- Farshchi, Z.S.; Chan, S.; Gupta, V.; Khalaf, D.; Lutynski, A.; Minden, M.D.; Rostom, A.; Rydlewski, A.; Schuh, A.C.; Sibai, H.; et al. Remissions after third induction chemotherapy for primary non-responders with acute myeloid leukemia (AML) are uncommon and short-lived. Leuk. Lymphoma 2018, 59, 237–240. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, Y.; Chen, B. Mechanisms of drug resistance in acute myeloid leukemia. OncoTargets Ther. 2019, 12, 1937–1945. [Google Scholar] [CrossRef] [Green Version]
- Breier, A.; Gibalova, L.; Seres, M.; Barancik, M.; Sulova, Z. New insight into p-glycoprotein as a drug target. Anti-Cancer Agents Med. Chem. 2013, 13, 159–170. [Google Scholar] [CrossRef]
- Bell, C.C.; Fennell, K.A.; Chan, Y.C.; Rambow, F.; Yeung, M.M.; Vassiliadis, D.; Lara, L.; Yeh, P.; Martelotto, L.G.; Rogiers, A.; et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat. Commun. 2019, 10, 2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikhodeyev, O.N. The mechanisms of epigenetic inheritance: How diverse are they? Biol. Rev. Camb. Philos. Soc. 2018, 93, 1987–2005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Pre-mirna. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 102. [Google Scholar]
- Santhekadur, P.K.; Kumar, D.P. Risc assembly and post-transcriptional gene regulation in hepatocellular carcinoma. Genes Dis. 2020, 7, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Bracht, J.; Hunter, S.; Massirer, K.; Holtz, J.; Eachus, R.; Pasquinelli, A.E. Regulation by let-7 and lin-4 MIRNAS results in target mRNA degradation. Cell 2005, 122, 553–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of micrornas (mirnas) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol. Hematol. 2012, 81, 103–122. [Google Scholar] [CrossRef]
- Sun, E.; Shi, Y. Micrornas: Small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 2015, 268, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Sontheimer, E.J.; Carthew, R.W. Silence from within: Endogenous SIRNAS and MIRNAS. Cell 2005, 122, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.P.; Glasner, M.E.; Yekta, S.; Burge, C.B.; Bartel, D.P. Vertebrate microrna genes. Science 2003, 299, 1540. [Google Scholar] [CrossRef] [Green Version]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. Micrornas associated with the different human argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian rnai. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef] [Green Version]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human argonaute2 mediates rna cleavage targeted by mirnas and sirnas. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of micrornas on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan micrornas. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- An, X.; Sarmiento, C.; Tan, T.; Zhu, H. Regulation of multidrug resistance by micrornas in anti-cancer therapy. Acta Pharm. Sin. B 2017, 7, 38–51. [Google Scholar] [CrossRef]
- Liang, Y.; Liang, Q.; Qiao, L.; Xiao, F. Micrornas modulate drug resistance-related mechanisms in hepatocellular carcinoma. Front. Oncol. 2020, 10, 920. [Google Scholar] [CrossRef]
- Medarova, Z.; Pantazopoulos, P.; Yoo, B. Screening of potential mirna therapeutics for the prevention of multi-drug resistance in cancer cells. Sci. Rep. 2020, 10, 1970. [Google Scholar] [CrossRef] [Green Version]
- Brady, S.P.; Monosson, E.; Matson, C.W.; Bickham, J.W. Evolutionary toxicology: Toward a unified understanding of life’s response to toxic chemicals. Evol. Appl. 2017, 10, 745–751. [Google Scholar] [CrossRef] [Green Version]
- Torres-Martinez, Z.; Delgado, Y.; Ferrer-Acosta, Y.; Suarez-Arroyo, I.J.; Joaquin-Ovalle, F.M.; Delinois, L.J.; Griebenow, K. Key genes and drug delivery systems to improve the efficiency of chemotherapy. Cancer Drug Resist. 2021, 4, 163–191. [Google Scholar] [CrossRef]
- Cree, I.A.; Charlton, P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer 2017, 17, 10. [Google Scholar] [CrossRef] [Green Version]
- Obligacion, R.; Murray, M.; Ramzan, I. Drug-metabolizing enzymes and transporters: Expression in the human prostate and roles in prostate drug disposition. J. Androl. 2006, 27, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, C.Y.; Kong, A.N. Induction of phase i, ii and iii drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Raunio, H.; Kuusisto, M.; Juvonen, R.O.; Pentikainen, O.T. Modeling of interactions between xenobiotics and cytochrome p450 (cyp) enzymes. Front. Pharm. 2015, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Wikvall, K.; Miller, W.L. Human cytochromes p450 in health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120431. [Google Scholar] [PubMed]
- Meijerman, I.; Beijnen, J.H.; Schellens, J.H.M. Combined action and regulation of phase ii enzymes and multidrug resistance proteins in multidrug resistance in cancer. Cancer Treat. Rev. 2008, 34, 505–520. [Google Scholar] [CrossRef]
- Chakraborty, S.; Hosen, M.I.; Ahmed, M.; Shekhar, H.U. Onco-multi-omics approach: A new frontier in cancer research. Biomed. Res. Int. 2018, 2018, 9836256. [Google Scholar] [CrossRef] [Green Version]
- Garnis, C.; Buys, T.P.; Lam, W.L. Genetic alteration and gene expression modulation during cancer progression. Mol. Cancer 2004, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; Akiyama, Y.; Yuasa, Y. Multiple-to-multiple relationships between micrornas and target genes in gastric cancer. PLoS ONE 2013, 8, e62589. [Google Scholar] [CrossRef] [Green Version]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of micrornas in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting oncomirnas and mimicking tumor suppressor mirnas: Nuew trends in the development of mirna therapeutic strategies in oncology (review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Lotterman, C.D.; Kent, O.A.; Mendell, J.T. Functional integration of micrornas into oncogenic and tumor suppressor pathways. Cell Cycle 2008, 7, 2493–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drakaki, A.; Iliopoulos, D. Microrna gene networks in oncogenesis. Curr. Genom. 2009, 10, 35–41. [Google Scholar] [CrossRef] [Green Version]
- Fuziwara, C.S.; Kimura, E.T. Insights into regulation of the mir-17-92 cluster of mirnas in cancer. Front. Med. 2015, 2, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, H.; Takeuchi, T.; Nishikawa, E.; Yanagisawa, K.; Hayashita, Y.; Ebi, H.; Yamada, H.; Suzuki, M.; Nagino, M.; Nimura, Y.; et al. Apoptosis induction by antisense oligonucleotides against mir-17-5p and mir-20a in lung cancers overexpressing mir-17-92. Oncogene 2007, 26, 6099–6105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a myc-activated microrna cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.; Nagarkatti, P.S.; Nagarkatti, M. Delta(9) tetrahydrocannabinol attenuates staphylococcal enterotoxin b-induced inflammatory lung injury and prevents mortality in mice by modulation of mir-17-92 cluster and induction of t-regulatory cells. Br. J. Pharm. 2015, 172, 1792–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-micrornas with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Kim, S.; Lee, E.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.J.; Lee, H.J.; Chae, S.Y.; Jeon, S.M.; et al. Microrna-155 positively regulates glucose metabolism via pik3r1-foxo3a-cmyc axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef] [Green Version]
- Frezzetti, D.; De Menna, M.; Zoppoli, P.; Guerra, C.; Ferraro, A.; Bello, A.M.; De Luca, P.; Calabrese, C.; Fusco, A.; Ceccarelli, M.; et al. Upregulation of mir-21 by ras in vivo and its role in tumor growth. Oncogene 2011, 30, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic micrornas: Key players in malignant transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef]
- Buscaglia, L.E.; Li, Y. Apoptosis and the target genes of microrna-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microrna-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.X.; Lu, B.B.; Wang, H.; Cheng, Z.X.; Yin, Y.M. Microrna-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting pten. Arch. Med. Res. 2011, 42, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Lu, Q.; Wu, D.; Li, P.; Xu, B.; Qing, W.; Wang, M.; Zhang, Z.; Zhang, W. Microrna-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol. Rep. 2011, 25, 1721–1729. [Google Scholar] [PubMed]
- Barancik, M.; Bohacova, V.; Sedlak, J.; Sulova, Z.; Breier, A. Ly294,002, a specific inhibitor of pi3k/akt kinase pathway, antagonizes p-glycoprotein-mediated multidrug resistance. Eur. J. Pharm. Sci 2006, 29, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Lowenberg, B.; Ossenkoppele, G.J.; van Putten, W.; Schouten, H.C.; Graux, C.; Ferrant, A.; Sonneveld, P.; Maertens, J.; Jongen-Lavrencic, M.; von Lilienfeld-Toal, M.; et al. High-dose daunorubicin in older patients with acute myeloid leukemia. N. Engl. J. Med. 2009, 361, 1235–1248. [Google Scholar] [CrossRef]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T.; et al. Microrna-21 induces resistance to 5-fluorouracil by down-regulating human DNA muts homolog 2 (hmsh2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Li, Y.; Shen, H.; Li, H.; Long, L.; Hui, L.; Xu, W. Mir-137 restoration sensitizes multidrug-resistant mcf-7/adm cells to anticancer agents by targeting yb-1. Acta Biochim. Biophys. Sin. 2013, 45, 80–86. [Google Scholar] [CrossRef] [Green Version]
- Messingerova, L.; Imrichova, D.; Kavcova, H.; Seres, M.; Sulova, Z.; Breier, A. A decrease in cellular microrna-27a content is involved in azacytidine-induced p-glycoprotein expression in skm-1 cells. Toxicol. Vitr. 2016, 36, 81–88. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, T.; Huang, C.; Zhang, L.; Lv, X.; Xu, T.; Hu, T.; Li, J. Mir-27a modulates the mdr1/p-glycoprotein expression by inhibiting fzd7/beta-catenin pathway in hepatocellular carcinoma cells. Cell Signal. 2013, 25, 2693–2701. [Google Scholar] [CrossRef]
- Zhu, H.; Wu, H.; Liu, X.; Evans, B.R.; Medina, D.J.; Liu, C.G.; Yang, J.M. Role of microrna mir-27a and mir-451 in the regulation of mdr1/p-glycoprotein expression in human cancer cells. Biochem. Pharm. 2008, 76, 582–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drayton, R.M.; Dudziec, E.; Peter, S.; Bertz, S.; Hartmann, A.; Bryant, H.E.; Catto, J.W. Reduced expression of mirna-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger slc7a11. Clin. Cancer Res. 2014, 20, 1990–2000. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.Y.; Cui, S.Y.; Chen, Y.T.; Song, H.Z.; Huang, G.C.; Feng, B.; Sun, M.; De, W.; Wang, R.; Chen, L.B. Microrna-650 was a prognostic factor in human lung adenocarcinoma and confers the docetaxel chemoresistance of lung adenocarcinoma cells via regulating bcl-2/bax expression. PLoS ONE 2013, 8, e72615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.X.; Gao, S.; Pan, Y.Z.; Yu, C.; Sun, C.Y. Overexpression of microrna-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Mol. Med. Rep. 2014, 10, 995–1002. [Google Scholar] [CrossRef] [Green Version]
- Kastl, L.; Brown, I.; Schofield, A.C. Mirna-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Ru, P.; Steele, R.; Hsueh, E.C.; Ray, R.B. Anti-mir-203 upregulates socs3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer 2011, 2, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Hasskarl, J. Sorafenib: Targeting multiple tyrosine kinases in cancer. In Recent Results Cancer Res; Springer: Berlin/Heidelberg, Germany, 2014; Volume 201, pp. 145–164. [Google Scholar]
- Metibemu, D.S.; Akinloye, O.A.; Akamo, A.J.; Ojo, D.A.; Okeowo, O.T.; Omotuyi, I.O. Exploring receptor tyrosine kinases-inhibitors in cancer treatments. Egypt. J. Med. Hum. Genet. 2019, 20, 35. [Google Scholar] [CrossRef] [Green Version]
- Procopio, G.; Verzoni, E.; Testa, I.; Nicolai, N.; Salvioni, R.; Debraud, F. Experience with sorafenib in the treatment of advanced renal cell carcinoma. Ther. Adv. Urol. 2012, 4, 303–313. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, J.; He, C.; Xu, K.; Liu, J.; Sun, J.; Wu, G.; Tan, C.; Zeng, Y.; Wang, J.; et al. Restoration of mir-193b sensitizes hepatitis b virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014, 352, 245–252. [Google Scholar] [CrossRef]
- Hano, M.; Tomasova, L.; Seres, M.; Pavlikova, L.; Breier, A.; Sulova, Z. Interplay between p-glycoprotein expression and resistance to endoplasmic reticulum stressors. Molecules 2018, 23, 337. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-relevant abc transporter for anti-cancer drug resistance. Front. Pharm. 2021, 12, 648407. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zheng, Z.M.; Li, X.N.; Li, Z.F.; Wang, Y.; Geng, Y.F.; Bai, L.; Zhang, X.B. Mir-223 modulates multidrug resistance via downregulation of abcb1 in hepatocellular carcinoma cells. Exp. Biol. Med. 2013, 238, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, T.; Guo, R.; Yang, X.; Yin, J.; Yu, J.; Xiang, Q.; Pan, X.; Tang, H.; Lei, X. Involvement of mir-133a and mir-326 in adm resistance of hepg2 through modulating expression of abcc1. J. Drug Target 2015, 23, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Aldossary, S.A. Review on pharmacology of cisplatin: Clinical use, toxicity and mechanism of resistance of cisplatin. Biomed. Pharm. J. 2019, 12, 7–15. [Google Scholar] [CrossRef]
- Zhan, M.; Qu, Q.; Wang, G.; Zhou, H. Let-7c sensitizes acquired cisplatin-resistant a549 cells by targeting abcc2 and bcl-xl. Pharmazie 2013, 68, 955–961. [Google Scholar]
- Dong, Z.; Zhong, Z.; Yang, L.; Wang, S.; Gong, Z. Microrna-31 inhibits cisplatin-induced apoptosis in non-small cell lung cancer cells by regulating the drug transporter abcb9. Cancer Lett. 2014, 343, 249–257. [Google Scholar] [CrossRef]
- Wang, Q.; Zhong, M.; Liu, W.; Li, J.; Huang, J.; Zheng, L. Alterations of micrornas in cisplatin-resistant human non-small cell lung cancer cells (a549/ddp). Exp. Lung Res. 2011, 37, 427–434. [Google Scholar] [CrossRef]
- Ning, F.L.; Wang, F.; Li, M.L.; Yu, Z.S.; Hao, Y.Z.; Chen, S.S. Microrna-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting pdcd4. Diagn Pathol. 2014, 9, 143. [Google Scholar] [CrossRef] [Green Version]
- Lei, L.; Huang, Y.; Gong, W. Mir-205 promotes the growth, metastasis and chemoresistance of nsclc cells by targeting pten. Oncol. Rep. 2013, 30, 2897–2902. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhu, L.J.; Yang, Y.C.; Wang, Z.X.; Wang, R. Mir-224 promotes the chemoresistance of human lung adenocarcinoma cells to cisplatin via regulating g(1)/s transition and apoptosis by targeting p21(waf1/cip1). Br. J. Cancer 2014, 111, 339–354. [Google Scholar] [CrossRef]
- Ma, Y.; Li, X.; Cheng, S.; Wei, W.; Li, Y. Microrna-106a confers cisplatin resistance in non-small cell lung cancer a549 cells by targeting adenosine triphosphatase-binding cassette a1. Mol. Med. Rep. 2015, 11, 625–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, L.; Yao, Q.; Tao, Z. Mir-15b regulates cisplatin resistance and metastasis by targeting pebp4 in human lung adenocarcinoma cells. Cancer Gene Ther. 2015, 22, 108–114. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Song, Y.; Fu, Z.; Yu, W. Mir-27a regulates cisplatin resistance and metastasis by targeting rkip in human lung adenocarcinoma cells. Mol. Cancer 2014, 13, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhu, J.; Xing, R.; Tie, Y.; Fu, H.; Zheng, X.; Yu, B. Mir-513a-3p sensitizes human lung adenocarcinoma cells to chemotherapy by targeting gstp1. Lung Cancer 2012, 77, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Zhong, N.; Chen, G.; Huang, B.; Wu, S. Downregulation of pebp4, a target of mir-34a, sensitizes drug-resistant lung cancer cells. Tumour. Biol. 2014, 35, 10341–10349. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Guan, Y.; Liu, X.; Meng, Q.; Guo, Q. Mir-92b regulates the cell growth, cisplatin chemosensitivity of a549 non small cell lung cancer cell line and target pten. Biochem. Biophys. Res. Commun. 2013, 440, 604–610. [Google Scholar] [CrossRef]
- Das, M.K.; Evensen, H.S.F.; Furu, K.; Haugen, T.B. Mirna-302s may act as oncogenes in human testicular germ cell tumours. Sci. Rep. 2019, 9, 9189. [Google Scholar] [CrossRef] [Green Version]
- Echevarria-Vargas, I.M.; Valiyeva, F.; Vivas-Mejia, P.E. Upregulation of mir-21 in cisplatin resistant ovarian cancer via jnk-1/c-jun pathway. PLoS ONE 2014, 9, e97094. [Google Scholar] [CrossRef]
- Nishida, N.; Yamashita, S.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Yamamoto, H.; Ishii, H.; Doki, Y.; Mori, M. Microrna-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann. Surg. Oncol. 2012, 19, 3065–3071. [Google Scholar] [CrossRef]
- Chai, H.; Liu, M.; Tian, R.; Li, X.; Tang, H. Mir-20a targets bnip2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma sw480 and sw620 cell lines. Acta Biochim. Biophys. Sin. 2011, 43, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Boyerinas, B.; Park, S.M.; Murmann, A.E.; Gwin, K.; Montag, A.G.; Zillhardt, M.; Hua, Y.J.; Lengyel, E.; Peter, M.E. Let-7 modulates acquired resistance of ovarian cancer to taxanes via imp-1-mediated stabilization of multidrug resistance 1. Int. J. Cancer 2012, 130, 1787–1797. [Google Scholar] [CrossRef] [Green Version]
- Messingerova, L.; Imrichova, D.; Coculova, M.; Zelina, M.; Pavlikova, L.; Kavcova, H.; Seres, M.; Bohacova, V.; Lakatos, B.; Sulova, Z.; et al. Different mechanisms of drug resistance in myelodysplastic syndromes and acute myeloid leukemia. In Myelodysplastic Syndromes; Fusch, O., Ed.; Intech: Rijeka, Croatia, 2016; pp. 181–200. [Google Scholar]
- Ishikawa, T.; Yoshikawa, M. ABC transporters: A new approach to toxicogenomics. In Toxicogenomics; Inoue, T., Pennie, W.T., Eds.; Springer: Tokyo, Janpan, 2003; pp. 109–114. [Google Scholar]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of abc transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Antona, C.; Ingelman-Sundberg, M. Cytochrome p450 pharmacogenetics and cancer. Oncogene 2006, 25, 1679–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guengerich, F.P. Mechanisms of cytochrome p450-catalyzed oxidations. ACS Catal. 2018, 8, 10964–10976. [Google Scholar] [CrossRef] [PubMed]
- Srejber, M.; Navratilova, V.; Paloncyova, M.; Bazgier, V.; Berka, K.; Anzenbacher, P.; Otyepka, M. Membrane-attached mammalian cytochromes p450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners. J. Inorg. Biochem. 2018, 183, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Rendic, S.; Guengerich, F.P. Survey of human oxidoreductases and cytochrome p450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem. Res. Toxicol. 2015, 28, 38–42. [Google Scholar] [CrossRef] [Green Version]
- Rydberg, P.; Olsen, L.; Ryde, U. Quantum-mechanical studies of reactions performed by cytochrome p450 enzymes. Curr. Inorg. Chem. 2012, 2, 292–315. [Google Scholar] [CrossRef] [Green Version]
- Hakkola, J.; Bernasconi, C.; Coecke, S.; Richert, L.; Andersson, T.B.; Pelkonen, O. Cytochrome p450 induction and xeno-sensing receptors pregnane x receptor, constitutive androstane receptor, aryl hydrocarbon receptor and peroxisome proliferator-activated receptor alpha at the crossroads of toxicokinetics and toxicodynamics. Basic Clin. Pharmacol. Toxicol. 2018, 123 (Suppl. 5), 42–50. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Tolleson, W.H.; Yu, D.; Chen, S.; Guo, L.; Xiao, W.; Tong, W.; Ning, B. Regulation of cytochrome p450 expression by micrornas and long noncoding rnas: Epigenetic mechanisms in environmental toxicology and carcinogenesis. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2019, 37, 180–214. [Google Scholar] [CrossRef]
- Guo, J.; Zhu, X.; Badawy, S.; Ihsan, A.; Liu, Z.; Xie, C.; Wang, X. Metabolism and mechanism of human cytochrome p450 enzyme 1a2. Curr. Drug Metab. 2021, 22, 40–49. [Google Scholar] [CrossRef]
- Thorn, C.F.; Aklillu, E.; Klein, T.E.; Altman, R.B. Pharmgkb summary: Very important pharmacogene information for cyp1a2. Pharm. Genom. 2012, 22, 73–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, Z.B.; Ye, M.; Cheng, Y.; Wu, W.Z. Four polymorphisms in the cytochrome p450 1a2 (cyp1a2) gene and lung cancer risk: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 5673–5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elfaki, I.; Mir, R.; Almutairi, F.M.; Duhier, F.M.A. Cytochrome p450: Polymorphisms and roles in cancer, diabetes and atherosclerosis. Asian Pac. J. Cancer Prev. 2018, 19, 2057–2070. [Google Scholar] [PubMed]
- Chen, Y.; Zeng, L.; Wang, Y.; Tolleson, W.H.; Knox, B.; Chen, S.; Ren, Z.; Guo, L.; Mei, N.; Qian, F.; et al. The expression, induction and pharmacological activity of cyp1a2 are post-transcriptionally regulated by microrna hsa-mir-132-5p. Biochem. Pharmacol. 2017, 145, 178–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackburn, H.L.; Ellsworth, D.L.; Shriver, C.D.; Ellsworth, R.E. Role of cytochrome p450 genes in breast cancer etiology and treatment: Effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control. 2015, 26, 319–332. [Google Scholar] [CrossRef]
- Shimada, T. Inhibition of carcinogen-activating cytochrome p450 enzymes by xenobiotic chemicals in relation to antimutagenicity and anticarcinogenicity. Toxicol. Res. 2017, 33, 79–96. [Google Scholar] [CrossRef]
- Martinez, V.G.; O’Connor, R.; Liang, Y.; Clynes, M. Cyp1b1 expression is induced by docetaxel: Effect on cell viability and drug resistance. Br. J. Cancer 2008, 98, 564–570. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Nakajima, M.; Takagi, S.; Taniya, T.; Yokoi, T. Microrna regulates the expression of human cytochrome p450 1b1. Cancer Res. 2006, 66, 9090–9098. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Hu, C.; Zhang, H.; Qu, Z.; Cen, J.; Qiu, Z.; Li, C.; Ren, H.; Li, Y.; He, X.; et al. Mir-27b synergizes with anticancer drugs via p53 activation and cyp1b1 suppression. Cell Res. 2015, 25, 477–495. [Google Scholar] [CrossRef] [Green Version]
- Chang, I.; Mitsui, Y.; Fukuhara, S.; Gill, A.; Wong, D.K.; Yamamura, S.; Shahryari, V.; Tabatabai, Z.L.; Dahiya, R.; Shin, D.M.; et al. Loss of mir-200c up-regulates cyp1b1 and confers docetaxel resistance in renal cell carcinoma. Oncotarget 2015, 6, 7774–7787. [Google Scholar] [CrossRef] [Green Version]
- Mao, M.; Wu, Z.; Chen, J. Microrna-187-5p suppresses cancer cell progression in non-small cell lung cancer (nsclc) through down-regulation of cyp1b1. Biochem. Biophys. Res. Commun. 2016, 478, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Rieger, J.K.; Reutter, S.; Hofmann, U.; Schwab, M.; Zanger, U.M. Inflammation-associated microrna-130b down-regulates cytochrome p450 activities and directly targets cyp2c9. Drug Metab. Dispos. 2015, 43, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, L.; Skilving, I.; Ovesjo, M.L.; Aklillu, E.; Nylen, H.; Rane, A.; Diczfalusy, U.; Bjorkhem-Bergman, L. Mirna-27b levels are associated with cyp3a activity in vitro and in vivo. Pharm. Res. Perspect. 2015, 3, e00192. [Google Scholar] [CrossRef]
- Pan, Y.Z.; Gao, W.; Yu, A.M. Micrornas regulate cyp3a4 expression via direct and indirect targeting. Drug Metab. Dispos. 2009, 37, 2112–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.E.; Ren, B.; Tang, L.; Tang, Q.J.; Liu, X.Y.; Li, X.; Bai, X.; Zhong, W.P.; Meng, J.X.; Lin, H.M.; et al. The independent contribution of mirnas to the missing heritability in cyp3a4/5 functionality and the metabolism of atorvastatin. Sci. Rep. 2016, 6, 26544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, H.M.; Fang, Z.; Sun, W.; Clark, L.H.; Stine, J.E.; Tran, A.Q.; Sullivan, S.A.; Gilliam, T.P.; Zhou, C.; Bae-Jump, V.L. Atorvastatin exhibits anti-tumorigenic and anti-metastatic effects in ovarian cancer in vitro. Am. J. Cancer Res. 2017, 7, 2478–2490. [Google Scholar]
- Takagi, S.; Nakajima, M.; Mohri, T.; Yokoi, T. Post-transcriptional regulation of human pregnane x receptor by micro-rna affects the expression of cytochrome p450 3a4. J. Biol. Chem. 2008, 283, 9674–9680. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chen, C.; Yang, S.; Gong, W.; Wang, Y.; Cianflone, K.; Tang, J.; Wang, D.W. Let-7b inhibits human cancer phenotype by targeting cytochrome p450 epoxygenase 2j2. PLoS ONE 2012, 7, e39197. [Google Scholar] [CrossRef] [Green Version]
- Jančová, P.; Šiller, M. Phase II Drug Metabolism. In Topics on drug metabolism; Paxton, J., Ed.; InTech: Houston, TX, USA, 2012. [Google Scholar]
- Hodges, R.E.; Minich, D.M. Modulation of metabolic detoxification pathways using foods and food-derived components: A scientific review with clinical application. J. Nutr. Metab. 2015, 2015, 760689. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, J.; Zhang, X.; Bian, X. Micrornas as key mediators of hepatic detoxification. Toxicology 2016, 368–369, 80–90. [Google Scholar] [CrossRef]
- Nakano, M.; Nakajima, M. Current knowledge of microrna-mediated regulation of drug metabolism in humans. Expert. Opin. Drug Metab. Toxicol. 2018, 14, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Duffel, M.W.; Marshal, A.D.; McPhie, P.; Sharma, V.; Jakoby, W.B. Enzymatic aspects of the phenol (aryl) sulfotransferases. Drug Metab. Rev. 2001, 33, 369–395. [Google Scholar] [CrossRef] [PubMed]
- Mercer, K.E.; Apostolov, E.O.; Gamboa da Costa, G.; Yu, X.; Lang, P.; Roberts, D.W.; Davis, W.; Basnakian, A.G.; Kadlubar, F.F.; Kadlubar, S.A. Expression of sulfotransferase isoform 1a1 (sult1a1) in breast cancer cells significantly increases 4-hydroxytamoxifen-induced apoptosis. Int. J. Mol. Epidemiol. Genet. 2010, 1, 92–103. [Google Scholar] [PubMed]
- Yu, X.; Dhakal, I.B.; Beggs, M.; Edavana, V.K.; Williams, S.; Zhang, X.; Mercer, K.; Ning, B.; Lang, N.P.; Kadlubar, F.F.; et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1a1 (sult1a1) and their effect on enzymatic activity. Toxicol. Sci. 2010, 118, 391–403. [Google Scholar] [CrossRef] [Green Version]
- McLellan, L.I.; Wolf, C.R. Glutathione and glutathione-dependent enzymes in cancer drug resistance. Drug Resist. Updat. 1999, 2, 153–164. [Google Scholar] [CrossRef]
- Sawers, L.; Ferguson, M.J.; Ihrig, B.R.; Young, H.C.; Chakravarty, P.; Wolf, C.R.; Smith, G. Glutathione s-transferase p1 (gstp1) directly influences platinum drug chemosensitivity in ovarian tumour cell lines. Br. J. Cancer 2014, 111, 1150–1158. [Google Scholar] [CrossRef]
- Cagala, M.; Pavlikova, L.; Seres, M.; Kadlecikova, K.; Breier, A.; Sulova, Z. Development of resistance to endoplasmic reticulum stress-inducing agents in mouse leukemic l1210 cells. Molecules 2020, 25, 2517. [Google Scholar] [CrossRef]
- Gibalova, L.; Seres, M.; Rusnak, A.; Ditte, P.; Labudova, M.; Uhrik, B.; Pastorek, J.; Sedlak, J.; Breier, A.; Sulova, Z. P-glycoprotein depresses cisplatin sensitivity in l1210 cells by inhibiting cisplatin-induced caspase-3 activation. Toxicol. Vitr. 2012, 26, 435–444. [Google Scholar] [CrossRef]
- Moriya, Y.; Nohata, N.; Kinoshita, T.; Mutallip, M.; Okamoto, T.; Yoshida, S.; Suzuki, M.; Yoshino, I.; Seki, N. Tumor suppressive microrna-133a regulates novel molecular networks in lung squamous cell carcinoma. J. Hum. Genet. 2012, 57, 38–45. [Google Scholar] [CrossRef] [Green Version]
- Papageorgiou, I.; Court, M.H. Identification and validation of micrornas directly regulating the udp-glucuronosyltransferase 1a subfamily enzymes by a functional genomics approach. Biochem. Pharm. 2017, 137, 93–106. [Google Scholar] [CrossRef]
- Meng, C.L.; Zhao, W.; Zhong, D.N. Epigenetics and micrornas in ugt1as. Hum. Genom. 2021, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.C.; Bartels, H.; Viviani, R.; Lehmann, M.L.; Brockmoller, J. Relevance of udp-glucuronosyltransferase polymorphisms for drug dosing: A quantitative systematic review. Pharmacol. Ther. 2014, 141, 92–116. [Google Scholar] [CrossRef] [PubMed]
- Dluzen, D.F.; Sutliff, A.K.; Chen, G.; Watson, C.J.; Ishmael, F.T.; Lazarus, P. Regulation of ugt2b expression and activity by mir-216b-5p in liver cancer cell lines. J. Pharmacol. Exp. Ther. 2016, 359, 182–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijayakumara, D.D.; Mackenzie, P.I.; McKinnon, R.A.; Hu, D.G.; Meech, R. Regulation of udp-glucuronosyltransferases ugt2b4 and ugt2b7 by micrornas in liver cancer cells. J. Pharmacol. Exp. Ther. 2017, 361, 386–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, G.; Morris, M.E. Overview of drug transporter families. In Drug Transporters: Molecular Characterization and Role in Drug Disposition, 2nd ed.; You, G., Morris, M.E., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2014; Volume 4, pp. 1–6. [Google Scholar]
- Breier, A.; Gibalova, L.; Seres, M.; Barancik, M.; Sulova, Z. P-Glycoprotein Mediated Multidrug Resistance of Cancer Tissue: Implication for Cancer Chemotherapy; PETRUS Publisher: Bratislava, Slovakia, 2012. [Google Scholar]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of atp-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human atp-binding cassette (abc) transporter family. Hum. Genom. 2009, 3, 281–290. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Ling, V. The molecular basis of multidrug resistance in cancer: The early years of p-glycoprotein research. FEBS Lett. 2006, 580, 998–1009. [Google Scholar] [CrossRef] [Green Version]
- Juliano, R.L.; Ling, V. A surface glycoprotein modulating drug permeability in chinese hamster ovary cell mutants. Biochim. Biophys. Acta 1976, 455, 152–162. [Google Scholar] [CrossRef]
- Vaidyanathan, A.; Sawers, L.; Gannon, A.L.; Chakravarty, P.; Scott, A.L.; Bray, S.E.; Ferguson, M.J.; Smith, G. Abcb1 (mdr1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 2016, 115, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Yan, L.J.; Wu, Q. Abcb1 (c1236t) polymorphism affects p-glycoprotein-mediated transport of methotrexate, doxorubicin, actinomycin d, and etoposide. DNA Cell Biol. 2019, 38, 485–490. [Google Scholar] [CrossRef]
- Tecza, K.; Pamula-Pilat, J.; Lanuszewska, J.; Grzybowska, E. Genetic polymorphisms and response to 5-fluorouracil, doxorubicin and cyclophosphamide chemotherapy in breast cancer patients. Oncotarget 2016, 7, 66790–66808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.R.; Jamieson, D.; Thomas, H.D.; Brown, C.D.; Boddy, A.V.; Veal, G.J. Characterisation of the roles of abcb1, abcc1, abcc2 and abcg2 in the transport and pharmacokinetics of actinomycin d in vitro and in vivo. Biochem. Pharm. 2013, 85, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakanishi, T.; Menju, T.; Nishikawa, S.; Takahashi, K.; Miyata, R.; Shikuma, K.; Sowa, T.; Imamura, N.; Hamaji, M.; Motoyama, H.; et al. The synergistic role of atp-dependent drug efflux pump and focal adhesion signaling pathways in vinorelbine resistance in lung cancer. Cancer Med. 2018, 7, 408–419. [Google Scholar] [CrossRef] [Green Version]
- Gromicho, M.; Dinis, J.; Magalhaes, M.; Fernandes, A.R.; Tavares, P.; Laires, A.; Rueff, J.; Rodrigues, A.S. Development of imatinib and dasatinib resistance: Dynamics of expression of drug transporters abcb1, abcc1, abcg2, mvp, and slc22a1. Leuk. Lymphoma 2011, 52, 1980–1990. [Google Scholar] [CrossRef] [PubMed]
- Seres, M.; Pavlikova, L.; Bohacova, V.; Kyca, T.; Borovska, I.; Lakatos, B.; Breier, A.; Sulova, Z. Overexpression of grp78/bip in p-glycoprotein-positive l1210 cells is responsible for altered response of cells to tunicamycin as a stressor of the endoplasmic reticulum. Cells 2020, 9, 890. [Google Scholar] [CrossRef] [Green Version]
- Seres, M.; Polakova, E.; Krizanova, O.; Hudecova, S.; Klymenko, S.V.; Breier, A.; Sulova, Z. Overexpression of p-glycoprotein in l1210/vcr cells is associated with changes in several endoplasmic reticulum proteins that may be partially responsible for the lack of thapsigargin sensitivity. Gen. Physiol. Biophys. 2008, 27, 211–221. [Google Scholar]
- Sulova, Z.; Ditte, P.; Kurucova, T.; Polakova, E.; Rogozanova, K.; Gibalova, L.; Seres, M.; Skvarkova, L.; Sedlak, J.; Pastorek, J.; et al. The presence of p-glycoprotein in l1210 cells directly induces down-regulation of cell surface saccharide targets of concanavalin a. Anticancer Res. 2010, 30, 3661–3668. [Google Scholar]
- Sulova, Z.; Mislovicova, D.; Gibalova, L.; Vajcnerova, Z.; Polakova, E.; Uhrik, B.; Tylkova, L.; Kovarova, A.; Sedlak, J.; Breier, A. Vincristine-induced overexpression of p-glycoprotein in l1210 cells is associated with remodeling of cell surface saccharides. J. Proteome Res. 2009, 8, 513–520. [Google Scholar] [CrossRef]
- Wang, F.; Li, T.; Zhang, B.; Li, H.; Wu, Q.; Yang, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. Microrna-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem. Biophys. Res. Commun. 2013, 434, 688–694. [Google Scholar] [CrossRef]
- Li, N.; Yang, L.; Wang, H.; Yi, T.; Jia, X.; Chen, C.; Xu, P. Mir-130a and mir-374a function as novel regulators of cisplatin resistance in human ovarian cancer a2780 cells. PLoS ONE 2015, 10, e0128886. [Google Scholar] [CrossRef]
- Jin, J.; Yao, J.; Yue, F.; Jin, Z.; Li, D.; Wang, S. Decreased expression of microrna-214 contributes to imatinib mesylate resistance of chronic myeloid leukemia patients by upregulating abcb1 gene expression. Exp. Ther. Med. 2018, 16, 1693–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Boya, V.K.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. Mirna-205 nanoformulation sensitizes prostate cancer cells to chemotherapy. Cancers 2018, 10, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Lu, F.Y.; Shi, R.H.; Feng, Y.D.; Zhao, X.D.; Lu, Z.P.; Xiao, L.; Zhou, G.Q.; Qiu, J.M.; Cheng, C.E. Mir-26b regulates 5-fu-resistance in human colorectal cancer via down-regulation of PGP. Am. J. Cancer Res. 2018, 8, 2518–2527. [Google Scholar] [PubMed]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microrna-451 in resistance of the mcf-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef] [Green Version]
- Feng, D.D.; Zhang, H.; Zhang, P.; Zheng, Y.S.; Zhang, X.J.; Han, B.W.; Luo, X.Q.; Xu, L.; Zhou, H.; Qu, L.H.; et al. Down-regulated mir-331-5p and mir-27a are associated with chemotherapy resistance and relapse in leukaemia. J. Cell Mol. Med. 2011, 15, 2164–2175. [Google Scholar] [CrossRef] [Green Version]
- Gromnicova, R.; Romero, I.; Male, D. Transcriptional control of the multi-drug transporter abcb1 by transcription factor sp3 in different human tissues. PLoS ONE 2012, 7, e48189. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, X.; Chen, J.; Wei, W.; Yu, C.; Yan, H.; Pu, M.; Li, Y.; Miao, L.; Li, C.; et al. The mir-491-3p/sp3/abcb1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett. 2017, 408, 102–111. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, L.; Hu, J.; Ruan, J. Mir-138 might reverse multidrug resistance of leukemia cells. Leuk Res. 2010, 34, 1078–1082. [Google Scholar] [CrossRef]
- Bao, L.; Hazari, S.; Mehra, S.; Kaushal, D.; Moroz, K.; Dash, S. Increased expression of p-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by mir-298. Am. J. Pathol. 2012, 180, 2490–2503. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Ohms, S.J.; Li, Z.; Wang, Q.; Gong, G.; Hu, Y.; Mao, Z.; Shannon, M.F.; Fan, J.Y. Changes in the expression of mir-381 and mir-495 are inversely associated with the expression of the mdr1 gene and development of multi-drug resistance. PLoS ONE 2013, 8, e82062. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Zhang, Z.; Liu, Z.; Feng, B.; Ren, G.; Li, K.; Zhou, L.; Sun, Y.; Li, M.; Zhou, J.; et al. Mir-508-5p regulates multidrug resistance of gastric cancer by targeting abcb1 and znrd1. Oncogene 2014, 33, 3267–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, L.; Ross, D.D. Multidrug resistance mediated by the breast cancer resistance protein bcrp (abcg2). Oncogene 2003, 22, 7340–7358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The role of abc transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polgar, O.; Robey, R.W.; Bates, S.E. Abcg2: Structure, function and role in drug response. Expert. Opin. Drug Metab. Toxicol. 2008, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Haimeur, A.; Conseil, G.; Deeley, R.G.; Cole, S.P. The mrp-related and bcrp/abcg2 multidrug resistance proteins: Biology, substrate specificity and regulation. Curr. Drug Metab. 2004, 5, 21–53. [Google Scholar] [CrossRef]
- To, K.K.; Zhan, Z.; Litman, T.; Bates, S.E. Regulation of abcg2 expression at the 3’ untranslated region of its mrna through modulation of transcript stability and protein translation by a putative microrna in the s1 colon cancer cell line. Mol. Cell Biol. 2008, 28, 5147–5161. [Google Scholar] [CrossRef] [Green Version]
- Li, W.Q.; Li, Y.M.; Tao, B.B.; Lu, Y.C.; Hu, G.H.; Liu, H.M.; He, J.; Xu, Y.; Yu, H.Y. Downregulation of abcg2 expression in glioblastoma cancer stem cells with mirna-328 may decrease their chemoresistance. Med. Sci. Monit. 2010, 16, HY27–HY30. [Google Scholar]
- Pan, Y.Z.; Morris, M.E.; Yu, A.M. Microrna-328 negatively regulates the expression of breast cancer resistance protein (bcrp/abcg2) in human cancer cells. Mol. Pharm. 2009, 75, 1374–1379. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.; Robey, R.W.; Knutsen, T.; Zhan, Z.; Ried, T.; Bates, S.E. Escape from hsa-mir-519c enables drug-resistant cells to maintain high expression of abcg2. Mol. Cancer Ther. 2009, 8, 2959–2968. [Google Scholar] [CrossRef] [Green Version]
- Liao, R.; Sun, J.; Zhang, L.; Lou, G.; Chen, M.; Zhou, D.; Chen, Z.; Zhang, S. Micrornas play a role in the development of human hematopoietic stem cells. J. Cell. Biochem. 2008, 104, 805–817. [Google Scholar] [CrossRef]
- Wang, F.; Xue, X.; Wei, J.; An, Y.; Yao, J.; Cai, H.; Wu, J.; Dai, C.; Qian, Z.; Xu, Z.; et al. Hsa-mir-520h downregulates abcg2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br. J. Cancer 2010, 103, 567–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, X.; Zhao, L.; Ma, M.; Bai, X.; He, M.; Yan, Y.; Wang, Y.; Chen, Q.; Zhao, X.; Zhou, M.; et al. Mir-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (bcrp/abcg2). Breast. Cancer Res. Treat. 2013, 139, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Y.Z.; Seigel, G.M.; Hu, Z.H.; Huang, M.; Yu, A.M. Breast cancer resistance protein bcrp/abcg2 regulatory micrornas (hsa-mir-328, -519c and -520h) and their differential expression in stem-like abcg2+ cancer cells. Biochem. Pharm. 2011, 81, 783–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, M.T.; He, M.; Wang, Y.; Jiao, X.Y.; Zhao, L.; Bai, X.F.; Yu, Z.J.; Wu, H.Z.; Sun, M.L.; Song, Z.G.; et al. Mir-487a resensitizes mitoxantrone (mx)-resistant breast cancer cells (mcf-7/mx) to mx by targeting breast cancer resistance protein (bcrp/abcg2). Cancer Lett. 2013, 339, 107–115. [Google Scholar] [CrossRef]
- Turrini, E.; Haenisch, S.; Laechelt, S.; Diewock, T.; Bruhn, O.; Cascorbi, I. Microrna profiling in k-562 cells under imatinib treatment: Influence of mir-212 and mir-328 on abcg2 expression. Pharm. Genom. 2012, 22, 198–205. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S.; Zhang, L. Upregulated mir-132 in lgr5(+) gastric cancer stem cell-like cells contributes to cisplatin-resistance via sirt1/creb/abcg2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Jia, M.; Wei, Z.; Liu, P.; Zhao, X. Silencing of abcg2 by microrna-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. J. Korean Med. Sci. 2016, 31, 836–842. [Google Scholar] [CrossRef]
- Szentpetery, Z.; Kern, A.; Liliom, K.; Sarkadi, B.; Varadi, A.; Bakos, E. The role of the conserved glycines of atp-binding cassette signature motifs of mrp1 in the communication between the substrate-binding site and the catalytic centers. J. Biol. Chem. 2004, 279, 41670–41678. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Wu, H.; Xia, J.; Li, Y.; Zhang, Y.; Huang, K.; Wagar, N.; Yoon, Y.; Cho, H.T.; Scala, S.; et al. Involvement of mir-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem. Pharm. 2010, 79, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.Z.; Zhou, A.; Hu, Z.; Yu, A.M. Small nucleolar rna-derived microrna hsa-mir-1291 modulates cellular drug disposition through direct targeting of abc transporter abcc1. Drug Metab. Dispos. 2013, 41, 1744–1751. [Google Scholar] [CrossRef]
- Zhan, M.; Zhao, X.; Wang, H.; Chen, W.; Xu, S.; Wang, W.; Shen, H.; Huang, S.; Wang, J. Mir-145 sensitizes gallbladder cancer to cisplatin by regulating multidrug resistance associated protein 1. Tumour. Biol. 2016, 37, 10553–10562. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.M.; Ravicz, J.R.; Liu, S.; Chawla, S.P.; Hall, F.L. Cell cycle checkpoint control: The cyclin g1/mdm2/p53 axis emerges as a strategic target for broad-spectrum cancer gene therapy—A review of molecular mechanisms for oncologists. Mol. Clin. Oncol. 2018, 9, 115–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aylon, Y.; Oren, M. Living with p53, dying of p53. Cell 2007, 130, 597–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Leslie, P.L.; Zhang, Y. Life and death decision-making by p53 and implications for cancer immunotherapy. Trends. Cancer 2021, 7, 226–239. [Google Scholar] [CrossRef]
- Oren, M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003, 10, 431–442. [Google Scholar] [CrossRef]
- Hermeking, H. P53 enters the microrna world. Cancer Cell 2007, 12, 414–418. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. Microrna-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.S.; Su, J.L.; Hung, M.C. Dysregulation of micrornas in cancer. J. Biomed. Sci 2012, 19, 90. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Zhang, C.; Wu, R.; Hu, W. Tumor suppressor p53 meets micrornas. J. Mol. Cell Biol. 2011, 3, 44–50. [Google Scholar] [CrossRef]
- Fatt, M.P.; Cancino, G.I.; Miller, F.D.; Kaplan, D.R. P63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ. 2014, 21, 1546–1559. [Google Scholar] [CrossRef] [Green Version]
- Ory, B.; Ramsey, M.R.; Wilson, C.; Vadysirisack, D.D.; Forster, N.; Rocco, J.W.; Rothenberg, S.M.; Ellisen, L.W. A microrna-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J. Clin. Investig. 2011, 121, 809–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, K.; Fukushi, J.; Matsumoto, Y.; Oda, Y.; Takahashi, Y.; Fujiwara, T.; Fujiwara-Okada, Y.; Hatano, M.; Nabashima, A.; Kamura, S.; et al. Mir-125b develops chemoresistance in ewing sarcoma/primitive neuroectodermal tumor. Cancer Cell Int. 2013, 13, 21. [Google Scholar] [CrossRef] [Green Version]
- Fornari, F.; Gramantieri, L.; Giovannini, C.; Veronese, A.; Ferracin, M.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Tavolari, S.; et al. Mir-122/cyclin g1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009, 69, 5761–5767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of mir-34a on cell growth and chemoresistance in prostate cancer pc3 cells. Biochem. Biophys. Res. Commun. 2008, 377, 114–119. [Google Scholar] [CrossRef]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Korner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of mir-34a by aberrant cpg methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, B.; Wang, Y.; Xi, Y.; Kudo, K.; Bruheim, S.; Botchkina, G.I.; Gavin, E.; Wan, Y.; Formentini, A.; Kornmann, M.; et al. Mechanism of chemoresistance mediated by mir-140 in human osteosarcoma and colon cancer cells. Oncogene 2009, 28, 4065–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Geng, L.; Talmon, G.; Wang, J. Microrna-520g confers drug resistance by regulating p21 expression in colorectal cancer. J. Biol. Chem. 2015, 290, 6215–6225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cataldo, A.; Cheung, D.G.; Balsari, A.; Tagliabue, E.; Coppola, V.; Iorio, M.V.; Palmieri, D.; Croce, C.M. Mir-302b enhances breast cancer cell sensitivity to cisplatin by regulating e2f1 and the cellular DNA damage response. Oncotarget 2016, 7, 786–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. Microrna-221/222 confers tamoxifen resistance in breast cancer by targeting p27kip1. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Ding, K.; Zhang, G.; Yin, M.; Yao, G.; Tian, H.; Lian, J.; Liu, L.; Liang, M.; Zhu, T.; et al. Microrna-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting arpp-19 and errgamma. Sci. Rep. 2015, 5, 8735. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Liu, J.; Wang, C.; Wang, Y.; Jiang, Y.; Guo, M. Microrna-25 regulates small cell lung cancer cell development and cell cycle through cyclin e2. Int. J. Clin. Exp. Pathol. 2014, 7, 7726–7734. [Google Scholar] [PubMed]
- Ivanov, S.V.; Goparaju, C.M.; Lopez, P.; Zavadil, J.; Toren-Haritan, G.; Rosenwald, S.; Hoshen, M.; Chajut, A.; Cohen, D.; Pass, H.I. Pro-tumorigenic effects of mir-31 loss in mesothelioma. J. Biol. Chem. 2010, 285, 22809–22817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, K.J.; Tait, S.W.G. Targeting bcl-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Liu, X.; Li, D.; Wang, Q.; Zhang, W.; Li, L. Tumor suppressor genes associated with drug resistance in ovarian cancer (review). Oncol. Rep. 2013, 30, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.N.; Chen, J.J.; Wang, L.X.; Xiao, R.Z.; Liu, L.L.; Fang, Z.G.; Liu, Q.; Long, Z.J.; Lin, D.J. Inhibition of c-myc overcomes cytotoxic drug resistance in acute myeloid leukemia cells by promoting differentiation. PLoS ONE 2014, 9, e105381. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- rna genes mir15 and mir16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. Mir-15 and mir-16 induce apoptosis by targeting bcl2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [Green Version]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. Bcl2 and mir-15/16: From gene discovery to treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. Mir-15b and mir-16 modulate multidrug resistance by targeting bcl2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef]
- Dong, J.; Zhao, Y.P.; Zhou, L.; Zhang, T.P.; Chen, G. Bcl-2 upregulation induced by mir-21 via a direct interaction is associated with apoptosis and chemoresistance in mia paca-2 pancreatic cancer cells. Arch. Med. Res. 2011, 42, 8–14. [Google Scholar] [CrossRef]
- Singh, R.; Saini, N. Downregulation of bcl2 by mirnas augments drug-induced apoptosis--a combined computational and experimental approach. J. Cell Sci. 2012, 125, 1568–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Wu, J.; Jiao, K.; Wu, Q.; Ma, J.; Chen, D.; Kang, J.; Zhao, G.; Shi, Y.; Fan, D.; et al. Microrna-495-3p inhibits multidrug resistance by modulating autophagy through grp78/mtor axis in gastric cancer. Cell Death Dis. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Sacconi, A.; Biagioni, F.; Canu, V.; Mori, F.; Di Benedetto, A.; Lorenzon, L.; Ercolani, C.; Di Agostino, S.; Cambria, A.M.; Germoni, S.; et al. Mir-204 targets bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012, 3, e423. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Hidaka, H.; Majid, S.; Saini, S.; Arora, S.; Deng, G.; Shahryari, V.; Chang, I.; et al. Genistein up-regulates tumor suppressor microrna-574-3p in prostate cancer. PLoS ONE 2013, 8, e58929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Tian, W.; Chen, H.; Deng, Y. Microrna-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting mcl-1. Mol. Med. Rep. 2016, 13, 1923–1929. [Google Scholar] [CrossRef] [Green Version]
- Romano, G.; Acunzo, M.; Garofalo, M.; Di Leva, G.; Cascione, L.; Zanca, C.; Bolon, B.; Condorelli, G.; Croce, C.M. Mir-494 is regulated by erk1/2 and modulates trail-induced apoptosis in non-small-cell lung cancer through bim down-regulation. Proc. Natl. Acad. Sci. USA 2012, 109, 16570–16575. [Google Scholar] [CrossRef] [Green Version]
- Hamada, S.; Masamune, A.; Miura, S.; Satoh, K.; Shimosegawa, T. Mir-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein shc1 and pro-apoptotic regulator bax. Cell Signal. 2014, 26, 179–185. [Google Scholar] [CrossRef]
- Mishra, P.J. The mirna-drug resistance connection: A new era of personalized medicine using noncoding rna begins. Pharmacogenomics 2012, 13, 1321–1324. [Google Scholar] [CrossRef] [Green Version]
- Sohrabi, B.; Dayeri, B.; Zahedi, E.; Khoshbakht, S.; Nezamabadi Pour, N.; Ranjbar, H.; Davari Nejad, A.; Noureddini, M.; Alani, B. Mesenchymal stem cell (msc)-derived exosomes as novel vehicles for delivery of mirnas in cancer therapy. Cancer Gene Therapy 2022. [Google Scholar] [CrossRef]
- Dilsiz, N. Role of exosomes and exosomal micrornas in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Zhou, C.R.; Bai, M.J.; Zhu, D.; Chen, J.W.; Wang, H.F.; Li, M.A.; Wu, C.; Li, Z.R.; Huang, M.S. Exosome-mediated mirna delivery promotes liver cancer emt and metastasis. Am. J. Transl. Res. 2020, 12, 1080–1095. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlíková, L.; Šereš, M.; Breier, A.; Sulová, Z. The Roles of microRNAs in Cancer Multidrug Resistance. Cancers 2022, 14, 1090. https://doi.org/10.3390/cancers14041090
Pavlíková L, Šereš M, Breier A, Sulová Z. The Roles of microRNAs in Cancer Multidrug Resistance. Cancers. 2022; 14(4):1090. https://doi.org/10.3390/cancers14041090
Chicago/Turabian StylePavlíková, Lucia, Mário Šereš, Albert Breier, and Zdena Sulová. 2022. "The Roles of microRNAs in Cancer Multidrug Resistance" Cancers 14, no. 4: 1090. https://doi.org/10.3390/cancers14041090
APA StylePavlíková, L., Šereš, M., Breier, A., & Sulová, Z. (2022). The Roles of microRNAs in Cancer Multidrug Resistance. Cancers, 14(4), 1090. https://doi.org/10.3390/cancers14041090