HPRT1 Promotes Chemoresistance in Oral Squamous Cell Carcinoma via Activating MMP1/PI3K/Akt Signaling Pathway
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Patients and Tumor Tissue Samples
2.3. Cell Culture
2.4. RNA Isolation and Quantitative Real-Time Reverse Transcription PCR
2.5. Constructs, Transfection, and Retroviral Infection
2.6. Total RNA Isolation, RNA-Seq Library Preparation, and Sequencing
2.7. Western Blotting
2.8. Immunohistochemistry
2.9. Annexin V-Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI)-Staining Assay
2.10. Cell Viability Assay
2.11. Colony Formation Assay
2.12. TUNEL Assay
2.13. Bioinformatics Analysis
3. Statistical Analysis
4. Results
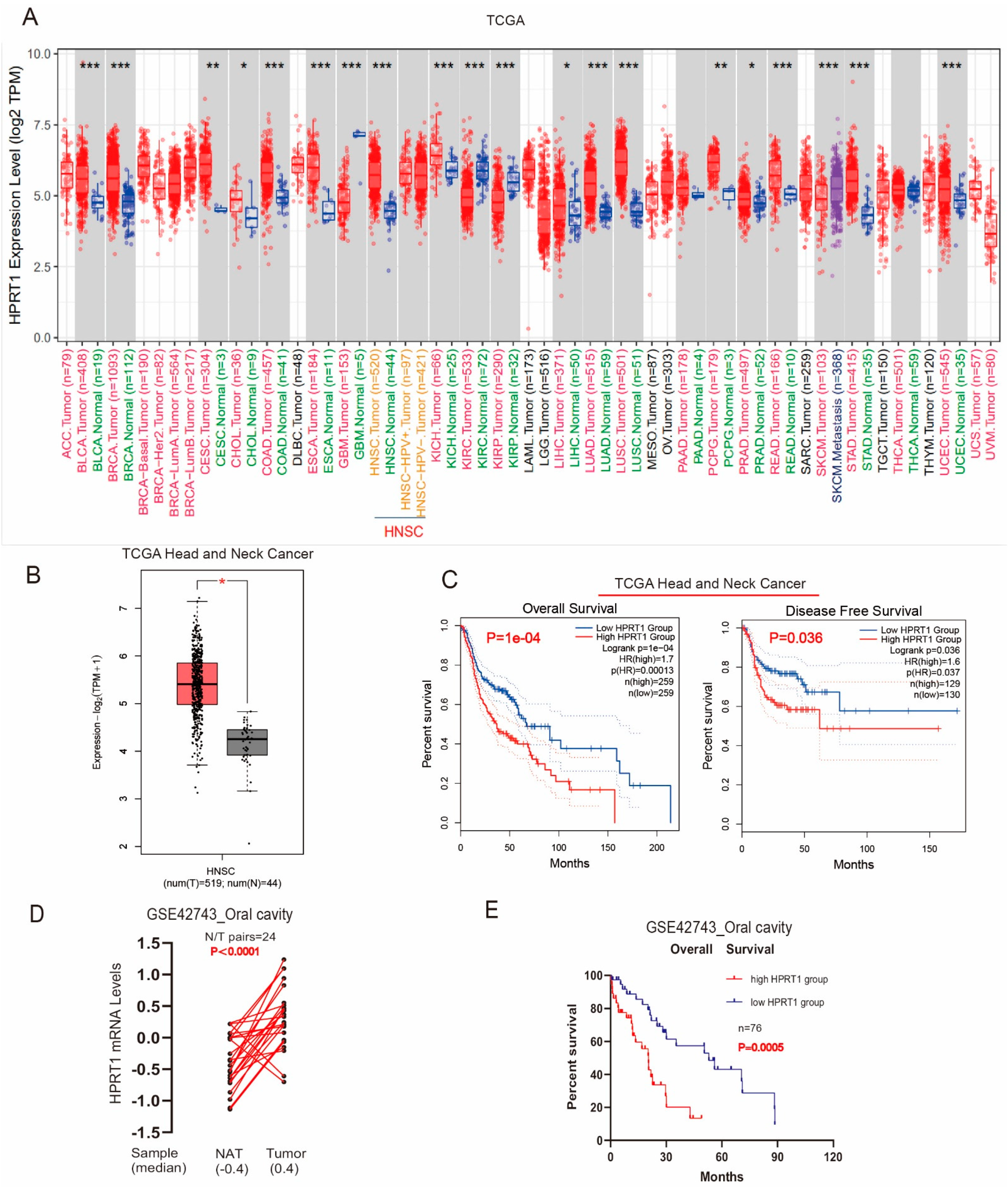
4.1. Overexpressed HPRT1 Predicted a Poor Prognosis in OSCC Patients in TCGA
4.2. HPRT1 Is a Potential Prognostic Indicator in OSCC
4.3. Knockdown of HPRT1 Reduced Cisplatin Resistance of Oral Cancer Cells In Vitro
4.4. HPRT1 Positively Regulates the Expression of MMP1 in Oral Cancer Cells
4.5. HPRT1 Promote Chemoresistance In Vivo
4.6. HPRT1 Promoted CDDP Chemoresistance in OSCC by Activating the PI3K/AKT Pathway
5. Clinical Value of HPRT1 in Other Cancer Types
6. Discussion
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HPRT1 | hypoxanthine phosphoribosyl transferase 1 |
| ANT | adjacent non-carcinoma tissues |
| OSCC | oral squamous cell carcinoma |
| CDDP | cisplatin |
| HNSCC | head and neck squamous cell carcinoma |
| ECM | extracellular matrix |
| MMPs | matrix metalloproteinases |
| BRCA | breast invasive carcinoma |
| CESC | cervical squamous cell carcinoma and endocervical adenocarcinoma |
| COAD | colon adenocarcinoma |
| ESCA | esophageal carcinoma |
| DLBC | lymphoid neoplasm diffuse large B-cell lymphoma |
| LUAD | lung adenocarcinoma |
| SKCM | skin cutaneous Melanoma |
| STAD | stomach adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| PAAD | pancreatic adenocarcinoma |
| THYM | thymoma |
| UCEC | uterine corpus endometrial carcinoma |
| KIRP | kidney renal papillary cell carcinoma |
| LGG | brain lower grade glioma |
| LIHC | liver hepatocellular carcinoma |
| UCS | uterine carcinosarcoma |
| LUAD | lung adenocarcinoma |
| MESO | mesothelioma |
| UVM | uveal melanoma |
| ACC | adrenocortical carcinoma |
| SARC | sarcoma |
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [Green Version]
- Colevas, A.D. Chemotherapy options for patients with metastatic or recurrent squamous cell carcinoma of the head and neck. J. Clin. Oncol. 2006, 24, 2644–2652. [Google Scholar] [CrossRef]
- Molin, Y.; Fayette, J. Current chemotherapies for recurrent/metastatic head and neck cancer. Anticancer Drugs 2011, 22, 621–625. [Google Scholar] [CrossRef]
- Monnat, R.J., Jr.; Chiaverotti, T.A.; Hackmann, A.F.; Maresh, G.A. Molecular structure and genetic stability of human hypoxanthine phosphoribosyltransferase (HPRT) gene duplications. Genomics 1992, 13, 788–796. [Google Scholar] [CrossRef]
- Torres, R.J.; Puig, J.G. Hypoxanthine-guanine phosophoribosyltransferase (HPRT) deficiency: Lesch Nyhan syndrome. Orphanet J. Rare Dis. 2007, 2, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedano, M.J.; Ramos, E.I.; Choudhari, R.; Harrison, A.L.; Subramani, R.; Lakshmanaswamy, R.; Zilaie, M.; Gadad, S.S. Hypoxanthine Phosphoribosyl Transferase 1 Is Upregulated, Predicts Clinical Outcome and Controls Gene Expression in Breast Cancer. Cancers 2020, 12, 1522. [Google Scholar] [CrossRef]
- Ahmadi, M.; Kenzerki, M.E.; Akrami, S.M.; Pashangzadeh, S.; Hajiesmaeili, F.; Rahnavard, S.; Habibipour, L.; Saffarzadeh, N.; Mousavi, P. Overexpression of HPRT1 is associated with poor prognosis in head and neck squamous cell carcinoma. FEBS Open Bio. 2021, 11, 2525–2540. [Google Scholar] [CrossRef] [PubMed]
- Townsend, M.H.; Ence, Z.E.; Felsted, A.M.; Parker, A.C.; Piccolo, S.R.; Robison, R.A.; O’Neill, K.L. Potential new biomarkers for endometrial cancer. Cancer Cell Int. 2019, 19, 19. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Shang, D.; Yu, F.; Liu, W.; Zhang, Y.; Feng, C.; Wang, Q.; Xu, Y.; Liu, Y.; et al. Characterizing and optimizing human anticancer drug targets based on topological properties in the context of biological pathways. J. Biomed. Inform. 2015, 54, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Stokes, A.; Joutsa, J.; Ala-Aho, R.; Pitchers, M.; Pennington, C.J.; Martin, C.; Premachandra, D.J.; Okada, Y.; Peltonen, J.; Grénman, R.; et al. Expression profiles and clinical correlations of degradome omponents in the tumor microenvironment of head and neck squamous cell carcinoma. Clin. Cancer Res. 2010, 16, 2022–2035. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. S2), W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pendleton, K.P.; Grandis, J.R. Cisplatin-Based Chemotherapy Options for Recurrent and/or Metastatic Squamous Cell Cancer of the Head and Neck. Clin. Med. Insights Ther. 2013, 5, CMT-S10409. [Google Scholar] [CrossRef] [Green Version]
- Townsend, M.H.; Freitas, C.M.T.; Larsen, D.; Piccolo, S.R.; Weber, S.; Robison, R.; O’Neill, K.L. Hypoxanthine Guanine Phosphoribosyltransferase expression is negatively correlated with immune activity through its regulation of purine synthesis. Immunobiology 2020, 225, 151931. [Google Scholar] [CrossRef] [PubMed]
- Gabasa, M.; Radisky, E.S.; Ikemori, R.; Bertolini, G.; Arshakyan, M.; Hockla, A.; Duch, P.; Rondinone, O.; Llorente, A.; Maqueda, M.; et al. MMP1 drives tumor progression in large cell carcinoma of the lung through fibroblast senescence. Cancer Lett. 2021, 507, 1–12. [Google Scholar] [CrossRef]
- Harati, R.; Hafezi, S.; Mabondzo, A.; Tlili, A. Silencing miR-202-3p increases MMP-1 and promotes a brain invasive phenotype in metastatic breast cancer cells. PLoS ONE 2020, 15, e0239292. [Google Scholar] [CrossRef]
- Wu, K.; Mao, Y.Y.; Han, N.N.; Wu, H.; Zhang, S. PLAU1 Facilitated Proliferation, Invasion, and Metastasis via Interaction With MMP1 in Head and Neck Squamous Carcinoma. Front. Oncol. 2021, 11, 574260. [Google Scholar] [CrossRef]
- Tepper, J.; Krasna, M.J.; Niedzwiecki, D.; Hollis, D.; Reed, C.E.; Goldberg, R.; Kiel, K.; Willett, C.; Sugarbaker, D.; Mayer, R. Phase III trial of trimodality therapy with cisplatin, fluorouracil, radiotherapy, and surgery compared with surgery alone for esophageal cancer: CALGB 9781. J. Clin. Oncol. 2008, 26, 1086–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussink, J.; van der Kogel, A.J.; Kaanders, J.H. Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008, 9, 288–296. [Google Scholar] [CrossRef]
- Zaryouh, H.; De Pauw, I.; Baysal, H.; Peeters, M.; Vermorken, J.B.; Lardon, F.; Wouters, A. Recent insights in the PI3K/Akt pathway as a promising therapeutic target in combination with EGFR-targeting agents to treat head and neck squamous cell carcinoma. Med. Res Rev. 2022, 42, 112–155. [Google Scholar] [CrossRef]
- Bassi, C.; Ho, J.; Srikumar, T.; Dowling, R.J.; Gorrini, C.; Miller, S.J.; Mak, T.W.; Neel, B.G.; Raught, B.; Stambolic, V. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 2013, 341, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Sun, C.; Li, N.; Shan, W.; Lu, H.; Guo, L.; Guo, E.; Xia, M.; Weng, D.; Meng, L. Cisplatin-induced CCL5 secretion from CAFs promotes cisplatin-resistance in ovarian cancer via regulation of the STAT3 and PI3K/Akt signaling pathways. Int. J. Oncol. 2016, 48, 2087–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Jiao, Z.; Li, Y.; Su, X.; Yao, F.; Peng, J.; Chen, W.; Yang, A. HPRT1 Promotes Chemoresistance in Oral Squamous Cell Carcinoma via Activating MMP1/PI3K/Akt Signaling Pathway. Cancers 2022, 14, 855. https://doi.org/10.3390/cancers14040855
Wu T, Jiao Z, Li Y, Su X, Yao F, Peng J, Chen W, Yang A. HPRT1 Promotes Chemoresistance in Oral Squamous Cell Carcinoma via Activating MMP1/PI3K/Akt Signaling Pathway. Cancers. 2022; 14(4):855. https://doi.org/10.3390/cancers14040855
Chicago/Turabian StyleWu, Tong, Zan Jiao, Yixuan Li, Xuan Su, Fan Yao, Jin Peng, Weichao Chen, and Ankui Yang. 2022. "HPRT1 Promotes Chemoresistance in Oral Squamous Cell Carcinoma via Activating MMP1/PI3K/Akt Signaling Pathway" Cancers 14, no. 4: 855. https://doi.org/10.3390/cancers14040855
APA StyleWu, T., Jiao, Z., Li, Y., Su, X., Yao, F., Peng, J., Chen, W., & Yang, A. (2022). HPRT1 Promotes Chemoresistance in Oral Squamous Cell Carcinoma via Activating MMP1/PI3K/Akt Signaling Pathway. Cancers, 14(4), 855. https://doi.org/10.3390/cancers14040855




