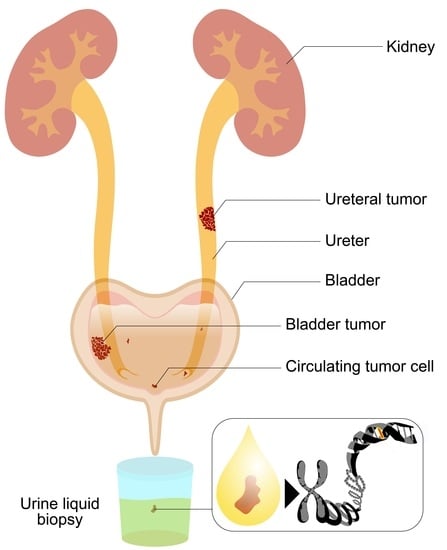
Detection of Cancer Mutations by Urine Liquid Biopsy as a Potential Tool in the Clinical Management of Bladder Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Healthy Volunteers and Patients
2.2. Study Design
2.2.1. Healthy Volunteers’ Sample Collection and Storage
2.2.2. Patients’ Sample Collection and Storage
2.3. DNA Isolation
2.3.1. DNA Isolation from Urine Samples
2.3.2. DNA Isolation from FFPE Samples
2.4. Sequencing
2.4.1. Next-Generation Sequencing of Urine DNA
2.4.2. Sanger Sequencing for Validation
2.4.3. 16S Sequencing
2.5. Bioinformatic Analysis
2.6. Prostate Cancer Tissue Analysis
2.7. Statistical Analysis
3. Results
3.1. Urine DNA Yield in Healthy Pools after Different Storage Periods
3.2. Patient Cohort
3.3. Variant Analysis
3.4. Concordance with Tumor Tissues
4. Discussion
4.1. Preoperative Urine Samples Harbor Tumor-Specific Mutations from Bladder Cancer
4.2. Matching Pre- and Postoperative Urine Eliminates Germline Mutations and Provides a Strategy to Assess Remaining Tumor Burden in a Patient
4.3. Technical Consideration in Urine Liquid Biopsy
4.4. Study Limitations and Future Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Freedman, N.D.; Silverman, D.T.; Hollenbeck, A.R.; Schatzkin, A.; Abnet, C.C. Association Between Smoking and Risk of Bladder Cancer Among Men and Women. JAMA 2011, 306, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Burger, M.; Catto, J.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and Risk Factors of Urothelial Bladder Cancer. Eur. Urol. 2012, 63, 234–241. [Google Scholar] [CrossRef]
- Egbers, L.; Grotenhuis, A.J.; Aben, K.K.; Witjes, J.A.; Kiemeney, L.A.; Vermeulen, S.H. The prognostic value of family history among patients with urinary bladder cancer. Int. J. Cancer 2014, 136, 1117–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.; Leiser, C.L.; O’Neil, B.; Gupta, S.; Lowrance, W.T.; Kohlmann, W.; Greenberg, S.; Pathak, P.; Smith, K.R.; A Hanson, H. Familial Cancer Clustering in Urothelial Cancer: A Population-Based Case–Control Study. JNCI: J. Natl. Cancer Inst. 2017, 110, 527–533. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.-Y.; Zhong, J.H.; Zhao, Z.; Liu, J.; Yu, H.-L.; Shi, R. Association between APE1 Asp148Glu polymorphism and the risk of urinary cancers: A meta-analysis of 18 case–control studies. OncoTargets Ther. 2016, 9, 1499–1510. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, D.; Gupta, A.; Canter, D.J.; Harrow, B.; Dobbs, R.; Kucherov, V.; A Mueller, E.; Streeper, N.; Uhlman, M.A.; Svatek, R.S.; et al. Microscopic haematuria at time of diagnosis is associated with lower disease stage in patients with newly diagnosed bladder cancer. Br. J. Urol. 2015, 117, 783–786. [Google Scholar] [CrossRef] [Green Version]
- Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Escrig, J.L.D.; Gontero, P.; Liedberg, F.; Masson-Lecomte, A.; Mostafid, A.H.; et al. European Association of Urology Guidelines on Non–muscle-invasive Bladder Cancer (Ta, T1, and Carcinoma in Situ). Eur. Urol. 2021, 81, 75–94. [Google Scholar] [CrossRef]
- Yafi, F.A.; Brimo, F.; Steinberg, J.; Aprikian, A.G.; Tanguay, S.; Kassouf, W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 66.e25–66.e31. [Google Scholar] [CrossRef]
- Raitanen, M.P.; Aine, R.; Rintala, E.; Kallio, J.; Rajala, P.; Juusela, H. Differences between local and review urinary cy-tology in diagnosis of bladder cancer. An interobserver multicenter analysis. Eur. Urol 2002, 41, 284–289. [Google Scholar] [CrossRef]
- Karakiewicz, P.I.; Benayoun, S.; Zippe, C.; Ludecke, G.; Boman, H.; Sanchez-Carbayo, M.; Casella, R.; Mian, C.; Friedrich, M.G.; Eissa, S.; et al. Institutional variability in the accuracy of urinary cytology for predicting recurrence of transitional cell carcinoma of the bladder. Br. J. Urol. 2006, 97, 997–1001. [Google Scholar] [CrossRef]
- Ng, K.; Stenzl, A.; Sharma, A.; Vasdev, N. Urinary biomarkers in bladder cancer: A review of the current landscape and future directions. In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Ferro, M.; Di Lorenzo, G.; Vartolomei, M.D.; Bruzzese, D.; Cantiello, F.; Lucarelli, G.; Musi, G.; Di Stasi, S.; Hurle, R.; Guazzoni, G.; et al. Absolute basophil count is associated with time to recurrence in patients with high-grade T1 bladder cancer receiving bacillus Calmette–Guérin after transurethral resection of the bladder tumor. World J. Urol. 2020, 38, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vartolomei, M.D.; Porav-Hodade, D.; Ferro, M.; Mathieu, R.; Abufaraj, M.; Foerster, B.; Kimura, S.; Shariat, S. Prognostic role of pretreatment neutrophil-to-lymphocyte ratio (NLR) in patients with non–muscle-invasive bladder cancer (NMIBC): A systematic review and meta-analysis. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Getzler, I.; Bahouth, Z.; Nativ, O.; Rubinstein, J.; Halachmi, S. Preoperative neutrophil to lymphocyte ratio improves recurrence prediction of non-muscle invasive bladder cancer. BMC Urol. 2018, 18, 90. [Google Scholar] [CrossRef] [Green Version]
- Vartolomei, M.D.; Ferro, M.; Cantiello, F.; Lucarelli, G.; Di Stasi, S.; Hurle, R.; Guazzoni, G.; Busetto, G.M.; De Berardinis, E.; Damiano, R.; et al. Validation of Neutrophil-to-lymphocyte Ratio in a Multi-institutional Cohort of Patients With T1G3 Non–muscle-invasive Bladder Cancer. Clin. Genitourin. Cancer 2018, 16, 445–452. [Google Scholar] [CrossRef]
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Yokoyama, H.; Amemiya, K.; Hagimoto, T.; Daimon, H.; Hosaka, K.; Oyama, T.; Mochizuki, H.; Omata, M. Genomic profile of urine has high diagnostic sensitivity compared to cytology in non-invasive urothelial bladder cancer. Cancer Sci. 2019, 110, 3235–3243. [Google Scholar] [CrossRef] [Green Version]
- Springer, S.U.; Chen, C.-H.; Pena, M.D.C.R.; Li, L.; Douville, C.; Wang, Y.; Cohen, J.D.; Taheri, D.; Silliman, N.; Schaefer, J.; et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 2018, 7, e32143. [Google Scholar] [CrossRef]
- Streleckiene, G.; Reid, H.M.; Arnold, N.; Bauerschlag, D.; Forster, M. Quantifying cell free DNA in urine: Comparison between commercial kits, impact of gender and inter-individual variation. BioTechniques 2018, 64, 225–230. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Wolf, B.; Kuonen, P.; Dandekar, T.; Atlan, D. DNAseq Workflow in a Diagnostic Context and an Example of a User Friendly Implementation. BioMed Res. Int. 2015, 2015, 403497. [Google Scholar] [CrossRef]
- Hendricks, A.; Amallraja, A.; Meißner, T.; Forster, P.; Rosenstiel, P.; Burmeister, G.; Schafmayer, C.; Franke, A.; Hinz, S.; Forster, M.; et al. Stage IV Colorectal Cancer Patients with High Risk Mutation Profiles Survived 16 Months Longer with Individualized Therapies. Cancers 2020, 12, 393. [Google Scholar] [CrossRef] [Green Version]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2017, 46, D1062–D1067. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium; Campbell, P.J.; Getz, G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Cancer 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Cancer 2014, 15, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Böhle, A.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Hernández, V.; Kaasinen, E.; Palou, J.; Rouprêt, M.; et al. EAU Guidelines on Non–Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur. Urol. 2016, 71, 447–461. [Google Scholar] [CrossRef] [PubMed]
- El Bali, L.; Diman, A.; Bernard, A.; Roosens, N.H.C.; De Keersmaecker, S.C.J. Comparative Study of Seven Commercial Kits for Human DNA Extraction from Urine Samples Suitable for DNA Biomarker-Based Public Health Studies. J. Biomol. Tech. JBT 2014, 25, 96–110. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.H.; Jeong, C.W.; Kwak, C.; Kim, H.H.; Choo, M.S.; Ku, J.H. Intravesical Chemotherapy after Radical Nephroureterectomy for Primary Upper Tract Urothelial Carcinoma: A Systematic Review and Network Meta-Analysis. J. Clin. Med. 2019, 8, 1059. [Google Scholar] [CrossRef] [Green Version]
- Mekayten, M.; Yutkin, V.; Duvdevani, M.; Pode, D.; Hidas, G.; Landau, E.H.; Youssef, F.; Gofrit, O.N. High frequency of bladder cancer after nephroureterectomy: Justification for adjuvant intravesical treatment? Res. Rep. Urol. 2018, 10, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Vu, N.T.; Chaturvedi, A.K.; Canfield, D.V. Genotyping for DQA1 and PM loci in urine using PCR-based amplification: Effects of sample volume, storage temperature, preservatives, and aging on DNA extraction and typing. Forensic Sci. Int. 1999, 102, 23–34. [Google Scholar] [CrossRef]
- Augustus, E.; Van Casteren, K.; Sorber, L.; Van Dam, P.; Roeyen, G.; Peeters, M.; Vorsters, A.; Wouters, A.; Raskin, J.; Rolfo, C.; et al. The art of obtaining a high yield of cell-free DNA from urine. PLoS ONE 2020, 15, e0231058. [Google Scholar] [CrossRef] [Green Version]
- Milde, A.; Haas-Rochholz, H. Improved DNA typing of human urine by adding EDTA. Int. J. Leg. Med. 1999, 112, 209–210. [Google Scholar] [CrossRef]
- Köhler, C.U.; Bonberg, N.; Ahrens, M.; Behrens, T.; Hovanec, J.; Eisenacher, M.; Noldus, J.; Deix, T.; Braun, K.; Gohlke, H.; et al. Noninvasive diagnosis of urothelial cancer in urine using DNA hypermethylation signatures—Gender matters. Int. J. Cancer 2019, 145, 2861–2872. [Google Scholar] [CrossRef] [PubMed]
- Perez-Carrasco, V.; Soriano-Lerma, A.; Soriano, M.; Gutiérrez-Fernández, J.; Garcia-Salcedo, J.A. Urinary Microbiome: Yin and Yang of the Urinary Tract. Front. Cell. Infect. Microbiol. 2021, 11, 617002. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Grellner, W.; Schmitt, C. DNA typing of urine samples following several years of storage. Int. J. Leg. Med. 1993, 106, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Del Giudice, F.; Carrieri, G.; Busetto, G.M.; Cormio, L.; Hurle, R.; Contieri, R.; Arcaniolo, D.; Sciarra, A.; Maggi, M.; et al. The Impact of SARS-CoV-2 Pandemic on Time to Primary, Secondary Resection and Adjuvant Intravesical Therapy in Patients with High-Risk Non-Muscle Invasive Bladder Cancer: A Retrospective Multi-Institutional Cohort Analysis. Cancers 2021, 13, 5276. [Google Scholar] [CrossRef]
- Mielczarek, Ł.; Zapała, P.; Krajewski, W.; Nowak, Ł.; Bajkowski, M.; Szost, P.; Szabłoński, W.; Zapała, Ł.; Poletajew, S.; Dybowski, B.; et al. Diagnostic and treatment delays among patients with primary bladder cancer in Poland: A survey study. Central Eur. J. Urol. 2020, 73, 152–159. [Google Scholar] [CrossRef]
- Roscigno, M.; Naspro, R.; Piccichè, A.; Muttin, F.; Angiolilli, D.; Deiana, G.; Pezzoli, F.; Da Pozzo, L.F. A Snapshot from the Department of Urology in Bergamo Evaluating the Timeline of the SARS-CoV-2 Outbreak: Which Patients Are We Missing? Eur. Urol. Focus 2020, 6, 1120–1123. [Google Scholar] [CrossRef]
- Richards, M.; Anderson, M.; Carter, P.; Ebert, B.L.; Mossialos, E. The impact of the COVID-19 pandemic on cancer care. Nat. Rev. Cancer 2020, 1, 565–567. [Google Scholar] [CrossRef]
- Schmidt, A.L.; Bakouny, Z.; Bhalla, S.; Steinharter, J.A.; Tremblay, D.A.; Awad, M.M.; Kessler, A.J.; Haddad, R.I.; Evans, M.; Busser, F.; et al. Cancer Care Disparities during the COVID-19 Pandemic: COVID-19 and Cancer Outcomes Study. Cancer Cell 2020, 38, 769–770. [Google Scholar] [CrossRef]
- Metzger, K.; Mrosek, J.; Zittel, S.; Pilz, M.; Held, T.; Adeberg, S.; Ristow, O.; Hoffmann, J.; Engel, M.; Freudlsperger, C.; et al. Treatment delay and tumor size in patients with oral cancer during the first year of the COVID-19 pandemic. Head Neck 2021, 43, 3493–3497. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | n |
|---|---|
| Total patients | 12 |
| Male | 7 |
| Female | 5 |
| Age (in years) | |
| Mean | 70.7 |
| Median | 72 |
| Range | 52–81 |
| Type of operation for tumor resection | |
| Cystectomy | 9 |
| Nephroureterectomy | 2 |
| Transurethral resection of bladder tumor (TURBT) | 1 |
| Immunohistochemistry markers | (positive/negative) |
| AMACR | 1/5 |
| CK7 | 1/5 |
| CK8 | 1/5 |
| CK20 | 5/1 |
| GATA3 | 2/4 |
| p63 | 1/5 |
| No available data | 6 |
| Pathologic staging of the tumor (pT-stage) | |
| Tis | 3 |
| T1 | 1 |
| T2 | 2 |
| T2a | 2 |
| T3a | 1 |
| T3b | 1 |
| T4a | 1 |
| No tumor found 1 | 1 |
| Regional lymph nodes (N-stage) | |
| NX | 4 |
| N0 | 6 |
| N1 | 1 |
| N2 | 1 |
| Presence of residual tumor (R-status) | |
| R0 | 11 |
| R2 | 1 |
| Lymphovascular invasion (LVI) | |
| Positive | 3 |
| Negative | 9 |
| Vascular invasion (VI) | |
| Positive | 3 |
| Negative | 9 |
| Perineural invasion (PNI) | |
| Positive | 1 |
| Negative | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, N.K.; Eraky, A.; Eggers, J.; Steiert, T.A.; Sebens, S.; Jünemann, K.-P.; Hendricks, A.; Bang, C.; Stanulla, M.; Franke, A.; et al. Detection of Cancer Mutations by Urine Liquid Biopsy as a Potential Tool in the Clinical Management of Bladder Cancer Patients. Cancers 2022, 14, 969. https://doi.org/10.3390/cancers14040969
Ibrahim NK, Eraky A, Eggers J, Steiert TA, Sebens S, Jünemann K-P, Hendricks A, Bang C, Stanulla M, Franke A, et al. Detection of Cancer Mutations by Urine Liquid Biopsy as a Potential Tool in the Clinical Management of Bladder Cancer Patients. Cancers. 2022; 14(4):969. https://doi.org/10.3390/cancers14040969
Chicago/Turabian StyleIbrahim, Nurul Khalida, Ahmed Eraky, Jan Eggers, Tim Alexander Steiert, Susanne Sebens, Klaus-Peter Jünemann, Alexander Hendricks, Corinna Bang, Martin Stanulla, Andre Franke, and et al. 2022. "Detection of Cancer Mutations by Urine Liquid Biopsy as a Potential Tool in the Clinical Management of Bladder Cancer Patients" Cancers 14, no. 4: 969. https://doi.org/10.3390/cancers14040969
APA StyleIbrahim, N. K., Eraky, A., Eggers, J., Steiert, T. A., Sebens, S., Jünemann, K. -P., Hendricks, A., Bang, C., Stanulla, M., Franke, A., Hamann, C., Röcken, C., Arnold, N., Hinze, L., & Forster, M. (2022). Detection of Cancer Mutations by Urine Liquid Biopsy as a Potential Tool in the Clinical Management of Bladder Cancer Patients. Cancers, 14(4), 969. https://doi.org/10.3390/cancers14040969