Understanding Aberrant Signaling to Elude Therapy Escape Mechanisms in Myeloproliferative Neoplasms
Abstract
Simple Summary
Abstract
1. Introduction
2. Mutational Landscape at a Glance
3. Resistance to JAK Inhibitors
3.1. Genetic Mechanisms of Resistance
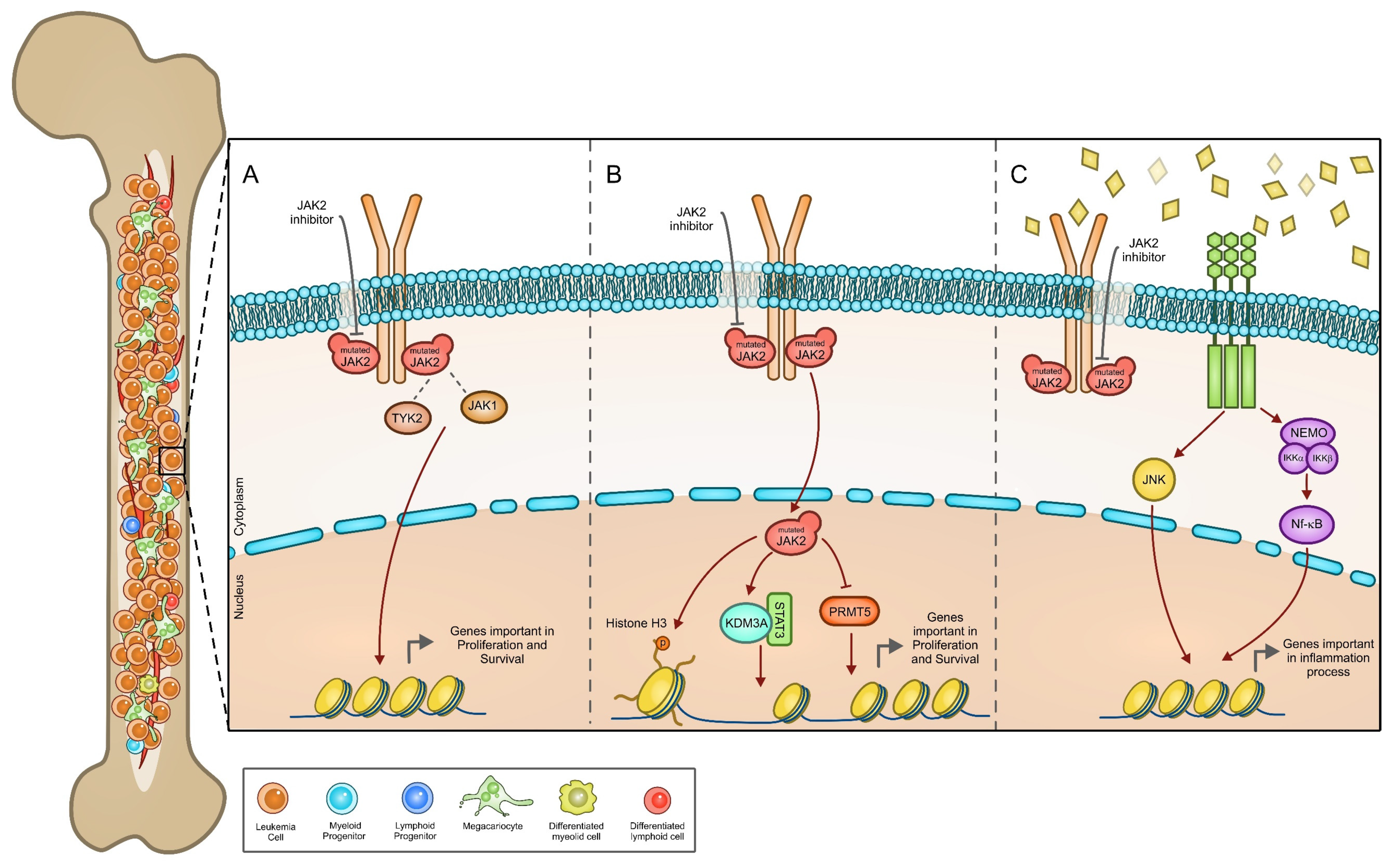
3.2. JAK2 Signaling
3.3. Cytokine Deregulation
3.4. Aurora A and ROCK
4. Drug Combinations: State of the Art
5. Multidrug Resistance: Lesson from Other Cancers
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- James, C.; Valerie, U.; Le Couedic, J.-P.; Staerk, J.; Delhommeau, F.; Lacout, C.; Garcon, L.; Raslova, H.; Berger, R.; Bennaceur-Griscelli, A.; et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005, 434, 1144–1148. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.-S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Pikman, Y.; Lee, B.H.; Mercher, T.; McDowell, E.; Ebert, B.L.; Gozo, M.; Cuker, A.; Wernig, G.; Moore, S.; Galinsky, I.; et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006, 3, e270. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, T.; Gisslinger, H.; Harutyunyan, A.S.; Nivarthi, H.; Rumi, E.; Milosevic, J.D.; Them, N.C.; Berg, T.; Gisslinger, B.; Pietra, D.; et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 2013, 369, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Nangalia, J.; Massie, C.E.; Baxter, E.J.; Nice, F.L.; Gundem, G.; Wedge, D.C.; Avezov, E.; Li, J.; Kollmann, K.; Kent, D.G.; et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013, 369, 2391–2405. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Roper, N.; Chaurasia, P.; Hoffman, R. Epigenetic abnormalities in myeloproliferative neoplasms: A target for novel therapeutic strategies. Clin. Epigenet. 2011, 2, 197–212. [Google Scholar] [CrossRef]
- Swierczek, S.I.; Yoon, D.; Bellanne-Chantelot, C.; Kim, S.J.; Saint-Martin, C.; Delhommeau, F.; Najman, A.; Prchal, J.T. Extent of hematopoietic involvement by TET2 mutations in JAK2V(6)(1)(7)F polycythemia vera. Haematologica 2011, 96, 775–778. [Google Scholar] [CrossRef]
- Hultcrantz, M.; Bjorkholm, M.; Dickman, P.W.; Landgren, O.; Derolf, A.R.; Kristinsson, S.Y.; Andersson, T.M.L. Risk for Arterial and Venous Thrombosis in Patients With Myeloproliferative Neoplasms: A Population-Based Cohort Study. Ann. Intern. Med. 2018, 168, 317–325. [Google Scholar] [CrossRef]
- Kennedy, J.A.; Atenafu, E.G.; Messner, H.A.; Craddock, K.J.; Brandwein, J.M.; Lipton, J.H.; Minden, M.D.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Treatment outcomes following leukemic transformation in Philadelphia-negative myeloproliferative neoplasms. Blood 2013, 121, 2725–2733. [Google Scholar] [CrossRef]
- Barbui, T.; Finazzi, G.; Carobbio, A.; Thiele, J.; Passamonti, F.; Rumi, E.; Ruggeri, M.; Rodeghiero, F.; Randi, M.L.; Bertozzi, I.; et al. Development and validation of an International Prognostic Score of thrombosis in World Health Organization-essential thrombocythemia (IPSET-thrombosis). Blood 2012, 120, 5128–5133, quiz 5252. [Google Scholar] [CrossRef]
- Gangat, N.; Caramazza, D.; Vaidya, R.; George, G.; Begna, K.; Schwager, S.; Van Dyke, D.; Hanson, C.; Wu, W.; Pardanani, A.; et al. DIPSS plus: A refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, F.; Cervantes, F.; Vannucchi, A.M.; Morra, E.; Rumi, E.; Pereira, A.; Guglielmelli, P.; Pungolino, E.; Caramella, M.; Maffioli, M.; et al. A dynamic prognostic model to predict survival in primary myelofibrosis: A study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment). Blood 2010, 115, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A. Primary myelofibrosis: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2021, 96, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Barbui, T. Polycythemia vera and essential thrombocythemia: 2019 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2019, 94, 133–143. [Google Scholar] [CrossRef]
- Tefferi, A.; Guglielmelli, P.; Nicolosi, M.; Mannelli, F.; Mudireddy, M.; Bartalucci, N.; Finke, C.M.; Lasho, T.L.; Hanson, C.A.; Ketterling, R.P.; et al. GIPSS: Genetically inspired prognostic scoring system for primary myelofibrosis. Leukemia 2018, 32, 1631–1642. [Google Scholar] [CrossRef]
- Barosi, G.; Birgegard, G.; Finazzi, G.; Griesshammer, M.; Harrison, C.; Hasselbalch, H.C.; Kiladjian, J.J.; Lengfelder, E.; McMullin, M.F.; Passamonti, F.; et al. Response criteria for essential thrombocythemia and polycythemia vera: Result of a European LeukemiaNet consensus conference. Blood 2009, 113, 4829–4833. [Google Scholar] [CrossRef]
- Barosi, G.; Mesa, R.; Finazzi, G.; Harrison, C.; Kiladjian, J.J.; Lengfelder, E.; McMullin, M.F.; Passamonti, F.; Vannucchi, A.M.; Besses, C.; et al. Revised response criteria for polycythemia vera and essential thrombocythemia: An ELN and IWG-MRT consensus project. Blood 2013, 121, 4778–4781. [Google Scholar] [CrossRef]
- Li, C.M.; Chen, Z. Autoimmunity as an Etiological Factor of Cancer: The Transformative Potential of Chronic Type 2 Inflammation. Front. Cell Dev. Biol. 2021, 9, 664305. [Google Scholar] [CrossRef]
- Di Battista, V.; Bochicchio, M.T.; Giordano, G.; Napolitano, M.; Lucchesi, A. Genetics and Pathogenetic Role of Inflammasomes in Philadelphia Negative Chronic Myeloproliferative Neoplasms: A Narrative Review. Int. J. Mol. Sci. 2021, 22, 561. [Google Scholar] [CrossRef]
- Skoda, R.C.; Duek, A.; Grisouard, J. Pathogenesis of myeloproliferative neoplasms. Exp. Hematol. 2015, 43, 599–608. [Google Scholar] [CrossRef]
- Grinfeld, J.; Nangalia, J.; Green, A.R. Molecular determinants of pathogenesis and clinical phenotype in myeloproliferative neoplasms. Haematologica 2017, 102, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.P.; Newberry, K.J.; Luthra, R.; Jabbour, E.; Pierce, S.; Cortes, J.; Singh, R.; Mehrotra, M.; Routbort, M.J.; Luthra, M.; et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood 2015, 126, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Tenedini, E.; Bernardis, I.; Artusi, V.; Artuso, L.; Roncaglia, E.; Guglielmelli, P.; Pieri, L.; Bogani, C.; Biamonte, F.; Rotunno, G.; et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia 2014, 28, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.P.S.; Getta, B.; Masarova, L.; Famulare, C.; Schulman, J.; Datoguia, T.S.; Puga, R.D.; Alves Paiva, R.M.; Arcila, M.E.; Hamerschlak, N.; et al. Prognostic impact of RAS-pathway mutations in patients with myelofibrosis. Leukemia 2020, 34, 799–810. [Google Scholar] [CrossRef]
- Winter, P.S.; Sarosiek, K.A.; Lin, K.H.; Meggendorfer, M.; Schnittger, S.; Letai, A.; Wood, K.C. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci. Signal. 2014, 7, ra122. [Google Scholar] [CrossRef]
- Hautin, M.; Mornet, C.; Chauveau, A.; Bernard, D.G.; Corcos, L.; Lippert, E. Splicing Anomalies in Myeloproliferative Neoplasms: Paving the Way for New Therapeutic Venues. Cancers 2020, 12, 2216. [Google Scholar] [CrossRef]
- Grinfeld, J.; Nangalia, J.; Baxter, E.J.; Wedge, D.C.; Angelopoulos, N.; Cantrill, R.; Godfrey, A.L.; Papaemmanuil, E.; Gundem, G.; MacLean, C.; et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N. Engl. J. Med. 2018, 379, 1416–1430. [Google Scholar] [CrossRef]
- Tefferi, A.; Vannucchi, A.M. Genetic Risk Assessment in Myeloproliferative Neoplasms. Mayo Clinic Proc. 2017, 92, 1283–1290. [Google Scholar] [CrossRef]
- Zhang, S.J.; Rampal, R.; Manshouri, T.; Patel, J.; Mensah, N.; Kayserian, A.; Hricik, T.; Heguy, A.; Hedvat, C.; Gonen, M.; et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012, 119, 4480–4485. [Google Scholar] [CrossRef]
- Stuckey, R.; Gomez-Casares, M.T. Recent Advances in the Use of Molecular Analyses to Inform the Diagnosis and Prognosis of Patients with Polycythaemia Vera. Int. J. Mol. Sci. 2021, 22, 5042. [Google Scholar] [CrossRef]
- Morotti, A.; Rocca, S.; Carra, G.; Saglio, G.; Brancaccio, M. Modeling myeloproliferative neoplasms: From mutations to mouse models and back again. Blood Rev. 2017, 31, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kent, D.G.; Chen, E.; Green, A.R. Mouse models of myeloproliferative neoplasms: JAK of all grades. Dis. Models Mech. 2011, 4, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Scroggins, B.T.; Robzyk, K.; Wang, D.; Marcu, M.G.; Tsutsumi, S.; Beebe, K.; Cotter, R.J.; Felts, S.; Toft, D.; Karnitz, L.; et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell 2007, 25, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Brkic, S.; Meyer, S.C. Challenges and Perspectives for Therapeutic Targeting of Myeloproliferative Neoplasms. HemaSphere 2021, 5, e516. [Google Scholar] [CrossRef]
- Meyer, S.C. Mechanisms of Resistance to JAK2 Inhibitors in Myeloproliferative Neoplasms. Hematol. Oncol. Clin. N. Am. 2017, 31, 627–642. [Google Scholar] [CrossRef]
- Verstovsek, S.; Gotlib, J.; Mesa, R.A.; Vannucchi, A.M.; Kiladjian, J.J.; Cervantes, F.; Harrison, C.N.; Paquette, R.; Sun, W.; Naim, A.; et al. Long-term survival in patients treated with ruxolitinib for myelofibrosis: COMFORT-I and -II pooled analyses. J. Hematol. Oncol. 2017, 10, 156. [Google Scholar] [CrossRef]
- Harrison, C.N.; Vannucchi, A.M.; Kiladjian, J.J.; Al-Ali, H.K.; Gisslinger, H.; Knoops, L.; Cervantes, F.; Jones, M.M.; Sun, K.; McQuitty, M.; et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia 2016, 30, 1701–1707, Correction in Leukemia 2017, 31, 775. [Google Scholar] [CrossRef]
- Mesa, R.A.; Kiladjian, J.J.; Catalano, J.V.; Devos, T.; Egyed, M.; Hellmann, A.; McLornan, D.; Shimoda, K.; Winton, E.F.; Deng, W.; et al. SIMPLIFY-1: A Phase III Randomized Trial of Momelotinib Versus Ruxolitinib in Janus Kinase Inhibitor-Naive Patients With Myelofibrosis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 3844–3850. [Google Scholar] [CrossRef]
- Harrison, C.N.; Vannucchi, A.M.; Platzbecker, U.; Cervantes, F.; Gupta, V.; Lavie, D.; Passamonti, F.; Winton, E.F.; Dong, H.; Kawashima, J.; et al. Momelotinib versus best available therapy in patients with myelofibrosis previously treated with ruxolitinib (SIMPLIFY 2): A randomised, open-label, phase 3 trial. Lancet Haematol. 2018, 5, e73–e81. [Google Scholar] [CrossRef]
- Vannucchi, A.M.; Kiladjian, J.J.; Griesshammer, M.; Masszi, T.; Durrant, S.; Passamonti, F.; Harrison, C.N.; Pane, F.; Zachee, P.; Mesa, R.; et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N. Engl. J. Med. 2015, 372, 426–435. [Google Scholar] [CrossRef]
- Mesa, R.A.; Vannucchi, A.M.; Mead, A.; Egyed, M.; Szoke, A.; Suvorov, A.; Jakucs, J.; Perkins, A.; Prasad, R.; Mayer, J.; et al. Pacritinib versus best available therapy for the treatment of myelofibrosis irrespective of baseline cytopenias (PERSIST-1): An international, randomised, phase 3 trial. Lancet Haematol. 2017, 4, e225–e236. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Hoffman, R.; Talpaz, M.; Gerds, A.T.; Stein, B.; Gupta, V.; Szoke, A.; Drummond, M.; Pristupa, A.; Granston, T.; et al. Pacritinib vs Best Available Therapy, Including Ruxolitinib, in Patients With Myelofibrosis: A Randomized Clinical Trial. JAMA Oncol. 2018, 4, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Pardanani, A.; Harrison, C.; Cortes, J.E.; Cervantes, F.; Mesa, R.A.; Milligan, D.; Masszi, T.; Mishchenko, E.; Jourdan, E.; Vannucchi, A.M.; et al. Safety and Efficacy of Fedratinib in Patients With Primary or Secondary Myelofibrosis: A Randomized Clinical Trial. JAMA Oncol. 2015, 1, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.N.; Schaap, N.; Vannucchi, A.M.; Kiladjian, J.J.; Tiu, R.V.; Zachee, P.; Jourdan, E.; Winton, E.; Silver, R.T.; Schouten, H.C.; et al. Janus kinase-2 inhibitor fedratinib in patients with myelofibrosis previously treated with ruxolitinib (JAKARTA-2): A single-arm, open-label, non-randomised, phase 2, multicentre study. Lancet Haematol. 2017, 4, e317–e324. [Google Scholar] [CrossRef]
- Sorensen, A.L.; Mikkelsen, S.U.; Knudsen, T.A.; Bjorn, M.E.; Andersen, C.L.; Bjerrum, O.W.; Brochmann, N.; Patel, D.A.; Gjerdrum, L.M.R.; El Fassi, D.; et al. Ruxolitinib and interferon-alpha2 combination therapy for patients with polycythemia vera or myelofibrosis: A phase II study. Haematologica 2020, 105, 2262–2272. [Google Scholar] [CrossRef]
- Harrison, C.N.; Gerds, A.T.; Kiladjian, J.-J.; Döhner, K.; Buckley, S.A.; Smith, J.A.; Craig, A.R.; Mascarenhas, J.; Verstovsek, S. Pacifica: A Randomized, Controlled Phase 3 Study of Pacritinib Vs. Physician’s Choice in Patients with Primary Myelofibrosis, Post Polycythemia Vera Myelofibrosis, or Post Essential Thrombocytopenia Myelofibrosis with Severe Thrombocytopenia (Platelet Count <50,000/mL). Blood 2019, 134, 4175. [Google Scholar] [CrossRef]
- Marit, M.R.; Chohan, M.; Matthew, N.; Huang, K.; Kuntz, D.A.; Rose, D.R.; Barber, D.L. Random mutagenesis reveals residues of JAK2 critical in evading inhibition by a tyrosine kinase inhibitor. PLoS ONE 2012, 7, e43437. [Google Scholar] [CrossRef][Green Version]
- Deshpande, A.; Reddy, M.M.; Schade, G.O.; Ray, A.; Chowdary, T.K.; Griffin, J.D.; Sattler, M. Kinase domain mutations confer resistance to novel inhibitors targeting JAK2V617F in myeloproliferative neoplasms. Leukemia 2012, 26, 708–715. [Google Scholar] [CrossRef]
- Marty, C.; Saint-Martin, C.; Pecquet, C.; Grosjean, S.; Saliba, J.; Mouton, C.; Leroy, E.; Harutyunyan, A.S.; Abgrall, J.F.; Favier, R.; et al. Germ-line JAK2 mutations in the kinase domain are responsible for hereditary thrombocytosis and are resistant to JAK2 and HSP90 inhibitors. Blood 2014, 123, 1372–1383. [Google Scholar] [CrossRef]
- Bhagwat, N.; Levine, R.L.; Koppikar, P. Sensitivity and resistance of JAK2 inhibitors to myeloproliferative neoplasms. Int. J. Hematol. 2013, 97, 695–702. [Google Scholar] [CrossRef]
- Downes, C.E.J.; McClure, B.J.; Bruning, J.B.; Page, E.; Breen, J.; Rehn, J.; Yeung, D.T.; White, D.L. Acquired JAK2 mutations confer resistance to JAK inhibitors in cell models of acute lymphoblastic leukemia. NPJ Precis. Oncol. 2021, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Nangalia, J.; Green, T.R. The evolving genomic landscape of myeloproliferative neoplasms. Hematol. Am. Soc. Hematol. Educ. Program. 2014, 2014, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Ortmann, C.A.; Kent, D.G.; Nangalia, J.; Silber, Y.; Wedge, D.C.; Grinfeld, J.; Baxter, E.J.; Massie, C.E.; Papaemmanuil, E.; Menon, S.; et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 2015, 372, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. Publ. Protein Soc. 2018, 27, 1984–2009. [Google Scholar] [CrossRef]
- Vainchenker, W.; Leroy, E.; Gilles, L.; Marty, C.; Plo, I.; Constantinescu, S.N. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Research 2018, 7, 82. [Google Scholar] [CrossRef]
- Schwartz, D.M.; Kanno, Y.; Villarino, A.; Ward, M.; Gadina, M.; O’Shea, J.J. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 2017, 17, 78. [Google Scholar] [CrossRef]
- Solimani, F.; Meier, K.; Ghoreschi, K. Emerging Topical and Systemic JAK Inhibitors in Dermatology. Front. Immunol. 2019, 10, 2847. [Google Scholar] [CrossRef]
- Salas, A.; Hernandez-Rocha, C.; Duijvestein, M.; Faubion, W.; McGovern, D.; Vermeire, S.; Vetrano, S.; Vande Casteele, N. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 323–337. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; O’Shea, J.J. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 2017, 18, 374–384. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Bar-Natan, M.; Nelson, E.A.; Xiang, M.; Frank, D.A. STAT signaling in the pathogenesis and treatment of myeloid malignancies. Jak-Stat 2012, 1, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Guijarro-Hernandez, A.; Vizmanos, J.L. A Broad Overview of Signaling in Ph-Negative Classic Myeloproliferative Neoplasms. Cancers 2021, 13, 984. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.M.; Suarez-Alvarez, B.; Lavin, J.L.; Ascension, A.M.; Gonzalez, M.; Lozano, J.J.; Raneros, A.B.; Bulnes, P.D.; Vidal-Castineira, J.R.; Huidobro, C.; et al. Signal Integration and Transcriptional Regulation of the Inflammatory Response Mediated by the GM-/M-CSF Signaling Axis in Human Monocytes. Cell Rep. 2019, 29, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Wicks, I.P.; Roberts, A.W. Targeting GM-CSF in inflammatory diseases. Nat. Rev. Rheumatol. 2016, 12, 37–48. [Google Scholar] [CrossRef]
- Comoglio, F.; Park, H.J.; Schoenfelder, S.; Barozzi, I.; Bode, D.; Fraser, P.; Green, A.R. Thrombopoietin signaling to chromatin elicits rapid and pervasive epigenome remodeling within poised chromatin architectures. Genome Res. 2018. [Google Scholar] [CrossRef]
- Dawson, M.A.; Bannister, A.J.; Gottgens, B.; Foster, S.D.; Bartke, T.; Green, A.R.; Kouzarides, T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 2009, 461, 819–822. [Google Scholar] [CrossRef]
- Girodon, F.; Steinkamp, M.P.; Cleyrat, C.; Hermouet, S.; Wilson, B.S. Confocal imaging studies cast doubt on nuclear localization of JAK2V617F. Blood 2011, 118, 2633–2634. [Google Scholar] [CrossRef]
- Behrmann, I.; Smyczek, T.; Heinrich, P.C.; Schmitz-Van de Leur, H.; Komyod, W.; Giese, B.; Muller-Newen, G.; Haan, S.; Haan, C. Janus kinase (Jak) subcellular localization revisited: The exclusive membrane localization of endogenous Janus kinase 1 by cytokine receptor interaction uncovers the Jak.receptor complex to be equivalent to a receptor tyrosine kinase. J. Biol. Chem. 2004, 279, 35486–35493. [Google Scholar] [CrossRef]
- Kim, H.; Kim, D.; Choi, S.A.; Kim, C.R.; Oh, S.K.; Pyo, K.E.; Kim, J.; Lee, S.H.; Yoon, J.B.; Zhang, Y.; et al. KDM3A histone demethylase functions as an essential factor for activation of JAK2-STAT3 signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, 11766–11771. [Google Scholar] [CrossRef]
- Branscombe, T.L.; Frankel, A.; Lee, J.H.; Cook, J.R.; Yang, Z.; Pestka, S.; Clarke, S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 2001, 276, 32971–32976. [Google Scholar] [CrossRef]
- Kim, H.; Ronai, Z.A. PRMT5 function and targeting in cancer. Cell Stress 2020, 4, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; Durant, S.T.; Cho, E.C.; Sheahan, S.; Edelmann, M.; Kessler, B.; La Thangue, N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008, 10, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef]
- Shan, Y.; Gnanasambandan, K.; Ungureanu, D.; Kim, E.T.; Hammaren, H.; Yamashita, K.; Silvennoinen, O.; Shaw, D.E.; Hubbard, S.R. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol. 2014, 21, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, S.; Hafer, M.; Vuorio, J.; Tucker, J.A.; Winkelmann, H.; Lochte, S.; Stanly, T.A.; Pulgar Prieto, K.D.; Poojari, C.; Sharma, V.; et al. Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 2020, 367, 643–652. [Google Scholar] [CrossRef]
- Rinaldi, C.R.; Rinaldi, P.; Alagia, A.; Gemei, M.; Esposito, N.; Formiggini, F.; Martinelli, V.; Senyuk, V.; Nucifora, G.; Pane, F. Preferential nuclear accumulation of JAK2V617F in CD34+ but not in granulocytic, megakaryocytic, or erythroid cells of patients with Philadelphia-negative myeloproliferative neoplasia. Blood 2010, 116, 6023–6026. [Google Scholar] [CrossRef]
- Yamada, Y.; Warren, A.J.; Dobson, C.; Forster, A.; Pannell, R.; Rabbitts, T.H. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 1998, 95, 3890–3895. [Google Scholar] [CrossRef]
- Morishima, T.; Krahl, A.C.; Nasri, M.; Xu, Y.; Aghaallaei, N.; Findik, B.; Klimiankou, M.; Ritter, M.; Hartmann, M.D.; Gloeckner, C.J.; et al. LMO2 activation by deacetylation is indispensable for hematopoiesis and T-ALL leukemogenesis. Blood 2019, 134, 1159–1175. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, X.; Perna, F.; Wang, L.; Koppikar, P.; Abdel-Wahab, O.; Harr, M.W.; Levine, R.L.; Xu, H.; Tefferi, A.; et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 2011, 19, 283–294. [Google Scholar] [CrossRef]
- Morotti, A.; Panuzzo, C.; Fava, C.; Saglio, G. Kinase-inhibitor-insensitive cancer stem cells in chronic myeloid leukemia. Expert Opin. Biol. Ther. 2014, 14, 287–299. [Google Scholar] [CrossRef]
- Stivala, S.; Codilupi, T.; Brkic, S.; Baerenwaldt, A.; Ghosh, N.; Hao-Shen, H.; Dirnhofer, S.; Dettmer, M.S.; Simillion, C.; Kaufmann, B.A.; et al. Targeting compensatory MEK/ERK activation increases JAK inhibitor efficacy in myeloproliferative neoplasms. J. Clin. Investig. 2019, 129, 1596–1611. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.M.; Babon, J.J.; Tvorogov, D.; Thomas, D. Persistence of myelofibrosis treated with ruxolitinib: Biology and clinical implications. Haematologica 2021, 106, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Harrison, C.N. Emerging treatments for classical myeloproliferative neoplasms. Blood 2017, 129, 693–703. [Google Scholar] [CrossRef] [PubMed]
- McLornan, D.; Harrison, C. Combination therapies in Myeloproliferative Neoplasms: Why do we need them and how to identify potential winners? J. Cell. Mol. Med. 2013, 17, 1410–1414. [Google Scholar] [CrossRef] [PubMed]
- Mondello, P.; Cuzzocrea, S.; Mian, M. Pim kinases in hematological malignancies: Where are we now and where are we going? J. Hematol. Oncol. 2014, 7, 95. [Google Scholar] [CrossRef]
- Plo, I. p53 at the crossroads of MPN treatment. Blood 2014, 124, 668–669. [Google Scholar] [CrossRef]
- Uras, I.Z.; Maurer, B.; Nivarthi, H.; Jodl, P.; Kollmann, K.; Prchal-Murphy, M.; Milosevic Feenstra, J.D.; Zojer, M.; Lagger, S.; Grausenburger, R.; et al. CDK6 coordinates JAK2 (V617F) mutant MPN via NF-kappaB and apoptotic networks. Blood 2019, 133, 1677–1690. [Google Scholar] [CrossRef]
- Dutta, A.; Nath, D.; Yang, Y.; Le, B.T.; Mohi, G. CDK6 Is a Therapeutic Target in Myelofibrosis. Cancer Res. 2021, 81, 4332–4345. [Google Scholar] [CrossRef]
- Nimmagadda, S.C.; Frey, S.; Muller, P.; Wolleschak, D.; Weinert, S.; Keller, U.; Edelmann, B.; Fischer, T. SDF1alpha-induced chemotaxis of JAK2-V617F-positive cells is dependent on Bruton tyrosine kinase and its downstream targets PI3K/AKT, PLCgamma1 and RhoA. Haematologica 2019, 104, e288–e292. [Google Scholar] [CrossRef]
- Koppikar, P.; Bhagwat, N.; Kilpivaara, O.; Manshouri, T.; Adli, M.; Hricik, T.; Liu, F.; Saunders, L.M.; Mullally, A.; Abdel-Wahab, O.; et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012, 489, 155–159. [Google Scholar] [CrossRef]
- Andraos, R.; Qian, Z.; Bonenfant, D.; Rubert, J.; Vangrevelinghe, E.; Scheufler, C.; Marque, F.; Regnier, C.H.; De Pover, A.; Ryckelynck, H.; et al. Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov. 2012, 2, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Tvorogov, D.; Thomas, D.; Liau, N.P.D.; Dottore, M.; Barry, E.F.; Lathi, M.; Kan, W.L.; Hercus, T.R.; Stomski, F.; Hughes, T.P.; et al. Accumulation of JAK activation loop phosphorylation is linked to type I JAK inhibitor withdrawal syndrome in myelofibrosis. Sci. Adv. 2018, 4, eaat3834. [Google Scholar] [CrossRef]
- Shah, R.R.; Redmond, J.M.; Mihut, A.; Menon, M.; Evans, J.P.; Murphy, J.A.; Bartholomew, M.A.; Coe, D.M. Hi-JAK-ing the ubiquitin system: The design and physicochemical optimisation of JAK PROTACs. Bioorg. Med. Chem. 2020, 28, 115326. [Google Scholar] [CrossRef] [PubMed]
- Kargbo, R.B. Degradation of Janus Kinase for Potential Application in Immune Response Therapeutics. ACS Med. Chem. Lett. 2021, 12, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Min, J.; Jarusiewicz, J.A.; Actis, M.; Yu-Chen Bradford, S.; Mayasundari, A.; Yang, L.; Chepyala, D.; Alcock, L.J.; Roberts, K.G.; et al. Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia. Blood 2021, 138, 2313–2326. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, G.; McPherson, S.; Smith, J.; Mead, A.; Harrison, C.; Mills, K.; McMullin, M.F. Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms. Cancers 2020, 12, 2669. [Google Scholar] [CrossRef]
- Tefferi, A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010, 24, 1128–1138. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Pardanani, A.; Rampal, R.; Lasho, T.L.; Levine, R.L.; Tefferi, A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia 2011, 25, 1219–1220. [Google Scholar] [CrossRef]
- Stegelmann, F.; Bullinger, L.; Schlenk, R.F.; Paschka, P.; Griesshammer, M.; Blersch, C.; Kuhn, S.; Schauer, S.; Dohner, H.; Dohner, K. DNMT3A mutations in myeloproliferative neoplasms. Leukemia 2011, 25, 1217–1219. [Google Scholar] [CrossRef]
- Walter, M.J.; Ding, L.; Shen, D.; Shao, J.; Grillot, M.; McLellan, M.; Fulton, R.; Schmidt, H.; Kalicki-Veizer, J.; O’Laughlin, M.; et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 2011, 25, 1153–1158. [Google Scholar] [CrossRef]
- Lundberg, P.; Karow, A.; Nienhold, R.; Looser, R.; Hao-Shen, H.; Nissen, I.; Girsberger, S.; Lehmann, T.; Passweg, J.; Stern, M.; et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014, 123, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- McPherson, S.; McMullin, M.F.; Mills, K. Epigenetics in Myeloproliferative Neoplasms. J. Cell. Mol. Med. 2017, 21, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Finke, C.M.; Elala, Y.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Pardanani, A. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016, 1, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Lasho, T.L.; Rotunno, G.; Score, J.; Mannarelli, C.; Pancrazzi, A.; Biamonte, F.; Pardanani, A.; Zoi, K.; Reiter, A.; et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: An international study of 797 patients. Leukemia 2014, 28, 1804–1810. [Google Scholar] [CrossRef]
- Vannucchi, A.M.; Lasho, T.L.; Guglielmelli, P.; Biamonte, F.; Pardanani, A.; Pereira, A.; Finke, C.; Score, J.; Gangat, N.; Mannarelli, C.; et al. Mutations and prognosis in primary myelofibrosis. Leukemia 2013, 27, 1861–1869. [Google Scholar] [CrossRef]
- Ramanathan, G.; Fleischman, A.G. The Microenvironment in Myeloproliferative Neoplasms. Hematol. Oncol. Clin. N. Am. 2021, 35, 205–216. [Google Scholar] [CrossRef]
- Wang, Y.; Zuo, X. Cytokines frequently implicated in myeloproliferative neoplasms. Cytokine X 2019, 1, 100005. [Google Scholar] [CrossRef]
- Fisher, D.A.C.; Fowles, J.S.; Zhou, A.; Oh, S.T. Inflammatory Pathophysiology as a Contributor to Myeloproliferative Neoplasms. Front. Immunol. 2021, 12, 683401. [Google Scholar] [CrossRef]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair 2019, 83, 102673. [Google Scholar] [CrossRef]
- Hasselbalch, H.C.; Bjorn, M.E. MPNs as Inflammatory Diseases: The Evidence, Consequences, and Perspectives. Mediat. Inflamm. 2015, 2015, 102476. [Google Scholar] [CrossRef]
- Vergara-Lluri, M.E.; Piatek, C.I.; Pullarkat, V.; Siddiqi, I.N.; O’Connell, C.; Feinstein, D.I.; Brynes, R.K. Autoimmune myelofibrosis: An update on morphologic features in 29 cases and review of the literature. Hum. Pathol. 2014, 45, 2183–2191. [Google Scholar] [CrossRef]
- Masselli, E.; Pozzi, G.; Gobbi, G.; Merighi, S.; Gessi, S.; Vitale, M.; Carubbi, C. Cytokine Profiling in Myeloproliferative Neoplasms: Overview on Phenotype Correlation, Outcome Prediction, and Role of Genetic Variants. Cells 2020, 9, 2136. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wu, B.; Ji, L.; Zhan, Y.; Li, F.; Cheng, L.; Cao, J.; Chen, H.; Ke, Y.; Min, Z.; et al. Cytokine Consistency Between Bone Marrow and Peripheral Blood in Patients With Philadelphia-Negative Myeloproliferative Neoplasms. Front. Med. 2021, 8, 598182. [Google Scholar] [CrossRef] [PubMed]
- Kleppe, M.; Kwak, M.; Koppikar, P.; Riester, M.; Keller, M.; Bastian, L.; Hricik, T.; Bhagwat, N.; McKenney, A.S.; Papalexi, E.; et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov. 2015, 5, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Tabarroki, A.; Rogers, H.J.; Visconte, V.; Hasrouni, E.; Advani, A.; Sekeres, M.A.; Duong, H.K.; Kalaycio, M.; Copelan, E.A.; Stein, B.L.; et al. The Molecular and Cytokine Profile of Triple-Negative (JAK2 V617F, JAK2 exon 12, MPL negative) Myelofibrosis, a Myeloproliferative Neoplasm with Distinct Clinico-Pathologic Characteristics. Blood 2012, 120, 3805. [Google Scholar] [CrossRef]
- Fisher, D.A.C.; Miner, C.A.; Engle, E.K.; Hu, H.; Collins, T.B.; Zhou, A.; Allen, M.J.; Malkova, O.N.; Oh, S.T. Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NFkappaB signaling. Leukemia 2019, 33, 1978–1995. [Google Scholar] [CrossRef] [PubMed]
- Sollazzo, D.; Forte, D.; Polverelli, N.; Romano, M.; Perricone, M.; Rossi, L.; Ottaviani, E.; Luatti, S.; Martinelli, G.; Vianelli, N.; et al. Crucial factors of the inflammatory microenvironment (IL-1beta/TNF-alpha/TIMP-1) promote the maintenance of the malignant hemopoietic clone of myelofibrosis: An in vitro study. Oncotarget 2016, 7, 43974–43988. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Vaidya, R.; Caramazza, D.; Finke, C.; Lasho, T.; Pardanani, A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: A comprehensive cytokine profiling study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1356–1363. [Google Scholar] [CrossRef]
- Fleischman, A.G.; Aichberger, K.J.; Luty, S.B.; Bumm, T.G.; Petersen, C.L.; Doratotaj, S.; Vasudevan, K.B.; LaTocha, D.H.; Yang, F.; Press, R.D.; et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood 2011, 118, 6392–6398. [Google Scholar] [CrossRef]
- Fisher, D.A.C.; Malkova, O.; Engle, E.K.; Miner, C.A.; Fulbright, M.C.; Behbehani, G.K.; Collins, T.B.; Bandyopadhyay, S.; Zhou, A.; Nolan, G.P.; et al. Mass cytometry analysis reveals hyperactive NF Kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia 2017, 31, 1962–1974. [Google Scholar] [CrossRef]
- Kleppe, M.; Koche, R.; Zou, L.; van Galen, P.; Hill, C.E.; Dong, L.; De Groote, S.; Papalexi, E.; Hanasoge Somasundara, A.V.; Cordner, K.; et al. Dual Targeting of Oncogenic Activation and Inflammatory Signaling Increases Therapeutic Efficacy in Myeloproliferative Neoplasms. Cancer Cell 2018, 33, 29–43.e27. [Google Scholar] [CrossRef] [PubMed]
- Hajmirza, A.; Emadali, A.; Gauthier, A.; Casasnovas, O.; Gressin, R.; Callanan, M.B. BET Family Protein BRD4: An Emerging Actor in NFkappaB Signaling in Inflammation and Cancer. Biomedicines 2018, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Yan, Y.; Wang, D.; Ding, D.; Ma, T.; Ye, Z.; Jimenez, R.; Wang, L.; Wu, H.; Huang, H. DUB3 Promotes BET Inhibitor Resistance and Cancer Progression by Deubiquitinating BRD4. Mol. Cell 2018, 71, 592–605.e594. [Google Scholar] [CrossRef]
- Lu, J.; Qian, Y.; Altieri, M.; Dong, H.; Wang, J.; Raina, K.; Hines, J.; Winkler, J.D.; Crew, A.P.; Coleman, K.; et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Saenz, D.T.; Fiskus, W.; Qian, Y.; Manshouri, T.; Rajapakshe, K.; Raina, K.; Coleman, K.G.; Crew, A.P.; Shen, A.; Mill, C.P.; et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary(s) AML cells. Leukemia 2017, 31, 1951–1961. [Google Scholar] [CrossRef]
- Fortunel, N.O.; Hatzfeld, A.; Hatzfeld, J.A. Transforming growth factor-beta: Pleiotropic role in the regulation of hematopoiesis. Blood 2000, 96, 2022–2036. [Google Scholar] [CrossRef] [PubMed]
- Chagraoui, H.; Komura, E.; Tulliez, M.; Giraudier, S.; Vainchenker, W.; Wendling, F. Prominent role of TGF-beta 1 in thrombopoietin-induced myelofibrosis in mice. Blood 2002, 100, 3495–3503. [Google Scholar] [CrossRef]
- Gastinne, T.; Vigant, F.; Lavenu-Bombled, C.; Wagner-Ballon, O.; Tulliez, M.; Chagraoui, H.; Villeval, J.L.; Lacout, C.; Perricaudet, M.; Vainchenker, W.; et al. Adenoviral-mediated TGF-beta1 inhibition in a mouse model of myelofibrosis inhibit bone marrow fibrosis development. Exp. Hematol. 2007, 35, 64–74. [Google Scholar] [CrossRef]
- Varricchio, L.; Iancu-Rubin, C.; Upadhyaya, B.; Zingariello, M.; Martelli, F.; Verachi, P.; Clementelli, C.; Denis, J.F.; Rahman, A.H.; Tremblay, G.; et al. TGF-beta1 protein trap AVID200 beneficially affects hematopoiesis and bone marrow fibrosis in myelofibrosis. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Zhao, M.; Perry, J.M.; Marshall, H.; Venkatraman, A.; Qian, P.; He, X.C.; Ahamed, J.; Li, L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 2014, 20, 1321–1326. [Google Scholar] [CrossRef]
- Malara, A.; Abbonante, V.; Zingariello, M.; Migliaccio, A.; Balduini, A. Megakaryocyte Contribution to Bone Marrow Fibrosis: Many Arrows in the Quiver. Mediterr. J. Hematol. Infect. Dis. 2018, 10, e2018068. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Norfo, R.; Pennucci, V.; Zini, R.; Manfredini, R. Genomic landscape of megakaryopoiesis and platelet function defects. Blood 2016, 127, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.B.; Allcock, R.J.; Mirzai, B.; Malherbe, J.A.; Choudry, F.A.; Frontini, M.; Chuah, H.; Liang, J.; Kavanagh, S.E.; Howman, R.; et al. Megakaryocytes in Myeloproliferative Neoplasms Have Unique Somatic Mutations. Am. J. Pathol. 2017, 187, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.J.; Yang, Q.; Goldenson, B.; Malinge, S.; Lasho, T.; Schneider, R.K.; Breyfogle, L.J.; Schultz, R.; Gilles, L.; Koppikar, P.; et al. Targeting megakaryocytic-induced fibrosis in myeloproliferative neoplasms by AURKA inhibition. Nat. Med. 2015, 21, 1473–1480. [Google Scholar] [CrossRef]
- Piszczatowski, R.T.; Steidl, U. Aurora Kinase A Inhibition: A Mega-Hit for Myelofibrosis Therapy? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4868–4870. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Crispino, J.D.; Wen, Q.J. Kinase signaling and targeted therapy for primary myelofibrosis. Exp. Hematol. 2017, 48, 32–38. [Google Scholar] [CrossRef]
- Avanzi, M.P.; Goldberg, F.; Davila, J.; Langhi, D.; Chiattone, C.; Mitchell, W.B. Rho kinase inhibition drives megakaryocyte polyploidization and proplatelet formation through MYC and NFE2 downregulation. Br. J. Haematol. 2014, 164, 867–876. [Google Scholar] [CrossRef]
- Lordier, L.; Jalil, A.; Aurade, F.; Larbret, F.; Larghero, J.; Debili, N.; Vainchenker, W.; Chang, Y. Megakaryocyte endomitosis is a failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood 2008, 112, 3164–3174. [Google Scholar] [CrossRef]
- Mali, R.S.; Ramdas, B.; Ma, P.; Shi, J.; Munugalavadla, V.; Sims, E.; Wei, L.; Vemula, S.; Nabinger, S.C.; Goodwin, C.B.; et al. Rho kinase regulates the survival and transformation of cells bearing oncogenic forms of KIT, FLT3, and BCR-ABL. Cancer Cell 2011, 20, 357–369. [Google Scholar] [CrossRef]
- Mali, R.S.; Kapur, S.; Kapur, R. Role of Rho kinases in abnormal and normal hematopoiesis. Curr. Opin. Hematol. 2014, 21, 271–275. [Google Scholar] [CrossRef][Green Version]
- Burthem, J.; Rees-Unwin, K.; Mottram, R.; Adams, J.; Lucas, G.S.; Spooncer, E.; Whetton, A.D. The rho-kinase inhibitors Y-27632 and fasudil act synergistically with imatinib to inhibit the expansion of ex vivo CD34(+) CML progenitor cells. Leukemia 2007, 21, 1708–1714. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Savino, A.; Panuzzo, C.; Rocca, S.; Familiari, U.; Piazza, R.; Crivellaro, S.; Carra, G.; Ferretti, R.; Fusella, F.; Giugliano, E.; et al. Morgana acts as an oncosuppressor in chronic myeloid leukemia. Blood 2015, 125, 2245–2253. [Google Scholar] [CrossRef] [PubMed]
- Rocca, S.; Carra, G.; Poggio, P.; Morotti, A.; Brancaccio, M. Targeting few to help hundreds: JAK, MAPK and ROCK pathways as druggable targets in atypical chronic myeloid leukemia. Mol. Cancer 2018, 17, 40. [Google Scholar] [CrossRef]
- Guilluy, C.; Bregeon, J.; Toumaniantz, G.; Rolli-Derkinderen, M.; Retailleau, K.; Loufrani, L.; Henrion, D.; Scalbert, E.; Bril, A.; Torres, R.M.; et al. The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nat. Med. 2010, 16, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Lobie, P.E. RhoA/ROCK activation by growth hormone abrogates p300/histone deacetylase 6 repression of Stat5-mediated transcription. J. Biol. Chem. 2004, 279, 32737–32750. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kong, D.; Byun, K.H.; Ye, C.; Koda, S.; Lee, D.H.; Oh, B.C.; Lee, S.W.; Lee, B.; Zabolotny, J.M.; et al. Rho-kinase regulates energy balance by targeting hypothalamic leptin receptor signaling. Nat. Neurosci. 2012, 15, 1391–1398. [Google Scholar] [CrossRef]
- Peelman, F.; Tavernier, J. ROCKing the JAKs. Jak-Stat 2013, 2, e24074. [Google Scholar] [CrossRef]
- Chen, W.; Nyuydzefe, M.S.; Weiss, J.M.; Zhang, J.; Waksal, S.D.; Zanin-Zhorov, A. ROCK2, but not ROCK1 interacts with phosphorylated STAT3 and co-occupies TH17/TFH gene promoters in TH17-activated human T cells. Sci. Rep. 2018, 8, 16636. [Google Scholar] [CrossRef]
- Fiskus, W.; Verstovsek, S.; Manshouri, T.; Rao, R.; Balusu, R.; Venkannagari, S.; Rao, N.N.; Ha, K.; Smith, J.E.; Hembruff, S.L.; et al. Heat shock protein 90 inhibitor is synergistic with JAK2 inhibitor and overcomes resistance to JAK2-TKI in human myeloproliferative neoplasm cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 7347–7358. [Google Scholar] [CrossRef]
- Marubayashi, S.; Koppikar, P.; Taldone, T.; Abdel-Wahab, O.; West, N.; Bhagwat, N.; Caldas-Lopes, E.; Ross, K.N.; Gonen, M.; Gozman, A.; et al. HSP90 is a therapeutic target in JAK2-dependent myeloproliferative neoplasms in mice and humans. J. Clin. Investig. 2010, 120, 3578–3593. [Google Scholar] [CrossRef]
- Evrot, E.; Ebel, N.; Romanet, V.; Roelli, C.; Andraos, R.; Qian, Z.; Dolemeyer, A.; Dammassa, E.; Sterker, D.; Cozens, R.; et al. JAK1/2 and Pan-deacetylase inhibitor combination therapy yields improved efficacy in preclinical mouse models of JAK2V617F-driven disease. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6230–6241. [Google Scholar] [CrossRef] [PubMed]
- Rampal, R.K.; Pinzon-Ortiz, M.; Somasundara, A.V.H.; Durham, B.; Koche, R.; Spitzer, B.; Mowla, S.; Krishnan, A.; Li, B.; An, W.; et al. Therapeutic Efficacy of Combined JAK1/2, Pan-PIM, and CDK4/6 Inhibition in Myeloproliferative Neoplasms. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 3456–3468. [Google Scholar] [CrossRef] [PubMed]
- Brkic, S.; Stivala, S.; Santopolo, A.; Szybinski, J.; Jungius, S.; Passweg, J.R.; Tsakiris, D.; Dirnhofer, S.; Hutter, G.; Leonards, K.; et al. Dual targeting of JAK2 and ERK interferes with the myeloproliferative neoplasm clone and enhances therapeutic efficacy. Leukemia 2021, 35, 2875–2884. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Kapur, R. Targeting phosphatidylinositol-3-kinase pathway for the treatment of Philadelphia-negative myeloproliferative neoplasms. Mol. Cancer 2015, 14, 118. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Barosi, G.; Rambaldi, A.; Marchioli, R.; Masciulli, A.; Tozzi, L.; Biamonte, F.; Bartalucci, N.; Gattoni, E.; Lupo, M.L.; et al. Safety and efficacy of everolimus, a mTOR inhibitor, as single agent in a phase 1/2 study in patients with myelofibrosis. Blood 2011, 118, 2069–2076. [Google Scholar] [CrossRef]
- Mazzacurati, L.; Lambert, Q.T.; Pradhan, A.; Griner, L.N.; Huszar, D.; Reuther, G.W. The PIM inhibitor AZD1208 synergizes with ruxolitinib to induce apoptosis of ruxolitinib sensitive and resistant JAK2-V617F-driven cells and inhibit colony formation of primary MPN cells. Oncotarget 2015, 6, 40141–40157. [Google Scholar] [CrossRef][Green Version]
- Yacoub, A.; Wang, E.S.; Rampal, R.K.; Borate, U.; Kremyanskaya, M.; Ali, H.; Hobbs, G.S.; O’Connell, C.; Assad, A.; Erickson-Viitanen, S.; et al. Abstract CT162: Addition of parsaclisib (INCB050465), a PI3Kδ inhibitor, in patients with suboptimal response to ruxolitinib: A phase 2 study in patients with myelofibrosis. Cancer Res. 2021, 81, CT162. [Google Scholar] [CrossRef]
- Tutt, A.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; Azambuja, E.d.; Fielding, A.; Gelpi, J.B.; et al. OlympiA: A phase III, multicenter, randomized, placebo-controlled trial of adjuvant olaparib after (neo)adjuvant chemotherapy in patients with germline BRCA1/2 mutations and high-risk HER2-negative early breast cancer. J. Clin. Oncol. 2021, 39, LBA1. [Google Scholar] [CrossRef]
- Choong, M.L.; Pecquet, C.; Pendharkar, V.; Diaconu, C.C.; Yong, J.W.; Tai, S.J.; Wang, S.F.; Defour, J.P.; Sangthongpitag, K.; Villeval, J.L.; et al. Combination treatment for myeloproliferative neoplasms using JAK and pan-class I PI3K inhibitors. J. Cell. Mol. Med. 2013, 17, 1397–1409. [Google Scholar] [CrossRef]
- Wang, J.C.; Chen, C.; Dumlao, T.; Naik, S.; Chang, T.; Xiao, Y.Y.; Sominsky, I.; Burton, J. Enhanced histone deacetylase enzyme activity in primary myelofibrosis. Leuk. Lymphoma 2008, 49, 2321–2327. [Google Scholar] [CrossRef]
- Skov, V.; Larsen, T.S.; Thomassen, M.; Riley, C.H.; Jensen, M.K.; Bjerrum, O.W.; Kruse, T.A.; Hasselbalch, H.C. Increased gene expression of histone deacetylases in patients with Philadelphia-negative chronic myeloproliferative neoplasms. Leuk. Lymphoma 2012, 53, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Verstovsek, S. Investigational histone deacetylase inhibitors (HDACi) in myeloproliferative neoplasms. Expert Opin. Investig. Drugs 2016, 25, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Guerini, V.; Barbui, V.; Spinelli, O.; Salvi, A.; Dellacasa, C.; Carobbio, A.; Introna, M.; Barbui, T.; Golay, J.; Rambaldi, A. The histone deacetylase inhibitor ITF2357 selectively targets cells bearing mutated JAK2(V617F). Leukemia 2008, 22, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Amaru Calzada, A.; Todoerti, K.; Donadoni, L.; Pellicioli, A.; Tuana, G.; Gatta, R.; Neri, A.; Finazzi, G.; Mantovani, R.; Rambaldi, A.; et al. The HDAC inhibitor Givinostat modulates the hematopoietic transcription factors NFE2 and C-MYB in JAK2(V617F) myeloproliferative neoplasm cells. Exp. Hematol. 2012, 40, 634–645.e610. [Google Scholar] [CrossRef]
- Akada, H.; Akada, S.; Gajra, A.; Bair, A.; Graziano, S.; Hutchison, R.E.; Mohi, G. Efficacy of vorinostat in a murine model of polycythemia vera. Blood 2012, 119, 3779–3789. [Google Scholar] [CrossRef]
- Pastore, F.; Bhagwat, N.; Pastore, A.; Radzisheuskaya, A.; Karzai, A.; Krishnan, A.; Li, B.; Bowman, R.L.; Xiao, W.; Viny, A.D.; et al. PRMT5 Inhibition Modulates E2F1 Methylation and Gene-Regulatory Networks Leading to Therapeutic Efficacy in JAK2(V617F)-Mutant MPN. Cancer Discov. 2020, 10, 1742–1757. [Google Scholar] [CrossRef]
- Huang, B.; Yang, X.D.; Zhou, M.M.; Ozato, K.; Chen, L.F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 2009, 29, 1375–1387. [Google Scholar] [CrossRef]
- Prescott, J.A.; Mitchell, J.P.; Cook, S.J. Inhibitory feedback control of NF-kappaB signalling in health and disease. Biochem. J. 2021, 478, 2619–2664. [Google Scholar] [CrossRef]
- Hofland, T.; de Weerdt, I.; Ter Burg, H.; de Boer, R.; Tannheimer, S.; Tonino, S.H.; Kater, A.P.; Eldering, E. Dissection of the Effects of JAK and BTK Inhibitors on the Functionality of Healthy and Malignant Lymphocytes. J. Immunol. 2019, 203, 2100–2109. [Google Scholar] [CrossRef]
- Marty, C.; Lacout, C.; Droin, N.; Le Couedic, J.P.; Ribrag, V.; Solary, E.; Vainchenker, W.; Villeval, J.L.; Plo, I. A role for reactive oxygen species in JAK2 V617F myeloproliferative neoplasm progression. Leukemia 2013, 27, 2187–2195. [Google Scholar] [CrossRef]
- Pilie, P.G.; Tang, C.; Mills, G.B.; Yap, T.A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2019, 16, 81–104. [Google Scholar] [CrossRef] [PubMed]
- Nieborowska-Skorska, M.; Maifrede, S.; Dasgupta, Y.; Sullivan, K.; Flis, S.; Le, B.V.; Solecka, M.; Belyaeva, E.A.; Kubovcakova, L.; Nawrocki, M.; et al. Ruxolitinib-induced defects in DNA repair cause sensitivity to PARP inhibitors in myeloproliferative neoplasms. Blood 2017, 130, 2848–2859. [Google Scholar] [CrossRef] [PubMed]
- Jayavelu, A.K.; Schnoder, T.M.; Perner, F.; Herzog, C.; Meiler, A.; Krishnamoorthy, G.; Huber, N.; Mohr, J.; Edelmann-Stephan, B.; Austin, R.; et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 2020, 588, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Xia, L.; Li, Y.; Wang, X.; Hoffman, R. The orally bioavailable MDM2 antagonist RG7112 and pegylated interferon alpha 2a target JAK2V617F-positive progenitor and stem cells. Blood 2014, 124, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, S.; Mascarenhas, J. Novel therapeutics in myeloproliferative neoplasms. J. Hematol. Oncol. 2020, 13, 162. [Google Scholar] [CrossRef]
- Vachani, P.; Lange, A.; Delgado, R.G.; Al-Ali, H.K.; Hernández-Rivas, J.M.; Kiladjian, J.-J.; Vannucchi, A.; Perkins, A.C.; Valmeekam, V.; Krejsa, C.M.; et al. Potential Disease-Modifying Activity of Navtemadlin (KRT-232), a First-in-Class MDM2 Inhibitor, Correlates with Clinical Benefits in Relapsed/Refractory Myelofibrosis (MF). Blood 2021, 138, 3581. [Google Scholar] [CrossRef]
- Yacoub, A.; Mascarenhas, J.; Kosiorek, H.; Prchal, J.T.; Berenzon, D.; Baer, M.R.; Ritchie, E.; Silver, R.T.; Kessler, C.; Winton, E.; et al. Pegylated interferon alfa-2a for polycythemia vera or essential thrombocythemia resistant or intolerant to hydroxyurea. Blood 2019, 134, 1498–1509. [Google Scholar] [CrossRef]
- Gisslinger, H.; Klade, C.; Georgiev, P.; Krochmalczyk, D.; Gercheva-Kyuchukova, L.; Egyed, M.; Rossiev, V.; Dulicek, P.; Illes, A.; Pylypenko, H.; et al. Ropeginterferon alfa-2b versus standard therapy for polycythaemia vera (PROUD-PV and CONTINUATION-PV): A randomised, non-inferiority, phase 3 trial and its extension study. Lancet Haematol. 2020, 7, e196–e208. [Google Scholar] [CrossRef]
- Bolli, N.; Biancon, G.; Moarii, M.; Gimondi, S.; Li, Y.; de Philippis, C.; Maura, F.; Sathiaseelan, V.; Tai, Y.T.; Mudie, L.; et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 2018, 32, 2604–2616. [Google Scholar] [CrossRef]
- Zhou, J.; Ji, Q.; Li, Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: Underlying mechanisms and reversal strategies. J. Exp. Clin. Cancer Res. CR 2021, 40, 328. [Google Scholar] [CrossRef]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimaraes, J.E.; Vasconcelos, M.H. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Stark, M.; Ofran, Y.; Assaraf, Y.G. Deciphering molecular mechanisms underlying chemoresistance in relapsed AML patients: Towards precision medicine overcoming drug resistance. Cancer Cell Int. 2021, 21, 53. [Google Scholar] [CrossRef] [PubMed]
- Cianfriglia, M. Targeting MDR1-P-glycoprotein (MDR1-Pgp) in immunochemotherapy of acute myeloid leukemia (AML). Ann. Ist. Super. Sanita 2013, 49, 190–208. [Google Scholar] [CrossRef] [PubMed]



| Clinical Trial | Type of Inhibitor | Setting of Disease | Reference |
|---|---|---|---|
| COMFORT-I | Ruxolitinib, (JAK1/2 inhibitor) | MF, PPV-MF, PET-MF | [36] |
| COMFORT-II | Ruxolitinib, (JAK1/2 inhibitor) | Intermediate-2 or high-risk MF, PPV-MF, PET-MF | [37] |
| SIMPLIFY-1 | Momelotinib, (JAK1/2 and ACVR1 inhibitor) | High-risk, intermediate-2-risk, or symptomatic intermediate-1-risk-naive MF | [38] |
| SIMPLIFY-II | Momelotinib, (JAK1/2 and ACVR1 inhibitor) | MF with suboptimal responses or haematological toxic effects with ruxolitinib | [39] |
| RESPONSE | Ruxolitinib, (JAK1/2 inhibitor) | Jak-inhibitor-naive PV | [40] |
| PERSIST-1 | Pacritinib, (JAK2, IRAK1 and FLT3 inhibitor) | High-risk MF | [41] |
| PERSIST-2 | Pacritinib, (JAK2, IRAK1 and FLT3 inhibitor) | Intermediate-1, intermediate-2, or high-risk primary or secondary MF | [42] |
| JAKARTA-1 | Fedratinib, (JAK2, RET and FLT3 inhibitor) | Primary or secondary MF | [43] |
| JAKARTA-2 | Fedratinib, (JAK2, RET and FLT3 inhibitor) | Intermediate- or high-risk MF, PPV-MF, or PET-MF previously treated with Ruxolitinib | [44] |
| COMBI | Ruxolitinib, (JAK1/2 inhibitor) and Interferon-α2 | MF and PV | [45] |
| PACIFICA | Pacritinib, (JAK2, IRAK1 and FLT3 inhibitor) | MF, PPV-MF, PET-MF | [46] |
| Agent | Disease Setting | Clinical Trial | Phase |
|---|---|---|---|
| Elotuzumab (anti CD319) | MF | NCT04517851 | Phase 2 |
| Selinexor (SINE inhibitor) | Naive MF | NCT04562389 | Phase 1/2 |
| CPI-0610 (BET inhibitor) | MF, PPV-MF, PET-MF | NCT04603495 | Phase 3 |
| Imetelstat (Telomerase inhibitor) | Intermediate-2- or high-risk MF refractory to JAK inhibitor | NCT04576156 | Phase 3 |
| Alisertib (AURKA inhibitor) | PMF | NCT02530619 | Pilot study |
| Navitoclax (Bcl-2 inhibitor) | MF/Relapsed/Refractory MF | NCT04454658/NCT04468984 | Phase 1/Phase 3 |
| TL-895 (BTK inhibitor) | MF | NCT04655118 | Phase 2 |
| Navtemadlin (MDM2 inhibitor) | MF, PPV-MF, PET-MF with suboptimal response to Ruxolitinib | NCT04485260 | Phase 1b/2 |
| Navtemadlin (MDM2 inhibitor) | MF, PPV-MF, PET-MF | NCT03662126 | Phase 2/3 |
| Navtemadlin (MDM2 inhibitor) + TL-895 (BTK inhibitor) | MF, PPV-MF, PET-MF | NCT04640532 | Phase 1/2 |
| Ruxolitinib (JAK1/2 inhibitor) + Parsaclisib (PI3Kδ ihibitor) | MF, PPV-MF, PET-MF | NCT04551066 | Phase 3 |
| JAK2 Downstream Targets | Function | Localization | Mechanism of Action |
|---|---|---|---|
| STATs | Signal transduction and activation of transcription | Cytoplasm and nucleus | STAT target gene transcription |
| PI3K/AKT/mTOR | Signal transduction | Cytoplasm | Increased cell survival and proliferation and regulation of cell metabolism [79] |
| ERK1/2 | Signal transduction | Cytoplasm | Cell survival and proliferation [69] |
| Histone H3 | Chromatin folding and accessibility | Nucleus | Chromatin decondensation and increased gene expression (i.e., lmo2) [66] |
| KDM3A | Histone demethylase | Nucleus | Enhanced STAT3 target gene transcription [69] |
| PRMT5 | Histone methyltransferase | Nucleus | Inhibition of PRMT5 methyltransferase function, gene transcription alteration [79] |
| YBX1 | Splicing factor | Cytoplasm and nucleus | Sustained ERK signaling and disease persistence [69] |
| PIM | Signal transduction | Cytoplasm and nucleus | Cell survival, proliferation, metabolism, and drug resistance [69,85] |
| MDM2 | Ubiquitin ligase | Cytoplasm | p53 degradation, increased cell survival, and proliferation [86] |
| CDK6 | Cyclin dependent kinases | Nucleus | Sustained NF-kB signaling, cytokine secretion [87,88] |
| BTK | Signal transduction | Cytoplasm and nucleus | Cell migration [89] |
| NLRP3 inflammasome | Cleavage of the precursors form of IL-1β and IL-18 | Cytoplasm | Maturation and secretion of pro-inflammatory IL-1β and IL-18 [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bochicchio, M.T.; Di Battista, V.; Poggio, P.; Carrà, G.; Morotti, A.; Brancaccio, M.; Lucchesi, A. Understanding Aberrant Signaling to Elude Therapy Escape Mechanisms in Myeloproliferative Neoplasms. Cancers 2022, 14, 972. https://doi.org/10.3390/cancers14040972
Bochicchio MT, Di Battista V, Poggio P, Carrà G, Morotti A, Brancaccio M, Lucchesi A. Understanding Aberrant Signaling to Elude Therapy Escape Mechanisms in Myeloproliferative Neoplasms. Cancers. 2022; 14(4):972. https://doi.org/10.3390/cancers14040972
Chicago/Turabian StyleBochicchio, Maria Teresa, Valeria Di Battista, Pietro Poggio, Giovanna Carrà, Alessandro Morotti, Mara Brancaccio, and Alessandro Lucchesi. 2022. "Understanding Aberrant Signaling to Elude Therapy Escape Mechanisms in Myeloproliferative Neoplasms" Cancers 14, no. 4: 972. https://doi.org/10.3390/cancers14040972
APA StyleBochicchio, M. T., Di Battista, V., Poggio, P., Carrà, G., Morotti, A., Brancaccio, M., & Lucchesi, A. (2022). Understanding Aberrant Signaling to Elude Therapy Escape Mechanisms in Myeloproliferative Neoplasms. Cancers, 14(4), 972. https://doi.org/10.3390/cancers14040972








