Simple Summary
Pancreatic ductal adenocarcinoma (PDAC) is currently one of the deadliest cancers. Despite the progress that has been made in the research of patient care and the understanding of pancreatic cancer, the survival rate remains mediocre. SMAD4, a tumor-suppressor gene, is specifically inactivated in 50–55% of pancreatic cancers. The role of SMAD4 protein loss in PDAC remains controversial, but seems to be associated with worse overall survival and metastasis. Here, we review the function of SMAD4 inactivation in the context of a specific biological process called epithelial–mesenchymal transition, as it has been increasingly associated with tumor formation, metastasis and resistance to therapy. By improving our understanding of these molecular mechanisms, we hope to find new targets for therapy and improve the care of patients with PDAC.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) presents a five-year survival rate of 10% and its incidence increases over the years. It is, therefore, essential to improve our understanding of the molecular mechanisms that promote metastasis and chemoresistance in PDAC, which are the main causes of death in these patients. SMAD4 is inactivated in 50% of PDACs and its loss has been associated with worse overall survival and metastasis, although some controversy still exists. SMAD4 is the central signal transducer of the transforming growth factor-beta (TGF-beta) pathway, which is notably known to play a role in epithelial–mesenchymal transition (EMT). EMT is a biological process where epithelial cells lose their characteristics to acquire a spindle-cell phenotype and increased motility. EMT has been increasingly studied due to its potential implication in metastasis and therapy resistance. Recently, it has been suggested that cells undergo EMT transition through intermediary states, which is referred to as epithelial–mesenchymal plasticity (EMP). The intermediary states are characterized by enhanced aggressiveness and more efficient metastasis. Therefore, this review aims to summarize and analyze the current knowledge on SMAD4 loss in patients with PDAC and to investigate its potential role in EMP in order to better understand its function in PDAC carcinogenesis.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death worldwide, with a five-year-survival rate at 10%, which is the lowest among all cancer types [1]. The incidence of pancreatic cancer has continued to increase over the years, and it is predicted to become the second leading cause of mortality related to cancer in 2030 [1,2].
Its extremely poor prognosis is due to several different factors. Pancreatic cancer is usually diagnosed at an advanced stage due to a lack of symptoms as well as diagnostic and prognostic tumor markers [3]. Consequently, only 10–20% of patients present with resectable pancreatic cancer at diagnosis [3,4,5].
For patients with unresectable or borderline resectable PDAC, the most commonly employed therapies include neoadjuvant chemotherapy and/or radiotherapy. Likewise, surgery is also usually followed by adjuvant chemotherapy in an attempt to improve overall survival (OS). Unfortunately, survival remains mediocre due to the intrinsic resistance of pancreatic cancer to conventional therapies, including chemotherapy, radiotherapy and targeted molecular therapy [3,5].
Although some progress has been made in the research of patient care and the understanding of the molecular and genetic background of pancreatic cancer, the improvement of OS for PDAC in the past two decades has been modest [6,7]. The accession of modified FOLFIRINOX as an adjuvant therapy might have modestly increased disease-free survival (DFS) and OS, but at the expense of a greater number of toxic effects [8].
More effective therapeutic strategies are, therefore, desperately needed. These include the implementation of better screening strategies and the discovery of tumor markers allowing for earlier detection. The development of targeted therapies and the identification of subgroups of patients who could benefit from individualized therapies are also required. Improving our understanding of pancreatic cancer carcinogenesis, namely of the molecular mechanisms that promote metastatic spread and chemoresistance, in an attempt to optimize the current treatments of pancreatic cancer, is indispensable.
PDAC has been extensively studied throughout the years and its genomic aberrations have been well-described [9,10,11,12,13,14,15]. However, despite the increasing precision and depth of coverage, the four main driver genes identified before the time of next-generation sequencing (NGS) remain on the forefront: the oncogene KRAS, which is mutated in almost 95% of PDACs together with alterations of the tumor-suppressor genes CDKN2A, TP53 and SMAD4 [9,16,17]. These mutations are acquired in a sequenced time of events, with KRAS and CDKN2A being affected early in the carcinogenesis of PDAC, while TP53 and SMAD4 characterize later events, predominantly occurring in invasive PDAC [18,19].
SMAD4 alterations are relatively specific to gastrointestinal cancers, notably pancreatic cancer, where SMAD4 is inactivated in 50–55% of cases [20,21,22,23,24]. SMAD4 is a member of the Smad family of transcription factor proteins and is the central signal transducer of the transforming growth factor-beta (TGF-beta) signaling pathway. This signaling pathway is well known for its role in inducing epithelial–mesenchymal transition (EMT).
EMT is the process through which cells with epithelial characteristics acquire a mesenchymal cell phenotype and behavior [25,26]. EMT has been increasingly studied in the last two decades for its role in carcinogenesis, metastasis and therapy resistance. However, recent research suggests that EMT is not a binary process, and that cells transition through a spectrum of intermediary states. Each state presents its own characteristics and behavior. The capacity of cells to shift through partial EMT states is referred to as epithelial–mesenchymal plasticity (EMP) [26].
The biological mechanisms underlying the role of SMAD4 in EMT and the metastatic process in PDAC remain unclear and are a subject of controversy.
This review aims to summarize the current knowledge concerning the role of SMAD4 alterations in patients with PDAC and to investigate its potential role in EMT, particularly in that of EMP, in order to better understand its function in terms of the aggressiveness of PDAC.
2. Literature Review
In order to investigate the published studies on the TGF-beta pathway, SMAD4 and EMT in patients with PDAC, a comprehensive literature search of the electronic database PubMed was performed up to January 2022. Studies were selected using the following search terms (non-exhaustive list): “pancreas”, “pancreatic cancer”, “ pancreatic ductal adenocarcinoma”, “PDAC”, “TGF-beta”, “SMAD4”, “epithelial-mesenchymal transition”, “EMT”, “hybrid EMT”, “partial EMT” and/or “epithelial-mesenchymal plasticity”.
3. Discovery and General Structure of SMAD4
The SMAD4 gene is located on the 18q21.2 human chromosome locus. It is composed of 12 exons, of which the second to twelfth are translated, with the first one containing just part of the 5′ untranslated region (5′UTR), and coding for a 552 amino acid sequence resulting in a 60 kDa protein [21,23,24,27].
SMAD4 was first identified as a tumor-suppressor gene by Hahn et al. in 1996. Its original name was initially designated as “Deleted in pancreatic cancer 4” (DPC4, homozygously deleted in pancreatic carcinoma, locus 4) [21]. Its current name, “Smad”, is derived from a combination of the names of the genes that code for two orthologous proteins: the “small” worm phenotype (sma) from Caenorhabditis elegans and Mothers against decapentaplegic (Mad) from Drosophila melanogaster. Indeed, the human protein sequence of SMAD4 presents similarities to the D. melanogaster Mad protein and to the C. elegans Mad homologs sma-2, sma-3, and sma-4, with SMAD4 being the human homolog of sma-4 [28,29].
The SMAD4 protein structure is composed of two functional sites: the N-terminal Mad homology 1 (MH1) domain, which essentially allows SMAD4 to bind to DNA, and the C-terminal Mad homology 2 (MH2) domain, which enables homo- and hetero-oligomerization with other Smad proteins. Both domains may interact with other proteins, but it is mainly the MH2 domain that can interact with transcription factors, co-activators and co-repressors. These two domains are connected by a linker region that serves predominately as a regulatory site [29,30].
4. SMAD4 and TGF-Beta Signaling Pathway
SMAD4 is the central mediator of the TGF-beta signaling pathway, which has key roles in the development and homeostasis of epithelial cells, stromal compartments and immune cells in the gastrointestinal system. However, the TGF-beta signaling pathway is also implicated in carcinogenesis, which is commonly called the “TGF-beta paradox” or the “TGF-beta switch” [31].
There are two branches in the TGF-beta family signaling pathway: the TGF-beta branch, represented by ligands such as TGF-beta, activin, nodal, or myostatin, and the bone morphogenetic protein (BMP) branch, represented by ligands such as BMP and growth and differentiation factor (GDF) [32,33]. In the present review, we will focus on the TGF-beta branch and the implication of SMAD4 in this pathway.
There are three different TGF-beta ligands (TGF-beta 1–3), which are almost identical and play virtually the same roles in vitro [25]. Three different types of TGF-beta receptors (TGFBR 1 to 3) also exist. TGFBR1/2 enable intracellular signaling through their intrinsic enzymatic activity, while TGFBR3 acts as a regulator of the TGF-beta pathway [23,25,30,34]. On the surface of human cells, TGFBR1 and TGFBR2 exist essentially as homodimers in the absence of a ligand. The TGF-beta ligand binds specifically to TGFBR2, which leads to the formation of tetra-heteromeric receptor complexes between TGFBR1 and TGFBR2. This enables the phosphorylation of TGFBR1 by TGFBR2 [23,30,34]. TGFBR1 subsequently phosphorylates the intracytoplasmic receptor-regulated Smad proteins (R-Smad), SMAD2 and SMAD3. They then proceed to form a heteromeric complex with the only known common-Smad protein (Co-Smad), SMAD4. This protein complex translocates to the nucleus to regulate gene transcription in association with different transcription factors (TFs) and co-activators/co-repressors, in a gene- and cell-specific manner [23,25,30,34]. The Smad protein complexes are incapable of direct recruitment of the basal transcription machinery to responsive promoters. They instead regulate gene transcription through chromatin remodeling and histone modifications [30]. When the TGF-beta signaling pathway acts through SMAD2/3, it is referred to as the “canonical TGF-beta pathway” (Figure 1).
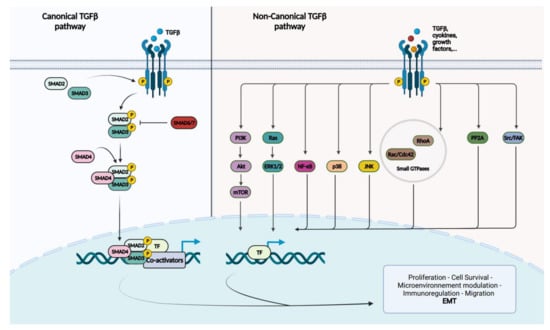
Figure 1.
Schematic representation of the canonical and non-canonical TGF-beta signaling pathways. TGF-beta or other types of ligands will bind to TGFBR2, which recruits and phosphorylates TGFBR1. In the canonical TGF-beta pathway, TGFBR1 phosphorylates R-Smads, SMAD2 and SMAD3. The Co-Smad, SMAD4, will form a heteromeric complex with the phosphorylated R-Smads and translocate to the nucleus. Once in the nucleus, Smad proteins interact with transcription factors (TFs) and co-activators/co-repressors to regulate gene transcription. I-Smads, SMAD6 and SMAD7 regulate TGF-beta signaling. The non-canonical TGF-beta signaling regroups MAP kinases including ERK1/2, p38 and JNK; the cell survival mediators PI3k/Akt/mTOR; inflammation mediators, such as NF-kB; small GTP-binding proteins, such as Ras, RhoA, Rac and Cdc42; and nonreceptor protein tyrosine kinases including PP2A, Src and FAK.
SMAD6 and 7 are inhibitory-Smads (I-Smads) that have a regulatory effect on the pathway. This regulation is possible by binding directly to R-Smads, preventing their recruitment or phosphorylation, but also by promoting the ubiquitin proteolysis of TGFBR1 and promoting its dephosphorylation [23,30,34].
The TGF-beta pathway can also function in the absence of SMAD2/3, which is called the “non-canonical TGF-beta pathway”. Besides Smad-mediated transcription, TGF-beta can activate the MAP kinases ERK1/2, JNK and p38MAPK; the cell survival mediators PI3K, AKT1/2 and mTOR; the inflammation mediators NF-kB, COX-2 and prostaglandins; small GTP-binding proteins such as Ras, RhoA, Rac1 and Cdc42; and the nonreceptor protein tyrosine kinases PP2A, Src, FAK and Abl [23,25,34]. Although the activation of these pathways are Smad independent, they can influence Smad-mediated pathways by regulating Smad activation, inducing or repressing Smad-mediated transcription.
5. The Role of the TGF-Beta Signaling Pathway in PDAC: In Brief
In epithelial cells, the TGF-beta pathway plays a dual role, functioning both as a tumor suppressor and a tumor promoter. It is not certain how this switch is operated, but it has been suggested that the canonical and non-canonical TGF-beta pathways play a tumor-suppressive role at the early stages of tumor development in pancreatic cancer. Indeed, in the early stages of PDAC, high TGF-beta1 expression may counteract proliferation and is associated with better OS [35]. In contrast, during the stages of disease progression, it will promote invasion, migration and metastasis [23,25,30,34,36]. Patients with high TGF-beta expression and advanced disease present significantly reduced survival [37,38].
TGF-beta has potent cytostatic effects and is known to regulate the cell cycle by inducing cyclin-dependent-kinase (CDK) inhibitors, such as CDKN2B and/or CDKN2A, depending on the cell type [33,39].
In a physiological setting, the TGF-beta pathway also triggers apoptosis, although the exact biological mechanism is still undefined. The TGF-beta signaling pathway may induce the expression of death-associated protein kinase (DAP-kinase) in a Smad-dependent manner [40]. Increased expression of SMAD4 has also been linked to apoptosis [41,42].
TGF-beta can restrict epithelial cell proliferation and tumor formation by acting on the surrounding stroma as well as the inflammatory cells. Indeed, TGF-beta will, for example, limit the expression of mitogenic factors secreted by the nearby fibroblasts, inhibiting epithelial growth [33,39]. However, when the TGF-beta switch takes place, the microenvironment is modulated in a manner that promotes epithelial cell proliferation. The composition of the tumor microenvironment may also change, enabling invasion and migration and inducing chemoresistance [25,33,39,43]. TGF-beta is also a potent immunosuppressor and promotes immune tolerance by regulating the activities of macrophages, natural killer (NK) and effector T lymphocytes [33,39]. Therefore, in the context of carcinogenesis, TGF-beta may permit the immune evasion of cancerous cells.
One of the most notable tumor-promoting mechanisms induced by the TGF-beta canonical and non-canonical pathways is EMT. The description of this process as well as its role in PDAC shall be thoroughly discussed later on in this review.
6. SMAD4 and Pancreatic Ductal Adenocarcinoma
In 2008, Jones et al. first introduced the concept of core signaling pathways based on high-throughput Sanger sequencing of 20,661 protein-coding genes. Their analysis demonstrated that the TGF-beta signaling pathway was affected in 100% of PDACs, with SMAD4 being the most frequently altered gene [9]. SMAD4 is mainly inactivated by two mechanisms, predominantly by homozygous deletion or mutations with the loss of the wild-type allele [21,44]. These somatic alterations include nonsense, missense and frameshift mutations, usually leading to a truncated and/or a non-functional protein. More than 80% of these alterations occur on the MH2 functional domain, with a hotspot of missense mutations occurring on codon 361 [21,24,45]. MH2 domain mutations can cause a failure to homo- and hetero-oligomerize, while MH1 domain mutations prevent DNA binding by SMAD4. [24,45,46]. It has also been demonstrated that some mutations in both the MH1 and MH2 domain can increase degradation via ubiquitin-mediated proteolysis [46,47,48]. It is interesting to note that larger genetic aberrations affect the SMAD4 gene; notably, a loss of heterozygosity (LOH) of the 18q (where SMAD4 is located) occurs in 90% of PDACs [21].
Immunohistochemistry (IHC) is a reliable method to evaluate the genetic status of SMAD4, due to it being a sensitive and specific marker (Figure 2) [27]. Wilentz et al. detected no difference in immunolabeling between PDACs presenting with homozygous deletion or intragenic mutations associated with allele loss. Additionally, a single wild-type allele was sufficient to exhibit “diffusely positive staining” [27]. Therefore, SMAD4 detection by IHC can be used in routine pathology to detect SMAD4 genetic alterations, which can be a useful marker in diagnosing PDAC on cytology, biopsy or surgery specimens [22,49,50].
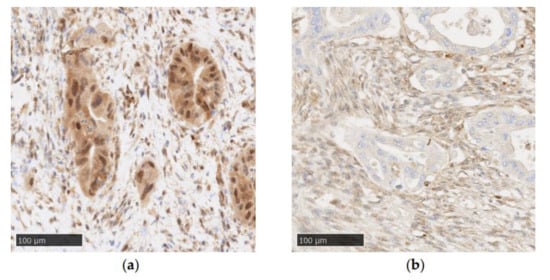
Figure 2.
Examples of SMAD4 immunohistochemistry in PDAC (Recombinant Anti-SMAD4 antibody (EP618Y), Abcam, original magnification: ×20): (a) positive SMAD4 staining with tumor cells showing cytoplasmic and nuclear staining; (b) negative SMAD4 staining characterized by a loss of nuclear and cytoplasmic staining in tumor cells.
In PDAC, a loss of SMAD4 expression has been associated with tumor size, peripancreatic extension, TNM staging, lymphatic invasion, nodal involvement and poor differentiation [51,52,53], influencing OS [54,55,56,57,58,59,60,61,62,63,64] and DFS [56,59,61,64].
However, the impact of SMAD4 loss remains controversial. Several large retrospective studies have not been able to statistically demonstrate a prognostic impact of SMAD4 alterations on OS and DFS [10,52,65,66,67,68,69,70,71]. In some studies, SMAD4 inactivation is a negative prognostic factor solely in a specific subset of patients. Hsieh et al. associated SMAD4 loss with worse DFS, but only in patients presenting with homozygous deletion and not SMAD4 mutations [72].
Similarly, SMAD4 deficiency alone was not always linked to prognosis, but its loss, associated with other biomarkers, could predict OS and/or DFS to a significant statistical degree [37,73,74]. For example, in a study conducted by Yokose et al., SMAD4 alterations alone could not predict OS or DFS, but the combination of KRAS and SMAD4 mutations was an independent poor prognostic factor in PDAC [75]. In the same way, Park et al. did not find any statistical difference in terms of OS in patients with SMAD4 loss, but poorly differentiated tumors that were SMAD4-negative were better predictors of survival [51].
The implication of SMAD4 in therapy resistance is equally debated. It has been suggested that patients with SMAD4-negative tumors present drug resistance and that they do not benefit form adjuvant chemotherapy [62,76]. Indeed, a recent meta-analysis focusing on SMAD4-related drug resistance suggests that the loss of SMAD4 is associated with worse OS and DFS in patients receiving adjuvant chemotherapy [77]. In a series of patients with PDAC receiving downstaging therapy, patients with intact SMAD4 tumors had better treatment response and tumor regression scores [78]. Similarly, patients with SMAD4-deficient locally advanced PDAC receiving radiofrequency ablation presented worse DFS after treatment. [79]. Furthermore, in in vitro and in vivo assays, SMAD4-negative PDAC cells were associated with increased radioresistance [80].
On the contrary, in certain studies, patients with SMAD4-negative tumors seemed to benefit from adjuvant chemotherapy and present longer OS or DFS [65,66,67]. Biankin et al. even indicated that SMAD4 loss co-segregated with resectability and that patients had better survival after surgery [10].
Most notably, SMAD4-negative status appeared to be correlated with the tendency to metastasize rather than to recur locally [56,74,76,81,82,83,84]. Yachida et al. identified the genetic subtype of PDAC characterized by a TP53 missense mutation and biallelic loss of SMAD4 which was linked with higher metastatic efficiency [85]. In an experimental setting, using genetically engineered mouse models (GEMMs), Bardeesy et al. evidenced an association between SMAD4 mutations and an increased number of liver metastases [86]. Similarly, functional SMAD4, by repressing specific genes such as FOSL1, suppresses metastatic colonization. If SMAD4 is lost, FOSL1 can promote migration and metastatic colonization [87]. Therefore, SMAD4 detection in PDAC could help in orienting patient treatment.
However, the propensity of SMAD4-negative tumors to metastasize widely is also controversial. In certain clinical studies, SMAD4 loss was not associated with a specific recurrence pattern. Indeed, SMAD4 deficiency did not correlate with locoregional nor distant recurrence [66,68,70].
In conclusion, further investigations are needed to understand the prognostic value of SMAD4 alterations in PDAC.
7. An Introduction to Epithelial–Mesenchymal Transition
In a physiological context, EMT is indispensable for development and wound healing. However, EMT has been increasingly associated with human disease, particularly in the cellular mechanisms of invasion, migration, metastasis and microenvironment remodeling in cancer [25].
EMT is the process through which cells transition from an epithelial state to a mesenchymal one. [25,26]. The epithelial state is defined by epithelial characteristics such as apical-to-basal polarization and tight cell-to-cell junctions in addition to the expression of epithelial markers, such as E-cadherin or cytokeratins [25,26]. In the mesenchymal state, cells acquire a fibroblast-like appearance and upregulate the expression of mesenchymal cell markers, including N-cadherin and vimentin [25,26].
One of the most important signaling pathways regulating EMT is the TGF-beta pathway [25,26,33,88,89,90]. TGF-beta signaling is known to induce EMT in a Smad-dependent and -independent manner through the expression of specific EMT transcription factors (EMT-TFs) such as Snail, Slug, Twist and ZEB1 [23,25,33,90].
For years, EMT has been described as a binary process, where cells transition from one state to the other completely and irreversibly. Recently, it was suggested that cells can pass through a variety of intermediary states, and express a mixture of epithelial and mesenchymal features, in a reversible manner [26,90]. Each intermediate state is defined by specific markers and behavior [26,90,91,92]. This type of EMT, previously called “hybrid” or “partial” EMT, is now preferably referred to as “epithelial-mesenchymal plasticity” (EMP) as it better describes the ability of cells to adopt mixed features and shift readily through the epithelial–mesenchymal spectrum [26]. Studies have indicated that cells transitioning through these intermediate states are characterized by higher tumor-initiation properties and metastatic potential than those on either side of the EMT spectrum, due to the co-expression of mesenchymal and epithelial markers [92,93,94].
8. Epithelial–Mesenchymal Transition in PDAC: What Is Known
Numerous studies have focused on the role of EMT in PDAC in order to understand the underlying mechanism of metastasis and drug resistance, mostly in in vitro and in vivo settings.
The detection of epithelial and mesenchymal markers using either IHC, immunofluorescence (IF) or RNA sequencing (RNA-Seq) has suggested the existence of EMT in PDAC [95,96,97,98,99,100,101,102]. The EMT signature appeared to be associated with a poor prognosis, having an impact on OS and DFS [96,97,98,100,101,102,103].
It has also been suggested that EMT may be an extremely early event in PDAC, playing an essential role in distant dissemination and patient prognosis. Indeed, in an experimental setting, the presence of EMT was detected in high-grade preneoplastic lesions and in circulating tumor cells (CTCs) before identifiable tumor formation [104,105].
However, the necessity of EMT in metastasis initiation is still a matter of debate. Using GEMM, studies have suggested that metastasis can occur in PDAC independently of EMT [106,107,108]. On the other hand, Krebs et al. indicated that the EMT-TF ZEB1 was indispensable for PDAC to metastasize; its knockout impacted PDAC tumorigenicity on multiple levels, notably that of cancer cell plasticity [109]. Additionally, the upregulation of EMT-TFs increased metastasis in vivo, suggesting a potential role of EMT in PDAC metastasis [110].
Another property of EMT is that this process could confer chemoresistance. In vitro studies demonstrated that chemoresistance of PDAC cells lines was associated with the expression of EMT markers such as ZEB1 [111,112]. Similarly, the loss of EMT-TFs Snail and Twist correlated with chemosensibility in vitro and in vivo [106].
To conclude, EMT is a complex cellular process that might influence the aggressiveness of PDAC. However, the exact underlying mechanisms are still not fully understood. We will, therefore, review the potential role of SMAD4 alterations in order to improve our understanding of their impact on EMT in PDAC.
9. SMAD4 Alterations and EMT in PDAC
Although SMAD4-dependent TGF-beta activation is associated with later stages of PDAC carcinogenesis and EMT, paradoxically, it is SMAD4 loss that appears to be linked to worse prognosis and metastasis. Therefore, it is not fully understood how TGF-beta-induced EMT operates in the absence of SMAD4.
Most studies suggest that EMT requires an intact TGF-beta signaling pathway, including SMAD4 [86,107,113,114,115,116,117,118]. For example, SMAD4-intact cell lines and tumors in GEMMs were capable of inducing EMT and acquiring a mesenchymal phenotype. On the other hand, SMAD4-deficient cell lines and GEMM tumors maintained a more epithelial identity when exposed to TGF-beta [86,117]. Similarly, EMT was identified in a subset of GEMMs with typical PDAC gene mutations (KRAS mutations + TP53 or CDKN2A alterations) but intact TGF-beta signaling [119,120]. Moreover, using RNA-Seq analysis on PDAC specimens, Chan-Seng-Yue et al. demonstrated that “Classical-like A/B” signatures, known to be associated with better prognosis and less EMT, correlated with SMAD4 inactivation. However, EMT markers were linked to PDACs presenting aggressive gene signatures, called “Basal-like A/B” [101].
Interestingly, in the early stages of carcinogenesis, it has been suggested that TGF-beta may have a tumor-suppressor effect by inducing a lethal EMT [113,118]. In SMAD4-intact cells, TGF-beta induced EMT and apoptosis via the EMT-TF Snail as well as the repression of KLF5 expression. In this situation, SOX4, known to be an important factor in tissue differentiation and present in progenitor pancreatic cells, will initiate the transcription of proapoptotic factors. In the absence of SMAD4, KFL5 is not repressed, and—conjointly with SOX4—will transcribe a protumorigenic gene program. This biological process might explain the recurring “TGF-beta switch” observed in PDAC and mirrors the empirical observations made in clinical studies.
In contrast, some studies indicate that tumors can exhibit EMT when SMAD4 is inactivated [52,56,121]. Indeed, in a retrospective clinical study that classified PDAC specimens in epithelial, hybrid and mesenchymal phenotypes using an IHC score, SMAD4 loss was found to be correlated with the mesenchymal phenotype, worse outcome and distant recurrence [56]. Moreover, Wartenberg et al. observed an association between SMAD4 inactivation and the “immune-escape” subtype, which was characterized by worse outcome and a high rate of EMT tumor budding [121]. Additionally, the SMAD4 Y353C missense mutation was linked with increased migration and EMT markers in vitro [52].
On a different note, Levy et al. suggested, using SMAD4 siRNA cell lines, that the loss of SMAD4 in cancer might contribute to cell cycle arrest and migration, but not EMT [122]. Indeed, it is well-known that some of the non-canonical TGF-beta pathways are capable of inducing EMT. Downstream kinases such as Ras, Src, PI3K, MAPK, Par6 and NF-kB have elicited Smad-independent EMT and have been extensively reviewed [123,124]. Therefore, it is possible that a certain redundancy in the TGF-beta pathway exists. If SMAD4 is inactivated, other pathways may be stimulated and promote TGF-beta-associated EMT (Table 1).

Table 1.
Review of the literature: role of the SMAD4 protein in EMT.
In conclusion, the biological function of SMAD4 alterations in PDAC EMT has not been completely elucidated.
10. Controversies Regarding SMAD4 Alterations and its Potential Role in PDAC EMP
As mentioned above, the prognostic value of SMAD4 alterations is still a subject of controversy and the exact biological function of SMAD4 in PDAC, specifically in the TGF-beta-induced EMT, is not yet fully understood.
Firstly, we think that the controversy around the clinicopathological and prognostic role of SMAD4 loss could be related to a number of different factors: (1) There is a lack of standardization in the histopathological study of SMAD4 IHC. Indeed, there is a high heterogeneity in the use of antibodies and the evaluation of SMAD4 IHC staining. This could explain the large difference in the proportion of SMAD4- negative tumors across studies. (2) Although SMAD4 IHC has been described as a reliable tool, discrepancies exist between protein expression and gene which do not always lead to a complete absence of staining [52,69,125]. Additionally, some studies have indicated that the nature of the genetic alterations has a significant impact on survival [72,125]. SMAD4 protein expression and genetic alterations may have to be analyzed simultaneously to determine specific PDAC phenotypes. (3) The discrepancies could also be linked to the patient selection regarding disease stage and/or treatment. The initial hypothesis that SMAD4 loss is related to an enhanced metastatic potential was evoked in an autopsy series by Iacobuzio-Donahue et al. in 2009, which mostly included patients with advanced disease [81]. Therefore, it is unclear if this hypothesis could be applied to patients who have undergone resection and received neoadjuvant and/or adjuvant therapy. The prognostic impact and pattern of recurrence of SMAD4 loss was also studied in patients with PDAC resection, but there was a great variety of adjuvant therapy protocols [10,53,58,60,61,65,66,74,76,82,83,125].
Secondly, diverging results were observed regarding the role of SMAD4 in PDAC EMT. One possible explanation is related to the lack of standardization and criteria to evaluate EMT, and more particularly EMP [26]. Indeed, the use of the term “EMT”, mainly in the study of cancer, has created a great number of discrepancies in data interpretation and disagreements as to whether the studied process is EMT or not [26]. Based on the Consensus Statement mediated by The EMT International Association (TEMTIA), there exist multiple issues concerning the characterization of EMT: (1) EMT cannot solely be defined by the expression of one or few biomarkers. EMT is characterized by the downregulation of epithelial markers to acquire a mesenchymal phenotype. However, diverse markers have been used to define EMT, which has been a major source of confusion. For example, in some studies in PDAC, EMT was defined only by the complete or partial loss of E-Cadherin and the upregulation of vimentin [56,100,117,126]. (2) There are no guidelines as how to define EMT based on the cellular and molecular markers. There exists a very important heterogeneity in the combination of the EMT-TFs and/or mesenchymal markers used to determine EMT and/or EMP states [95,96,97,99,101,102]. In addition, the up-regulation of EMT-TFs and other biomarkers may be context- and tissue-specific [110]. For example, the dysregulation of Wnt/beta-catenin plays an important role in EMT in colorectal carcinoma, but its role in pancreatic cancer is less clear and somewhat controversial [127]. Furthermore, the regulation of EMT also operates via the post-transcriptional regulation of EMT regulators at both the mRNA and protein levels [26,128,129]. (3) EMT is a complex process characterized by the modulation of the expression of diverse biomarkers, but also changes in cellular properties and characteristics. This is particularly true for studies that use RNA expression to exclusively survey EMT molecular markers [102]. Indeed, EMT is also defined as the loss of apical-to-basal polarity, tight cell junctions, the acquisition of spindle-cell-like characteristics and increased motility. These changes must also be analyzed and taken into account when defining EMT [25,26]. (4) It is equally important to note that EMT remains a dynamic process. A certain EMT phenotype could be a definitive state of the tumor or a transient state in time.
Finally, we hypothesize that the disagreements concerning the biological function of SMAD4 in PDAC might be due to its implication in the process of EMP. Indeed, the majority of research on SMAD4 alterations and EMT is focused on the binary model of EMT, rather than considering the context of EMP. However, the existence of EMP in pancreatic cancer has been evoked in various studies [56,108,130,131,132,133,134]. Aiello et al. demonstrated that PDAC cells can disseminate according to two different EMT programs: a partial and a complete EMT program [130]. The complete EMT program is characterized by cells that are derived from poorly differentiated tumors, repress epithelial markers, enhance mesenchymal markers and migrate in a single-cell manner. On the other hand, the partial EMT program is characterized by cells that derived from well-differentiated PDACs, co-express epithelial as well and mesenchymal markers and migrate in clusters of CTCs. It has been suggested that cells migrating in clusters, although less motile, have enhanced seeding capacity [135,136]. This would suggest that tumor cells in partial EMT states present a higher metastatic potential than cells that undergo a complete EMT. Indeed, the increased aggressivity of intermediate EMT states has already been evoked and mentioned above [92,93]. Interestingly, in a study conducted by Huang et al. using PDAC organoids, SMAD4 inactivation enabled a collective invasion program upon TGF-beta exposure, while organoids with wild-type SMAD4 invaded with a mesenchymal phenotype [108]. Moreover, in a GEMM of squamous cell carcinoma, later stages of partial EMT, closer to a mesenchymal phenotype, presented upregulation of SMAD2/3 [92]. Taken together, it is tempting to suggest that cells presenting SMAD4 alterations, even if SMAD2/3 are upregulated, acquire an early partial EMT state, closer to an epithelial phenotype, characterized by enhanced metastatic potential.
11. Conclusions
Despite the fact that progress has been made in the research of patient care and the molecular and genetic background of pancreatic cancer, the improvement of OS for PDAC is relatively modest and it remains one of the deadliest cancers of our time. It is, therefore, imperative to improve our understanding of pancreatic cancer carcinogenesis, namely of the molecular mechanisms that promote metastatic spread and chemoresistance, which are the main causes of death in pancreatic cancer.
It was previously demonstrated that the TGF-pathway is affected in all cases of PDACs, mainly by SMAD4 alteration [9]. Knowing that the TGF-beta pathway plays a crucial role in initiating EMT, we decided to explore the implication of SMAD4 in EMT and, more particularly, in the EMP of PDAC. Indeed, empirical observations have suggested that SMAD4 loss in PDAC is correlated with more metastasis.
However, it is important to note that SMAD4 signaling can be triggered by other impulses than TGF-beta. SMAD4 is also the central mediator of the BMP signaling pathway. While the TGF-β pathway is often disrupted in pancreatic cancer, alterations in the BMP pathway have not been frequently reported. The BMP pathway plays a role in tumorigenesis, but its role in pancreatic cancer remains poorly understood [86,137]. Concerning EMT, BMP proteins are capable of inducing EMT in pancreatic cell lines and are dependent on SMAD4 function [138,139]. However, the inactivation of BMP receptors and/or loss of SMAD4 have also been associated with increased proliferation, invasiveness and poor prognosis [140]. Seeing a similar controversy as that observed in the TGF-beta branch, it would therefore be interesting to study the different branches of the TGF-beta family and their potential mutual influence on EMT.
Although the biological function of SMAD4 alterations in EMT are not completely elucidated, SMAD4 inactivation may play an important role by inducing partial EMT states. These partial EMT states harbor both epithelial and mesenchymal traits and markers, and may have enhanced metastatic potential.
A better understanding of the molecular mechanisms underlying PDAC, SMAD4 and EMT could help in the development of effective targeted therapy, which is highly needed in PDAC [141].
Author Contributions
Writing—original draft preparation, M.-L.R.; writing—review and editing, M.-L.R., L.L., L.A., A.A.S., C.V.C., S.D.C., E.M.P., C.M., C.D. and N.D.; supervision, N.D. and I.S.; project administration, C.D. and I.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of grants awarded by the “Fonds Erasme pour la Recherche Médicale” (Brussels, Belgium).
Acknowledgments
This work was performed with the support of grants awarded by the “Fonds Erasme pour la Recherche Médicale” (Brussels, Belgium) and by the “Fonds Yvonne Boël” (Brussels, Belgium). C.D. is a Senior Research Associate with the F.N.R.S. (Belgian National Fund for Scientific Research). C.M.M.I. is supported by the European Regional Development Fund and the Walloon Region (Wallonia-biomed, #411132-957270, project “CMMI- ULB”). Images were created using Biorender.com, accessed on 18 January 2022.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 5’UTR | 5’ untranslated region |
| Beta-CAT | beta-catenin |
| BMP | bone morphogenic protein |
| CDK | cyclin-dependent kinase |
| ChIP | chromatin immunoprecipitation |
| Co-Smad | common-Smad |
| CTC | circulaing tumor cell |
| DFS | disease-free survival |
| DPC4 | deleted in pancreatic cancer, locus 4 |
| DAP-kinase | death-associated protein kinase |
| E-CAD | E-cadherin |
| EMP | epithelial–mesenchymal plasticity |
| EMT | epithelial–mesenchymal transition |
| EMT-TF | epithelial–mesenchymal transition transcription factor |
| GDF | growth and differentiation factor |
| GEMM | genetically engineered mouse model |
| IF | immunofluorescence |
| IHC | immunohistochemistry |
| I-Smads | inhibitory-Smads |
| CK19 | cytokeratin 19 |
| CK7 | cytokeratin 7 |
| CKAE1AE3 | cytokeratin AE1AE3 |
| LOH | loss of heterozygosity |
| Mad | Mothers against decapentaplegic |
| MH1 | Mad homology 1 |
| MH2 | Mad homology 2 |
| NB | Northern blot |
| N-CAD | N-Cadherin |
| NGS | next-generation sequencing |
| NK | natural killer |
| OS | overall survival |
| PDAC | pancreatic ductal adenocarcinoma |
| RNA-Seq | RNA Sequencing |
| R-Smad | receptor-regulated Smad |
| RT-PCR | reverse transcription polymerase chain reaction |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
| scRNA-Seq | single-cell RNA-Sequencing |
| shRNAi | short hairpin RNA interference |
| siRNA | small interfering RNA |
| sma | small worm phenotype |
| SMA | smooth muscle actin |
| shRNA | short hairpin RNA |
| TEMTIA | The Epithelial–Mesenchymal Transition International Association |
| TF | transcription factor |
| TGF-beta | transforming growth factor-beta |
| TGFBR | transforming growth factor-beta receptor |
| TMA | tissue microarray |
| VIM | vimentin |
| WB | Western blot |
| WGS | whole-genome sequencing |
| WTS | whole-transcriptiome sequencing |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primer 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Balaban, E.P.; Mangu, P.B.; Khorana, A.A.; Shah, M.A.; Mukherjee, S.; Crane, C.H.; Javle, M.M.; Eads, J.R.; Allen, P.; Ko, A.H.; et al. Locally Advanced, Unresectable Pancreatic Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 2654–2668. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.; Schwarz, L.; Borbath, I.; Henry, A.; Van Laethem, J.-L.; Malka, D.; Ducreux, M.; Conroy, T. An update on treatment options for pancreatic adenocarcinoma. Ther. Adv. Med. Oncol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Worni, M.; Guller, U.; White, R.R.; Castleberry, A.W.; Pietrobon, R.; Cerny, T.; Gloor, B.; Koeberle, D. Modest Improvement in Overall Survival for Patients With Metastatic Pancreatic Cancer: A Trend Analysis Using the Surveillance, Epidemiology, and End Results Registry From 1988 to 2008. Pancreas 2013, 42, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- StatBite, U.S. Pancreatic Cancer Rates. JNCI J. Natl. Cancer Inst. 2010, 102, 1822. [Google Scholar]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.-C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.-M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.-C.; Mansour, J.; Mollaee, M.; Wagner, K.-U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Chan-Seng-Yue, M.; Lemire, M.; Li, Y.; Wilson, G.W.; Connor, A.A.; Denroche, R.E.; Liang, S.-B.; Brown, A.M.K.; Kim, J.C.; et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016, 538, 378–382. [Google Scholar] [CrossRef]
- Australian Pancreatic Cancer Genome Initiative; Waddell, N.; Pajic, M.; Patch, A.-M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Connor, A.A.; Denroche, R.E.; Jang, G.H.; Lemire, M.; Zhang, A.; Chan-Seng-Yue, M.; Wilson, G.; Grant, R.C.; Merico, D.; Lungu, I.; et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell 2019, 35, 267.e7–282.e7. [Google Scholar] [CrossRef] [PubMed]
- Raphael, B.J.; Hruban, R.H.; Aguirre, A.J.; Moffitt, R.A.; Yeh, J.J.; Stewart, C.; Getz, G. Cancer Genome Atlas Research Network. Electronic address: Andrew_aguirre@dfci.harvard.edu, Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185.e13–203.e13. [Google Scholar] [CrossRef]
- Dreyer, S.; Chang, D.K.; Bailey, P.; Biankin, A.V. Pancreatic Cancer Genomes: Implications for Clinical Management and Therapeutic Development. Clin. Cancer Res. 2017, 23, 1638–1646. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Hong, J.; Iacobuzio-Donahue, C.A. The pancreatic cancer genome revisited. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, W.; Chianchiano, P.; Griffin, J.F.; Pittman, M.E.; Brosens, L.A.; Noë, M.; Yu, J.; Shindo, K.; Suenaga, M.; Rezaee, N.; et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J. Pathol. 2017, 242, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Wilentz, R.E.; Iacobuzio-Donahue, C.A.; Argani, P.; McCarthy, D.M.; Parsons, J.L.; Yeo, C.J.; Kern, S.E.; Hruban, R.H. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: Evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000, 60, 2002–2006. [Google Scholar] [PubMed]
- Ahmed, S.; Bradshaw, A.-D.; Gera, S.; Dewan, M.Z.; Xu, R. The TGF-β/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. J. Clin. Med. 2017, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.A.; Schutte, M.; Hoque, A.T.M.S.; Moskaluk, C.A.; da Costa, L.T.; Rozenblum, E.; Weinstein, C.L.; Fischer, A.; Yeo, C.J.; Hruban, R.H.; et al. DPC4, A Candidate Tumor Suppressor Gene at Human Chromosome 18q21.1. Science 1996, 271, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Cohen, C.; Little, J.V.; Sequeira, J.H.; Mosunjac, M.B.; Siddiqui, M.T. The utility of SMAD4 as a diagnostic immunohistochemical marker for pancreatic adenocarcinoma, and its expression in other solid tumors. Diagn. Cytopathol. 2007, 35, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Mishra, L.; Deng, C.X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef]
- Miyaki, M.; Kuroki, T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem. Biophys. Res. Commun. 2003, 306, 799–804. [Google Scholar] [CrossRef]
- Wendt, M.K.; Tian, M.; Schiemann, W.P. Deconstructing the mechanisms and consequences of TGF-β-induced EMT during cancer progression. Cell Tissue Res. 2011, 347, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352, Correction in Nat. Rev. Mol. Cell Biol. 2021, 22, 834. [Google Scholar] [CrossRef] [PubMed]
- Wilentz, R.E.; Su, G.; Le Dai, J.; Sparks, A.B.; Argani, P.; Sohn, T.A.; Yeo, C.J.; Kern, S.E.; Hruban, R.H. Immunohistochemical Labeling for Dpc4 Mirrors Genetic Status in Pancreatic Adenocarcinomas: A New Marker of DPC4 Inactivation. Am. J. Pathol. 2000, 156, 37–43. [Google Scholar] [CrossRef]
- Jj, S.; Sj, N.; La, R.; Eh, C.; Wm, G. Genetic Characterization and Cloning of Mothers against Dpp, a Gene Required for Decapentaplegic Function in Drosophila melanogaster. Genetics 1995, 139, 1347–1358. Available online: https://pubmed.ncbi.nlm.nih.gov/7768443/ (accessed on 4 January 2022).
- Xia, X.; Wu, W.; Huang, C.; Cen, G.; Jiang, T.; Cao, J.; Huang, K.; Qiu, Z. SMAD4 and its role in pancreatic cancer. Tumor Biol. 2014, 36, 111–119. [Google Scholar] [CrossRef]
- Schmierer, B.; Hill, C.S. TGFbeta-SMAD signal transduction: Molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007, 8, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Gough, N.R.; Xiang, X.; Mishra, L. TGF-β Signaling in Liver, Pancreas, and Gastrointestinal Diseases and Cancer. Gastroenterology 2021, 161, 434.e15–452.e15. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.-H. The regulation of TGFβ signal transduction. Development 2009, 136, 3699–3714. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–330. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Glazer, E.; Welsh, E.; Pimiento, J.M.; Teer, J.K.; Malafa, M.P. TGFβ1 overexpression is associated with improved survival and low tumor cell proliferation in patients with early-stage pancreatic ductal adenocarcinoma. Oncotarget 2016, 8, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, X.-Q.; Li, P. The Role of TGF-β and Its Receptors in Gastrointestinal Cancers. Transl. Oncol. 2019, 12, 475–484. [Google Scholar] [CrossRef]
- Javle, M.; Li, Y.; Tan, D.; Dong, X.; Chang, P.; Kar, S.; Li, D. Biomarkers of TGF-β Signaling Pathway and Prognosis of Pancreatic Cancer. PLoS ONE 2014, 9, e85942. [Google Scholar] [CrossRef]
- Park, H.; Bang, J.; Nam, A.; Park, J.E.; Jin, M.H.; Bang, Y.; Oh, D. The prognostic role of soluble TGF-beta and its dynamics in unresectable pancreatic cancer treated with chemotherapy. Cancer Med. 2019, 9, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Akhurst, R.J.; Balmain, A. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 2001, 29, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.-W.; Chen, C.-H.; Chen, C.-C.; Chen, J.; Su, Y.-H.; Chen, R.-H. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 2002, 4, 51–58. [Google Scholar] [CrossRef]
- Dai, J.L.; Bansal, R.K.; Kern, S.E. G1 cell cycle arrest and apoptosis induction by nuclear Smad4/Dpc4: Phenotypes reversed by a tumorigenic mutation. Proc. Natl. Acad. Sci. USA 1999, 96, 1427–1432. [Google Scholar] [CrossRef]
- Atfi, A.; Buisine, M.; Mazars, A.; Gespach, C. Induction of apoptosis by DPC4, a transcriptional factor regulated by transforming growth factor-beta through stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) signaling pathway. J. Biol. Chem. 1997, 272, 24731–24734. [Google Scholar] [CrossRef]
- Bulle, A.; Lim, K.-H. Beyond just a tight fortress: Contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther. 2020, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schutte, M.; Hruban, R.H.; Hedrick, L.; Cho, K.; Nadasdy, G.M.; Weinstein, C.L.; Bova, G.S.; Isaacs, W.B.; Cairns, P.; Nawroz, H.; et al. DPC4 gene in various tumor types. Cancer Res. 1996, 56, 2527–2530. [Google Scholar] [PubMed]
- Shi, Y.; Hata, A.; Lo, R.S.; Massagué, J.; Pavletich, N.P. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 1997, 388, 87–93. [Google Scholar] [CrossRef]
- Xu, J.; Attisano, L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor beta signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 2000, 97, 4820–4825. [Google Scholar] [CrossRef] [PubMed]
- Demagny, H.; De Robertis, E.M. Point mutations in the tumor suppressor Smad4/DPC4 enhance its phosphorylation by GSK3 and reversibly inactivate TGF-β signaling. Mol. Cell Oncol. 2016, 3, e1025181. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Huang, J.; Jhala, N.C.; Tytler, E.M.; Yang, L.; Vickers, S.M.; Tang, Y.; Lu, C.; Wang, N.; Cao, X. SCF(beta-TrCP1) controls Smad4 protein stability in pancreatic cancer cells. Am. J. Pathol. 2005, 166, 1379–1392. [Google Scholar] [CrossRef]
- Wang, J.-D.; Jin, K.; Chen, X.-Y.; Lv, J.-Q.; Ji, K.-W. Clinicopathological significance of SMAD4 loss in pancreatic ductal adenocarcinomas: A systematic review and meta-analysis. Oncotarget 2016, 8, 16704–16711. [Google Scholar] [CrossRef]
- Matsuda, Y.; Esaka, S.; Suzuki, A.; Hamashima, Y.; Imaizumi, M.; Matsukawa, M.; Fujii, Y.; Aida, J.; Takubo, K.; Ishiwata, T.; et al. Abnormal immunolabelling of SMAD 4 in cell block specimens to distinguish malignant and benign pancreatic cells. Cytopathology 2018, 30, 201–208. [Google Scholar] [CrossRef]
- Park, J.Y.; King, J.; Reber, H.; Hines, O.J.; Mederos, M.A.; Wang, H.L.; Dawson, D.; Wainberg, Z.; Donahue, T.; Girgis, M. Poorly differentiated histologic grade correlates with worse survival in SMAD4 negative pancreatic adenocarcinoma patients. J. Surg. Oncol. 2020, 123, 389–398. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Zhan, S.; Zhang, L.; Zhang, S.; Tang, Q.; Li, M.; Tan, Z.; Liu, S.; Xing, X. SMAD4 Y353C promotes the progression of PDAC. BMC Cancer 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z. Loss of DPC4 expression and its correlation with clinicopathological parameters in pancreatic carcinoma. World J. Gastroenterol. 2003, 9, 2764–2767. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.; Serrano, O.K.; Wolfgang, C.L.; Parmigiani, G.; Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Eshleman, J.R.; et al. SMAD4 Gene Mutations Are Associated with Poor Prognosis in Pancreatic Cancer. Clin. Cancer Res. 2009, 15, 4674–4679. [Google Scholar] [CrossRef]
- Jiang, H.; He, C.; Geng, S.; Sheng, H.; Shen, X.; Zhang, X.; Li, H.; Zhu, S.; Chen, X.; Yang, C.; et al. RhoT1 and Smad4 Are Correlated with Lymph Node Metastasis and Overall Survival in Pancreatic Cancer. PLoS ONE 2012, 7, e42234. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Fujii, T.; Shimoyama, Y.; Kanda, M.; Nakayama, G.; Sugimoto, H.; Koike, M.; Nomoto, S.; Fujiwara, M.; Nakao, A.; et al. SMAD4 Expression Predicts Local Spread and Treatment Failure in Resected Pancreatic Cancer. Pancreas 2015, 44, 660–664. [Google Scholar] [CrossRef]
- Held, T.; Verbeke, C.S.; Strobel, O.; Rutkowski, W.; Villard, C.; Moro, C.F.; Del Chiaro, M.; Büchler, M.; Heuchel, R.; Löhr, M. Immunohistochemical profiling of liver metastases and matched-pair analysis in patients with metastatic pancreatic ductal adenocarcinoma. Pancreatology 2019, 19, 963–970. [Google Scholar] [CrossRef]
- Tascilar, M.; Skinner, H.G.; Rosty, C.; Sohn, T.; Wilentz, R.E.; Offerhaus, G.J.; Adsay, V.; Abrams, R.A.; Cameron, J.L.; Kern, S.E.; et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2001, 7, 4115–4121. [Google Scholar] [PubMed]
- Shin, S.H.; Kim, H.J.; Hwang, D.W.; Lee, J.H.; Song, K.B.; Jun, E.; Shim, I.K.; Hong, S.; Kim, H.J.; Park, K.W.; et al. The DPC4/SMAD4 genetic status determines recurrence patterns and treatment outcomes in resected pancreatic ductal adenocarcinoma: A prospective cohort study. Oncotarget 2017, 8, 17945–17959. [Google Scholar] [CrossRef]
- Kadera, B.E.; Sunjaya, D.B.; Isacoff, W.H.; Li, L.; Hines, O.J.; Tomlinson, J.S.; Dawson, D.W.; Rochefort, M.M.; Donald, G.W.; Clerkin, B.M.; et al. Locally Advanced Pancreatic Cancer: Association Between Prolonged Preoperative Treatment and Lymph-Node Negativity and Overall Survival. JAMA Surg. 2014, 149, 145. [Google Scholar] [CrossRef]
- Oshima, M.; Okano, K.; Muraki, S.; Haba, R.; Maeba, T.; Suzuki, Y.; Yachida, S. Immunohistochemically Detected Expression of 3 Major Genes (CDKN2A/p16, TP53, and SMAD4/DPC4) Strongly Predicts Survival in Patients with Resectable Pancreatic Cancer. Ann. Surg. 2013, 258, 336–346. [Google Scholar] [CrossRef]
- Toga, T.; Nio, Y.; Hashimoto, K.; Higami, T.; Maruyama, R. The dissociated expression of protein and messenger RNA of DPC4 in human invasive ductal carcinoma of the pancreas and their implication for patient outcome. Anticancer Res. 2004, 24, 1173–1178. [Google Scholar] [PubMed]
- Ottenhof, N.A.; Morsink, F.H.M.; Kate, F.T.; Van Noorden, C.J.F.; Offerhaus, G.J.A. Multivariate analysis of immunohistochemical evaluation of protein expression in pancreatic ductal adenocarcinoma reveals prognostic significance for persistent Smad4 expression only. Cell. Oncol. 2012, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, L.; Xu, H.; Wu, C.; Xiang, J.; Xu, J.; Liu, C.; Long, J.; Ni, Q.; Yu, X. Infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br. J. Surg. 2016, 103, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Khorana, A.A.; Hu, Y.C.; Ryan, C.K.; Komorowski, R.A.; Hostetter, G.; Ahrendt, S.A. Vascular Endothelial Growth Factor and DPC4 Predict Adjuvant Therapy Outcomes in Resected Pancreatic Cancer. J. Gastrointest. Surg. 2005, 9, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Bachet, J.B.; Maréchal, R.; Demetter, P.; Bonnetain, F.; Couvelard, A.; Svrcek, M.; Bardier-Dupas, A.; Hammel, P.; Sauvanet, A.; Louvet, C.; et al. Contribution of CXCR4 and SMAD4 in predicting disease progression pattern and benefit from adjuvant chemotherapy in resected pancreatic adenocarcinoma. Ann. Oncol. 2012, 23, 2327–2335. [Google Scholar] [CrossRef] [PubMed]
- Ormanns, S.; Haas, M.; Remold, A.; Kruger, S.; Holdenrieder, S.; Kirchner, T.; Heinemann, V.; Boeck, S. The Impact of SMAD4 Loss on Outcome in Patients with Advanced Pancreatic Cancer Treated with Systemic Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1094. [Google Scholar] [CrossRef]
- Winter, J.M.; Tang, L.H.; Klimstra, D.S.; Liu, W.; Linkov, I.; Brennan, M.F.; D’Angelica, M.I.; DeMatteo, R.P.; Fong, Y.; Jarnagin, W.R.; et al. Failure Patterns in Resected Pancreas Adenocarcinoma: Lack of Predicted Benefit to SMAD4 Expression. Ann. Surg. 2013, 258, 331–335. [Google Scholar] [CrossRef]
- Qian, Z.R.; Rubinson, D.A.; Nowak, J.A.; Morales-Oyarvide, V.; Dunne, R.F.; Kozak, M.M.; Welch, M.W.; Brais, L.K.; Da Silva, A.; Li, T.; et al. Association of Alterations in Main Driver Genes With Outcomes of Patients With Resected Pancreatic Ductal Adenocarcinoma. JAMA Oncol. 2018, 4, e173420. [Google Scholar] [CrossRef]
- Pokataev, I.; Kudaibergenova, A.; Artemyeva, A.; Popova, A.; Rumyantsev, A.; Podluzhny, D.; Kudashkin, N.; Fedyanin, M.; Tryakin, A.; Tjulandin, S. Intratumoral Heterogeneity of SMAD4 Immunohistochemical Expression and Its Role in Prediction of Recurrence Pattern in Patients with Resectable Pancreatic Cancer. J. Gastrointest. Cancer 2019, 50, 478–484. [Google Scholar] [CrossRef]
- Lu, J.; Yu, R.; Liu, R.; Liang, X.; Sun, J.; Zhang, H.; Wu, H.; Zhang, Z.; Shao, Y.W.; Guo, J.; et al. Genetic aberrations in Chinese pancreatic cancer patients and their association with anatomic location and disease outcomes. Cancer Med. 2020, 10, 933–943. [Google Scholar] [CrossRef]
- Hsieh, Y.-Y.; Liu, T.-P.; Chou, C.-J.; Chen, H.-Y.; Lee, K.-H.; Yang, P.-M. Integration of Bioinformatics Resources Reveals the Therapeutic Benefits of Gemcitabine and Cell Cycle Intervention in SMAD4-Deleted Pancreatic Ductal Adenocarcinoma. Genes 2019, 10, 766. [Google Scholar] [CrossRef]
- Pen, S.-L.; Shan, Y.-S.; Hsiao, C.-F.; Liu, T.-W.; Chen, J.-S.; Ho, C.-L.; Chou, W.-C.; Hsieh, R.-K.; Chen, L.-T.; Ch’Ang, H.-J. High expression of krüppel-like factor 10 or Smad4 predicts clinical benefit of adjuvant chemoradiotherapy in curatively resected pancreatic adenocarcinoma: From a randomized phase III trial. Radiother. Oncol. 2021, 158, 146–154. [Google Scholar] [CrossRef]
- Cuneo, K.C.; Morgan, M.A.; Griffith, K.A.; Hawkins, P.G.; Greenson, J.K.; Ben-Josef, E.; Lawrence, T.S.; Zalupski, M.M. Prognostic Value of c-MET Expression in Patients With Pancreatic Cancer Receiving Adjuvant and Neoadjuvant Chemoradiation Therapy. Int. J. Radiat. Oncol. 2017, 100, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Yokose, T.; Kitago, M.; Matsuda, S.; Sasaki, Y.; Masugi, Y.; Nakamura, Y.; Shinoda, M.; Yagi, H.; Abe, Y.; Oshima, G.; et al. Combination of KRAS and SMAD4 mutations in formalin-fixed paraffin-embedded tissues as a biomarker for pancreatic cancer. Cancer Sci. 2020, 111, 2174–2182. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.M.; Jabbour, S.K.; Lin, S.H.; Deek, M.P.; Hsu, C.C.; Fishman, E.K.; Kim, S.; Cameron, J.L.; Chekmareva, M.; Laheru, D.A.; et al. Smad4 Loss Correlates With Higher Rates of Local and Distant Failure in Pancreatic Adenocarcinoma Patients Receiving Adjuvant Chemoradiation. Pancreas 2018, 47, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Lee, S.H.; Qiu, F.; Zhou, L.; Wang, X.; Ye, T.; Hu, X. Association of SMAD4 loss with drug resistance in clinical cancer patients: A systematic meta-analysis. PLoS ONE 2021, 16, e0250634. [Google Scholar] [CrossRef]
- Kassardjian, A.; Wang, H.L. SMAD4-Expressing Pancreatic Ductal Adenocarcinomas Have Better Response to Neoadjuvant Therapy and Significantly Lower Lymph Node Metastasis Rates. Pancreas 2020, 49, 1153–1160. [Google Scholar] [CrossRef]
- Paiella, S.; Malleo, G.; Cataldo, I.; Gasparini, C.; De Pastena, M.; De Marchi, G.; Marchegiani, G.; Rusev, B.; Scarpa, A.; Girelli, R.; et al. Radiofrequency ablation for locally advanced pancreatic cancer: SMAD4 analysis segregates a responsive subgroup of patients. Langenbeck’s Arch. Surg. 2017, 403, 213–220. [Google Scholar] [CrossRef]
- Wang, F.; Xia, X.; Yang, C.; Shen, J.; Mai, J.; Kim, H.-C.; Kirui, D.; Kang, Y.; Fleming, J.B.; Koay, E.J.; et al. SMAD4 Gene Mutation Renders Pancreatic Cancer Resistance to Radiotherapy through Promotion of Autophagy. Clin. Cancer Res. 2018, 24, 3176–3185. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; et al. DPC4 Gene Status of the Primary Carcinoma Correlates With Patterns of Failure in Patients With Pancreatic Cancer. J. Clin. Oncol. 2009, 27, 1806–1813. [Google Scholar] [CrossRef]
- Boone, B.A.; Sabbaghian, S.; Zenati, M.; Marsh, J.W.; Moser, A.J.; Zureikat, A.H.; Singhi, A.D.; Zeh, H.J., III; Krasinskas, A.M. Loss of SMAD4 staining in pre-operative cell blocks is associated with distant metastases following pancreaticoduodenectomy with venous resection for pancreatic cancer: Pre-Op SMAD4 in PDA With Venous Invasion. J. Surg. Oncol. 2014, 110, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Crane, C.H.; Varadhachary, G.R.; Yordy, J.S.; Staerkel, G.A.; Javle, M.M.; Safran, H.; Haque, W.; Hobbs, B.D.; Krishnan, S.; Fleming, J.B.; et al. Phase II Trial of Cetuximab, Gemcitabine, and Oxaliplatin Followed by Chemoradiation With Cetuximab for Locally Advanced (T4) Pancreatic Adenocarcinoma: Correlation of Smad4(Dpc4) Immunostaining With Pattern of Disease Progression. J. Clin. Oncol. 2011, 29, 3037–3043. [Google Scholar] [CrossRef] [PubMed]
- Gits, H.C.; Tang, A.H.; Harmsen, W.S.; Bamlet, W.R.; Graham, R.P.; Petersen, G.M.; Smyrk, T.C.; Mahipal, A.; Kowalchuk, R.O.; Ashman, J.B.; et al. Intact SMAD-4 is a predictor of increased locoregional recurrence in upfront resected pancreas cancer receiving adjuvant therapy. J. Gastrointest. Oncol. 2021, 12, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Iacobuzio-Donahue, C. Evolution and dynamics of pancreatic cancer progression. Oncogene 2013, 32, 5253–5260. [Google Scholar] [CrossRef] [PubMed]
- Bardeesy, N.; Cheng, K.-h.; Berger, J.H.; Chu, G.C.; Pahler, J.; Olson, P.; Hezel, A.F.; Horner, J.; Lauwers, G.Y.; Hanahan, D.; et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006, 20, 3130–3146. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Rennhack, J.P.; Arnoff, T.E.; Thaker, M.; Younger, S.T.; Doench, J.G.; Huang, A.Y.; Yang, A.; Aguirre, A.J.; Wang, B.; et al. SMAD4 represses FOSL1 expression and pancreatic cancer metastatic colonization. Cell Rep. 2021, 36, 109443. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Potts, J.D.; Runyan, R. Epithelial-mesenchymal cell transformation in the embryonic heart can be mediated, in part, by transforming growth factor β. Dev. Biol. 1989, 134, 392–401. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2018, 194, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2018, 24, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Bronsert, P.; Enderle-Ammour, K.; Bader, M.; Timme, S.; Kuehs, M.; Csanadi, A.; Kayser, G.; Kohler, I.; Bausch, D.; Hoeppner, J.; et al. Cancer cell invasion and EMT marker expression: A three-dimensional study of the human cancer-host interface: 3D cancer-host interface. J. Pathol. 2014, 234, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Kurahara, H.; Takao, S.; Maemura, K.; Mataki, Y.; Kuwahata, T.; Maeda, K.; Ding, Q.; Sakoda, M.; Iino, S.; Ishigami, S.; et al. Epithelial-mesenchymal transition and mesenchymal-epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer: EMT and MET in Pancreatic Cancer. J. Surg. Oncol. 2012, 105, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Galván, J.A.; Zlobec, I.; Wartenberg, M.; Lugli, A.; Gloor, B.; Perren, A.; Karamitopoulou, E. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer 2015, 112, 1944–1950. [Google Scholar] [CrossRef]
- Bronsert, P.; Kohler, I.; Timme, S.; Kiefer, S.; Werner, M.; Schilling, O.; Vashist, Y.; Makowiec, F.; Brabletz, T.; Hopt, U.T.; et al. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery 2014, 156, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Kohler, I.; Bronsert, P.; Timme, S.; Werner, M.; Brabletz, T.; Hopt, U.T.; Schilling, O.; Bausch, D.; Keck, T.; Wellner, U.F. Detailed analysis of epithelial-mesenchymal transition and tumor budding identifies predictors of long-term survival in pancreatic ductal adenocarcinoma. J. Gastroenterol. Hepatol. 2015, 30, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Masugi, Y.; Yamazaki, K.; Hibi, T.; Aiura, K.; Kitagawa, Y.; Sakamoto, M. Solitary cell infiltration is a novel indicator of poor prognosis and epithelial-mesenchymal transition in pancreatic cancer. Hum. Pathol. 2010, 41, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Chan-Seng-Yue, M.; Kim, J.C.; Wilson, G.W.; Ng, K.; Figueroa, E.F.; O’Kane, G.M.; Connor, A.A.; Denroche, R.E.; Grant, R.C.; McLeod, J.; et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020, 52, 231–240. [Google Scholar] [CrossRef]
- Lin, W.; Noel, P.; Borazanci, E.H.; Lee, J.; Amini, A.; Han, I.W.; Heo, J.S.; Jameson, G.S.; Fraser, C.; Steinbach, M.; et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-M.; Li, A.; Olino, K.; Wolfgang, C.L.; Herman, J.M.; Schulick, R.D.; Iacobuzio-Donahue, C.; Hruban, R.H.; Goggins, M. Loss of E-cadherin expression and outcome among patients with resectable pancreatic adenocarcinomas. Mod. Pathol. 2011, 24, 1237–1247. [Google Scholar] [CrossRef]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef]
- Handler, J.; Cullis, J.; Avanzi, A.; Vucic, E.A.; Bar-Sagi, D. Pre-neoplastic pancreas cells enter a partially mesenchymal state following transient TGF-β exposure. Oncogene 2018, 37, 4334–4342. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.C.; Izeradjene, K.; Rani, P.G.; Feng, L.; Carlson, M.A.; DelGiorno, K.E.; Wood, L.D.; Goggins, M.; Hruban, R.H.; Chang, A.E.; et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015, 161, 1345–1360. [Google Scholar] [CrossRef]
- Huang, W.; Navarro-Serer, B.; Jeong, Y.J.; Chianchiano, P.; Xia, L.; Luchini, C.; Veronese, N.; Dowiak, C.; Ng, T.; Trujillo, M.A.; et al. Pattern of Invasion in Human Pancreatic Cancer Organoids Is Associated with Loss of SMAD4 and Clinical Outcome. Cancer Res. 2020, 80, 2804–2817. [Google Scholar] [CrossRef]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Hotz, B.; Arndt, M.; Dullat, S.; Bhargava, S.; Buhr, H.-J.; Hotz, H.G. Epithelial to Mesenchymal Transition: Expression of the Regulators Snail, Slug, and Twist in Pancreatic Cancer. Clin. Cancer Res. 2007, 13, 4769–4776. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Summy, J.M.; Zhang, J.; Park, S.I.; Parikh, N.U.; Gallick, G.E. Development and Characterization of Gemcitabine-Resistant Pancreatic Tumor Cells. Ann. Surg. Oncol. 2007, 14, 3629–3637. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Ramachandran, V.; Fournier, K.F.; Wang, H.; Marquis, L.; Abbruzzese, J.L.; Gallick, G.E.; Logsdon, C.D.; McConkey, D.J.; Choi, W. Epithelial to Mesenchymal Transition Contributes to Drug Resistance in Pancreatic Cancer. Cancer Res. 2009, 69, 5820–5828. [Google Scholar] [CrossRef]
- David, C.J.; Huang, Y.-H.; Chen, M.; Su, J.; Zou, Y.; Bardeesy, N.; Iacobuzio-Donahue, C.A.; Massagué, J. TGF-β Tumor Suppression through a Lethal EMT. Cell 2016, 164, 1015–1030. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Venkatasubbarao, K.; Lazor, J.W.; Sperry, J.; Jin, C.; Cao, L.; Freeman, J.W. Inhibition of STAT3 Tyr705 phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Res. 2008, 68, 4221–4228. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Hsiao, P.-J.; Weng, C.-C.; Kuo, K.-K.; Kuo, T.-L.; Wu, D.-C.; Hung, W.-C.; Cheng, K.-H. SMAD4 Loss triggers the phenotypic changes of pancreatic ductal adenocarcinoma cells. BMC Cancer 2014, 14, 181. [Google Scholar] [CrossRef]
- Kang, Y.; Ling, J.; Suzuki, R.; Roife, D.; Chopin-Laly, X.; Truty, M.J.; Chatterjee, D.; Wang, H.; Thomas, R.M.; Katz, M.H.; et al. SMAD4 Regulates Cell Motility through Transcription of N-Cadherin in Human Pancreatic Ductal Epithelium. PLoS ONE 2014, 9, e107948. [Google Scholar] [CrossRef]
- Shichi, Y.; Sasaki, N.; Michishita, M.; Hasegawa, F.; Matsuda, Y.; Arai, T.; Gomi, F.; Aida, J.; Takubo, K.; Toyoda, M.; et al. Enhanced morphological and functional differences of pancreatic cancer with epithelial or mesenchymal characteristics in 3D culture. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Faheem, M.M.; Rasool, R.; Ahmad, S.M.; Jamwal, V.L.; Chakraborty, S.; Katoch, A.; Gandhi, S.G.; Bhagat, M.; Goswami, A. Par-4 mediated Smad4 induction in PDAC cells restores canonical TGF-β/Smad4 axis driving the cells towards lethal EMT. Eur. J. Cell Biol. 2020, 99, 151076. [Google Scholar]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Bardeesy, N.; Sinha, M.; Lopez, L.; Tuveson, D.A.; Horner, J.; Redston, M.S.; DePinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003, 17, 3112–3126. [Google Scholar] [CrossRef] [PubMed]
- Wartenberg, M.; Cibin, S.; Zlobec, I.; Vassella, E.; Eppenberger-Castori, S.; Terracciano, L.; Eichmann, M.; Worni, M.; Gloor, B.; Perren, A.; et al. Integrated Genomic and Immunophenotypic Classification of Pancreatic Cancer Reveals Three Distinct Subtypes with Prognostic/Predictive Significance. Clin. Cancer Res. 2018, 24, 4444–4454. [Google Scholar] [CrossRef]
- Levy, L.; Hill, C.S. Smad4 Dependency Defines Two Classes of Transforming Growth Factor β (TGF-β) Target Genes and Distinguishes TGF-β-Induced Epithelial-Mesenchymal Transition from Its Antiproliferative and Migratory Responses. Mol. Cell. Biol. 2005, 25, 8108–8125. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, J.J.; Rajasekaran, A.K. Reassessing Epithelial to Mesenchymal Transition as a Prerequisite for Carcinoma Invasion and Metastasis. Cancer Res. 2006, 66, 8319–8326. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Srinivasan, R.; Wig, J.D. SMAD4 Genetic Alterations Predict a Worse Prognosis in Patients With Pancreatic Ductal Adenocarcinoma. Pancreas 2012, 41, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Masugi, Y.; Yamazaki, K.; Emoto, K.; Effendi, K.; Tsujikawa, H.; Kitago, M.; Itano, O.; Kitagawa, Y.; Sakamoto, M. Upregulation of integrin β4 promotes epithelial–mesenchymal transition and is a novel prognostic marker in pancreatic ductal adenocarcinoma. Lab. Investig. 2015, 95, 308–319. [Google Scholar] [CrossRef]
- White, B.D.; Chien, A.J.; Dawson, D.W. Dysregulation of Wnt/β-Catenin Signaling in Gastrointestinal Cancers. Gastroenterology 2012, 142, 219–232. [Google Scholar] [CrossRef]
- Kang, E.; Seo, J.; Yoon, H.; Cho, S. The Post-Translational Regulation of Epithelial–Mesenchymal Transition-Inducing Transcription Factors in Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 3591. [Google Scholar] [CrossRef]
- Moreno-Bueno, G.; Portillo, F.; Cano, A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008, 27, 6958–6969. [Google Scholar] [CrossRef]
- Aiello, N.M.; Maddipati, R.; Norgard, R.J.; Balli, D.; Li, J.; Yuan, S.; Yamazoe, T.; Black, T.; Sahmoud, A.; Furth, E.E.; et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell 2018, 45, 681.e4–695.e4. [Google Scholar] [CrossRef]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; Karim, S.A.; Morton, J.; Hernández, A.D.R. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Li, W.; Shan, T.; Lin, W.R.; Ma, J.; Cui, X.; Yang, W.; Cao, G.; Li, Y.; et al. Conversion of epithelial-to-mesenchymal transition to mesenchymal-to-epithelial transition is mediated by oxygen concentration in pancreatic cancer cells. Oncol. Lett. 2018, 15, 7144–7152. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Chen, S.; Chen, X.; Lin, W.R.; Li, W.; Ma, J.; Wu, T.; Ji, H.; Li, Y.; Cui, X.; et al. Prometastatic mechanisms of CAF-mediated EMT regulation in pancreatic cancer cells. Int. J. Oncol. 2016, 50, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Zadran, S.; Arumugam, R.; Herschman, H.; Phelps, M.E.; Levine, R.D. Surprisal analysis characterizes the free energy time course of cancer cells undergoing epithelial-to-mesenchymal transition. Proc. Natl. Acad. Sci. USA 2014, 111, 13235–13240. [Google Scholar] [CrossRef]
- Whittle, M.C.; Hingorani, S.R. Disconnect between EMT and metastasis in pancreas cancer. Oncotarget 2015, 6, 30445–30446. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Kleeff, J.; Maruyama, H.; Ishiwata, T.; Sawhney, H.; Friess, H.; Büchler, M.W.; Korc, M. Bone morphogenetic protein 2 exerts diverse effects on cell growth in vitro and is expressed in human pancreatic cancer in vivo. Gastroenterology 1999, 116, 1202–1216. [Google Scholar] [CrossRef]
- Gordon, K.J.; Kirkbride, K.C.; How, T.; Blobe, G.C. Bone morphogenetic proteins induce pancreatic cancer cell invasiveness through a Smad1-dependent mechanism that involves matrix metalloproteinase-2. Carcinogenesis 2008, 30, 238–248. [Google Scholar] [CrossRef]
- Hamada, S.; Satoh, K.; Hirota, M.; Kimura, K.; Kanno, A.; Masamune, A.; Shimosegawa, T. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J. Cell. Physiol. 2007, 213, 768–774. [Google Scholar] [CrossRef]
- Voorneveld, P.W.; Stache, V.; Jacobs, R.J.; Smolders, E.; Sitters, A.I.; Liesker, A.; Korkmaz, K.S.; Lam, S.M.; De Miranda, N.F.C.C.; Morreau, H.; et al. Reduced expression of bone morphogenetic protein receptor IA in pancreatic cancer is associated with a poor prognosis. Br. J. Cancer 2013, 109, 1805–1812. [Google Scholar] [CrossRef][Green Version]
- Dardare, J.; Witz, A.; Merlin, J.-L.; Bochnakian, A.; Toussaint, P.; Gilson, P.; Harlé, A. Epithelial to Mesenchymal Transition in Patients with Pancreatic Ductal Adenocarcinoma: State-of-the-Art and Therapeutic Opportunities. Pharmaceuticals 2021, 14, 740. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).