Simple Summary
The present study selected four genes strongly related to autoimmunity. Their expression was found to be significantly altered in melanoma patients according to a multi-validation procedure carried out on 1948 patients. Such genes may represent suitable molecular targets to further investigate the role autoimmunity may play in melanoma setup and development. Our data suggest that autoimmunity may play a beneficial role in melanoma set up, at least to some extent.
Abstract
(1) Background. Immune response dysregulation plays a key role in melanoma, as suggested by the substantial prognosis improvement observed under immune-modulation therapy. Similarly, the role of autoimmunity is under large investigation in melanoma and other cancers. (2) Methods. Expression of 98 autoimmunity-related genes was investigated in 1948 individuals (1024 melanoma and 924 healthy controls). Data were derived from four independent databases, namely, GEO in the selection phase, and Ist Online, GEPIA2 and GENT2, in three sequential validation-steps. ROC analyses were performed to measure the ability to discriminate melanoma from controls. Principal Component Analysis (PCA) was used to combine expression data; survival analysis was carried out on the GEPIA2 platform. (3) Results. Expression levels of NOD2, BAX, IL-18 and ADRB2 were found to be significantly different in melanoma vs. controls and discriminate melanoma from controls in an extremely effective way, either as single molecules (AUC > 0.93 in all cases) or as a profile, according to the PCA analysis. Patients showing high-expression of NOD2 and of IL-18 also show a significant survival improvement as compared to low-expression patients. (4) Conclusions. Four genes strongly related to autoimmunity show a significant altered expression in melanoma samples, highlighting the role they may play in melanoma.
1. Introduction
Autoimmunity is activated when the immune system recognizes target cells as non-self. Under normal conditions and activity, the immune system reacts towards foreign molecules that are potentially harmful; for example, against molecules associated with viruses, bacteria and parasites. Under such conditions, structures recognized as self undergo immune tolerance [1]. On the other hand, if autologous cells or cellular components are recognized as non-self, the production of autoantibodies occurs, eventually leading toward autoimmune diseases. Autoantibodies may be directed against a single cell type or tissue type, and such autoantibodies cause organ-specific diseases such as Addison’s disease, celiac disease, Graves disease, Hashimoto thyroiditis and type I diabetes mellitus. Alternatively, autoantibodies may be directed against components common to all cells, causing systemic diseases, such as systemic lupus erythematosus or rheumatoid arthritis. The presence of autoantibodies has been demonstrated in patients with psoriasis, even in clinically silent phases [2], in essential hypertension [3] and pulmonary hypertension [4]. The possible connection between cancer and autoimmune disorders has been investigated for several years [5]. Although an autoimmune pathogenesis is not demonstrated in melanoma, a relation between the immune response to melanoma and the presence of autoimmune reactions is reported in different studies [6,7,8,9]. A study carried out on 20,482 cases of metastatic cutaneous melanoma showed that patients with a previous diagnosis of autoimmune disease (e.g., diabetes) have a 30% higher incidence of metastatic cutaneous melanoma in the first year of diagnosis, as compared to individuals without autoimmune disease [10]. Further, it is known that patients with Retinopathy Associated with Melanoma (MAR) have a high prevalence of autoimmune diseases in their families [11]. Furthermore, in melanoma patients undergoing therapy with interferon alpha, the appearance of autoantibodies is associated with a better prognosis [12,13], demonstrating that an autoimmune reaction is not neutral in melanoma progression. Although conflicting data are present in the literature on this aspect [14], such data demonstrate that melanoma onset and/or development as well as response to melanoma therapy has several relation points with autoimmunity. While specific mutations in autoimmunity-related genes are reported in melanoma, [15,16], gene expression levels have not been deeply investigated, under such respect. We therefore investigated the expression of autoimmunity related genes in melanoma patients as compared to healthy controls. To this aim the expression of 98 genes was investigated in 1948 individuals (1024 melanoma patients and 924 healthy controls), derived from four independent datasets, namely GDS1375 dataset from GEO database, IST Online, GEPIA2 and GENT2.
2. Results
2.1. The General Scheme of the Analysis Performed in The Present Study Is Reported
Figure 1 synthetizes the followed procedure.
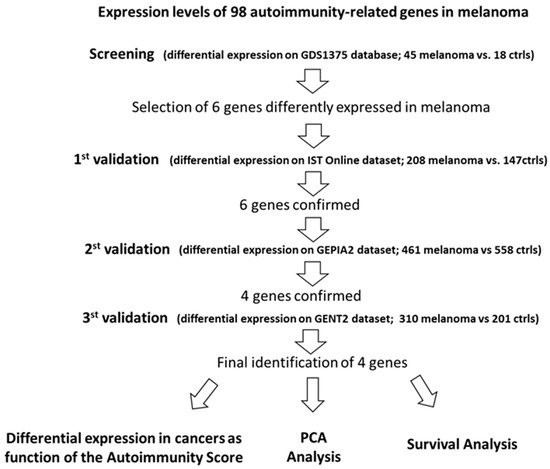
Figure 1.
Summary for the selection of the best autoimmunity-related candidate-genes in melanoma.
2.2. Knowledge-Based Autoimmunity Score (kb-AS) in Different Tumors
Melanoma is reported as being strongly related to autoimmune responses. In PubMed-indexed manuscripts we measured the occurrence of autoimmunity keywords in manuscripts related to 28 different cancer types, according to the methodology reported in Materials and Methods. Such occurrence is here reported as a “Knowledge-based Autoimmunity Score” (kb-AS). Table 1 reports the 28 cancer types sorted from the lowest to the highest kb-AS. This analysis indicates cancer types known to be mostly related to autoimmunity and confirmed that melanoma is in the top five group, along with Thymoma, Lymphoma, Thyroid cancer and Cholangiocarcinoma.

Table 1.
The “Knowledge-based Autoimmunity Score” (kb-AS) was calculated from the PubMed occurrence of autoimmunity-related words within cancer manuscripts. The kb-AS (column C) is expressed as a percentage of the co-occurrence (column B) on the total number of cancer-related manuscripts (column A). Cancer types were those present as cancer-datasets in the GEPIA2 database. The full names of each cancer type are reported in Material and Methods section. The occurrence is reported as number of manuscripts in PubMed, according to the search carried out on 15th November 2021 in the ALL-FIELDS field.
2.3. Expression of Autoimmunity-Related Genes in GEO Database, in Melanoma vs. Controls
Ninety-eight autoimmunity related-genes were selected from the literature [17,18,19] and their expression levels in melanoma vs. control samples were analyzed in the GDS1375 dataset in the GEO database (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS1375) accession date 8 April 2021. Table 2 reports the mean values in melanoma and in control samples. The fold change in melanoma vs. controls is also reported when it is <0.5 or >1.5. When this requirement is matched, the AUC (Area Under the Curve) is reported if >0.8, according to the ROC (Receiver Operating Characteristic) analysis; p value is also indicated. AUC indicates the ability to discriminate melanoma from nevi samples; its value ranges from 0.5 to 1, with 1 indicating 100% ability to discriminate controls from disease cases.

Table 2.
Mean expression values of the 98 autoimmunity-related genes in melanoma and control samples, according to the GDS1375 dataset. The fold change in melanoma vs. controls is reported when it is <0.5 or >1.5. In such cases, the AUC (Area Under the Curve) is indicated, if it is >0.8, according to the ROC (Receiver Operating Characteristic) analysis; also p values are reported in such cases (n.s = not significant).
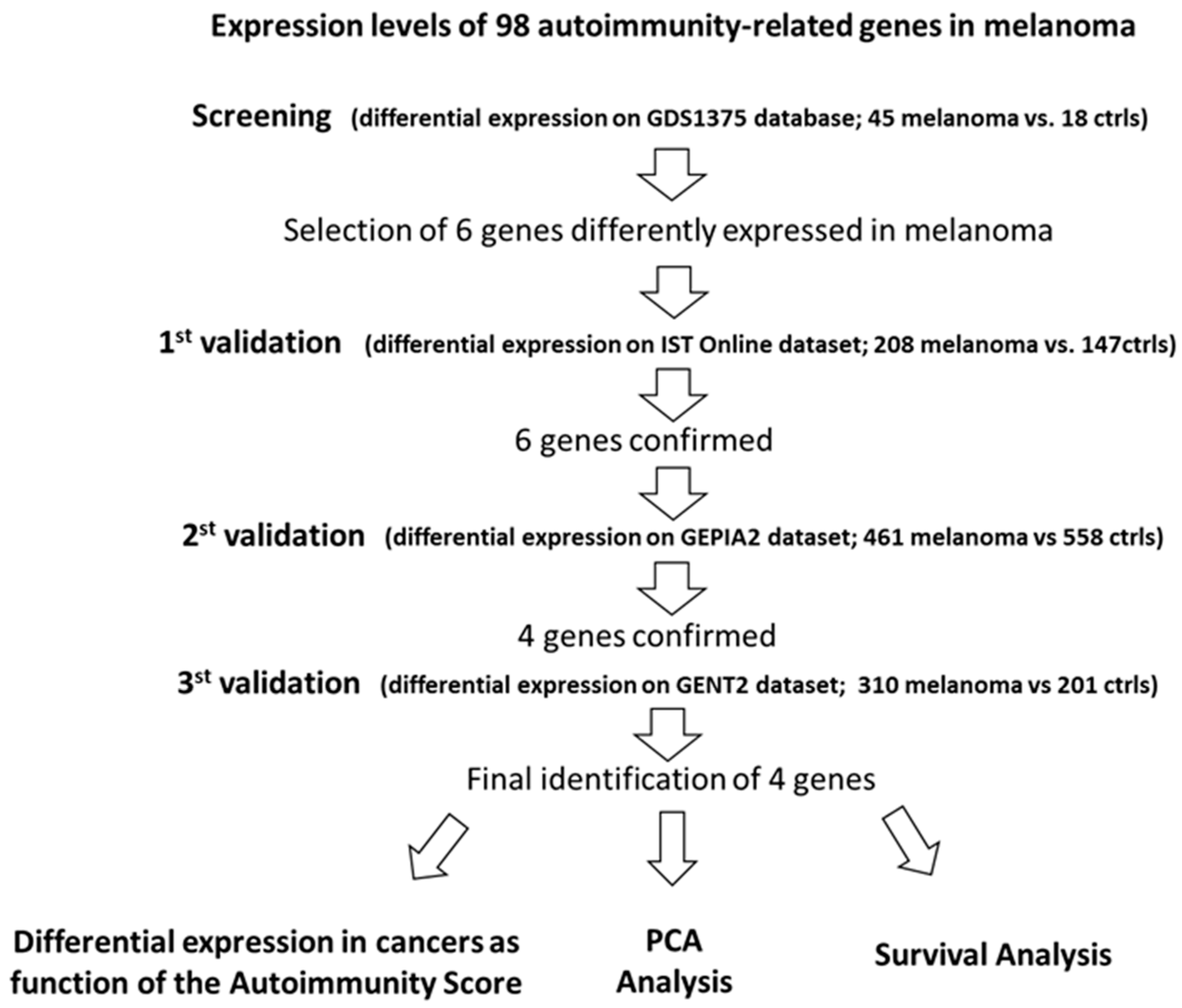
Severe requirements were selected from the values reported in Table 2 to identify differential expression in melanoma vs. controls able to effectively differentiate melanoma from control samples. Namely, the requirements were: fold change <0.5 or >1.5; AUC > 0.93, p < 0.0001. Six genes match these criteria, namely: NOD2, BAX, IL-18, ADRB2, ITGAV and MYO9B. The ROC analysis for each gene is reported in Figure 2, with the indication of AUC and p values. The AUC is > 0.93 in all cases, indicating an extremely high ability to differentiate melanoma samples from controls.
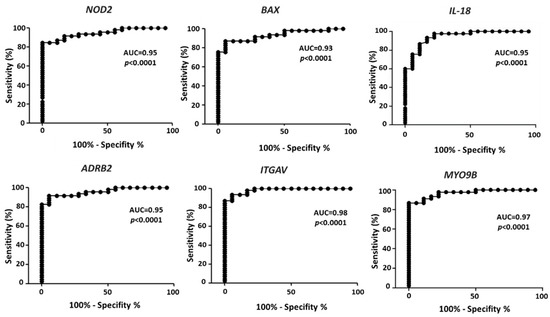
Figure 2.
ROC analysis on the expression values of the six selected genes, according to the expression values reported in GDS1375 from GEO Database. The area under the curve (AUC) is plotted as sensitivity% vs. 100-specificity%. The calculated AUC and p value are reported in each case.
2.4. First Validation: Gene Expression Analysis in IST Online Database
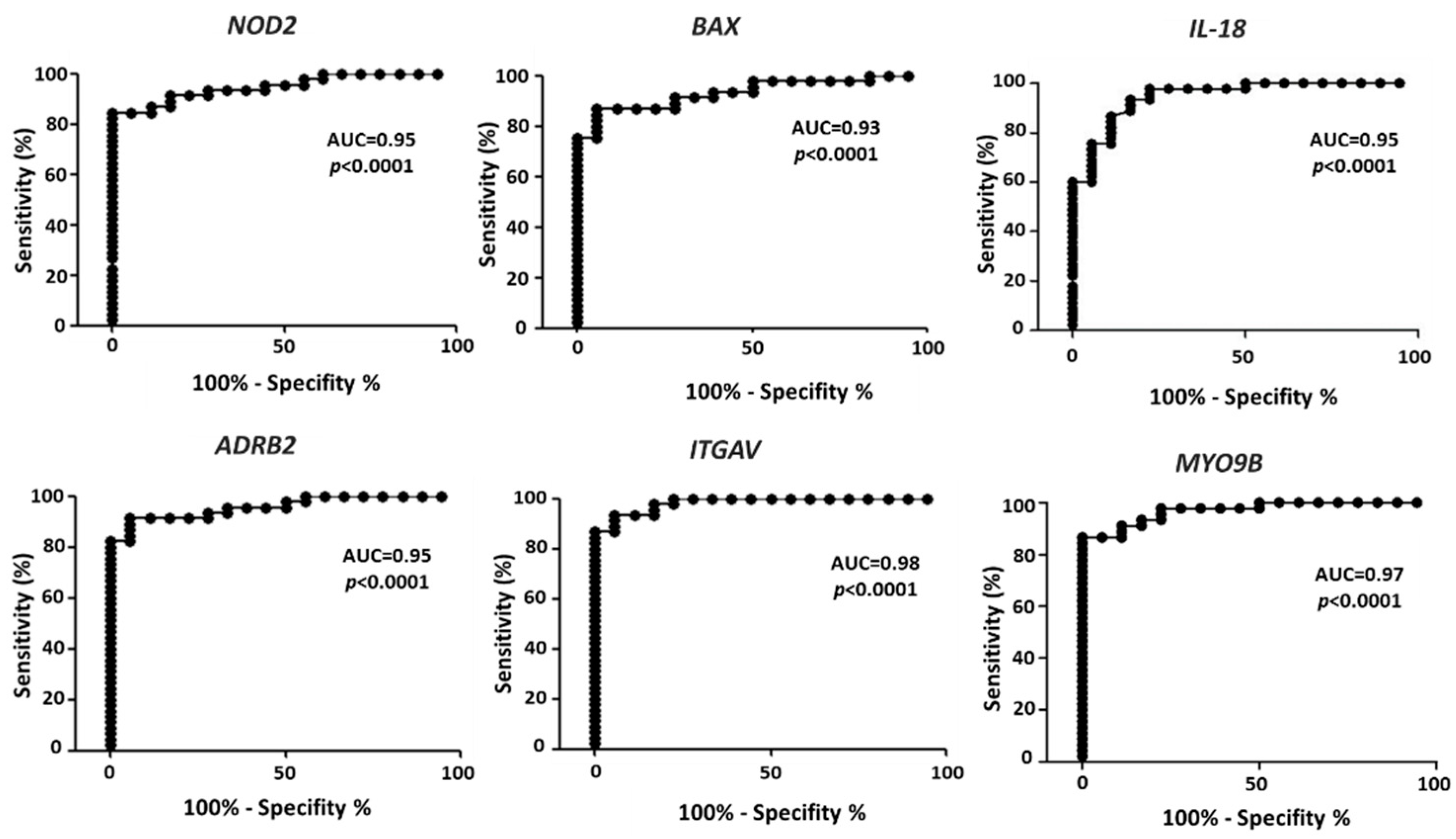
The expression of the six genes (NOD2, BAX, IL-18, ADRB2, ITGAV and MYO9B) was then investigated in an independent public database, namely IST Online. Such database contains several different cancers datasets and normal tissues datasets; the melanoma dataset contains 208 melanoma patients and the healthy skin dataset, used here as control, contains 147 healthy skin samples. Data collected on the GEO dataset were fully validated; in fact, all six genes confirmed significant different expression levels in melanoma vs. healthy skin, in the IST Online dataset (Figure 3), according to the Fisher’s exact test analysis (p < 0.0001). The used threshold values are indicated as dashed lines in Figure 3. We then concluded that all genes show a strongly different expression in melanoma vs. control biopsies, in both GDS1375 and IST Online datasets.
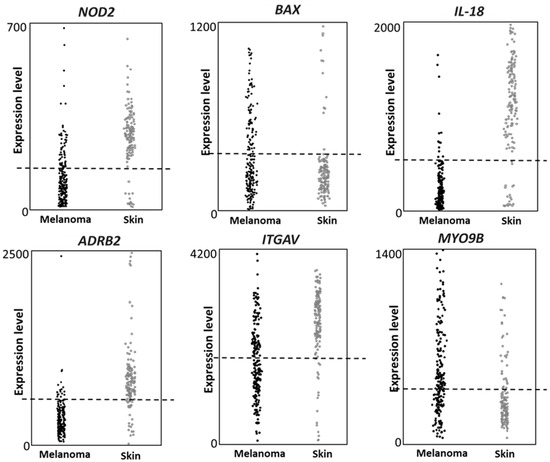
Figure 3.
Gene expression according to the IST online database. The six genes show different expression levels in melanoma vs. healthy skin. The expression level of each gene is reported in 208 melanoma biopsies and 147 healthy skin biopsies. The Fisher analysis shows for each gene a p value < 0.0001. The expression value threshold was: 200 for NOD2 and BAX; 500 for ADRB2, IL-18 and MYO9B; 2000 for ITGAV.
2.5. Second Validation: Gene Expression Analysis in GEPIA2 Database
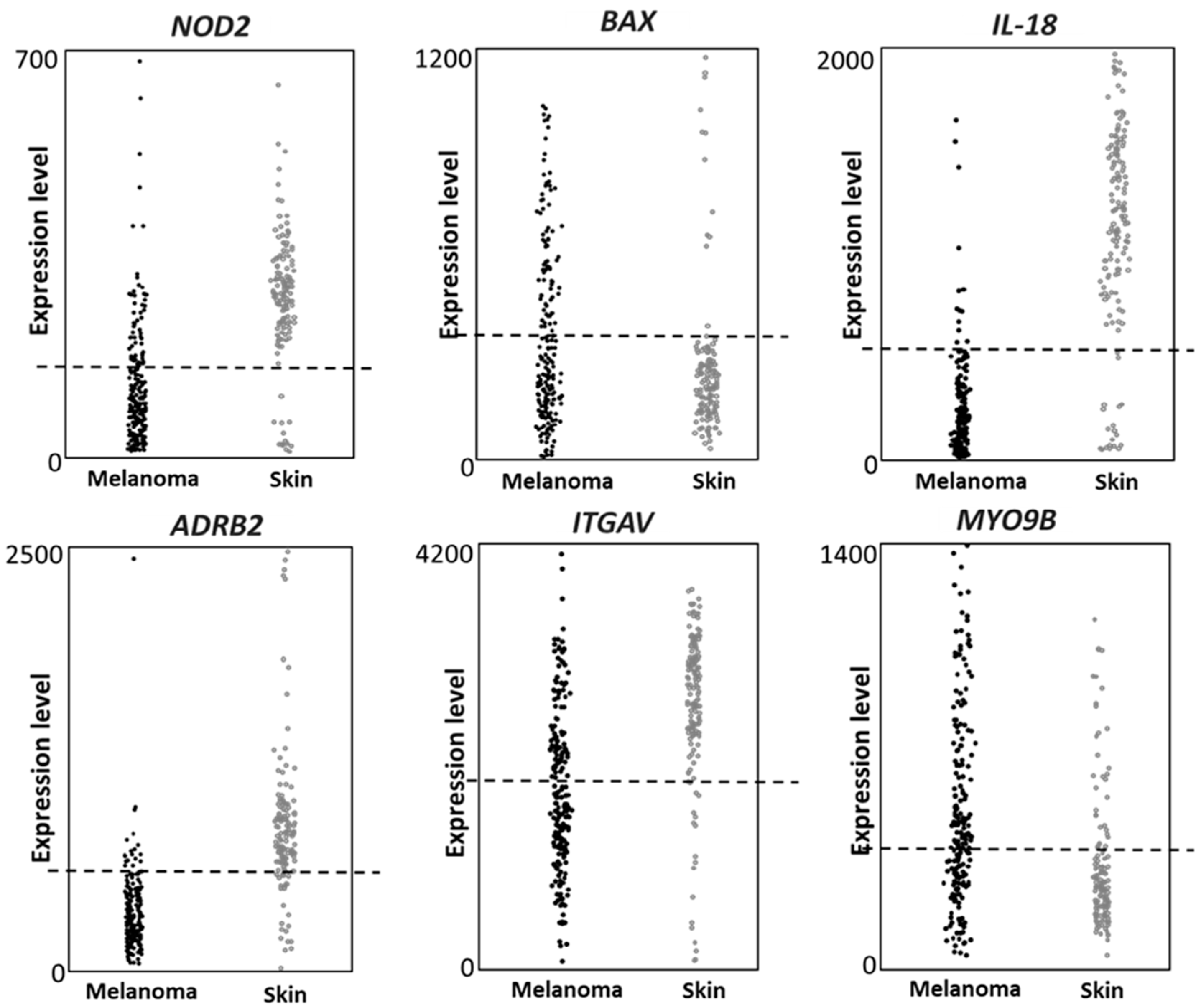
During the finalization of the present study, Ist Online database became not publicly available any longer. We then decided to validate the six genes in two additional public databases. As a second validation, the expression levels of NOD2, BAX, IL-18, ADRB2, ITGAV and MYO9B were analyzed in GEPIA2 database (available at http://gepia2.cancer-pku.cn/#index, accessed on 17 May 2021), a public database reporting expression data from 461 melanoma patients and 558 healthy skin controls. Four out of six genes showed a significantly different expression in melanoma vs. healthy skin, as depicted in Figure 4.
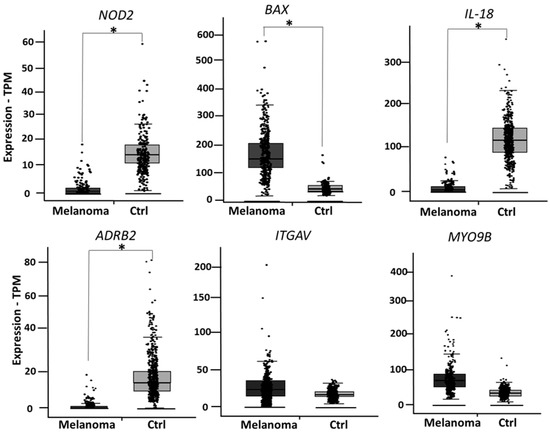
Figure 4.
According to GEPIA2 database, NOD2, BAX, IL-18 and ADRB2 show a significantly different expression in melanoma vs. healthy skin, while expression levels of ITGAV and MYOB9 are not significantly changed. * indicates p < 0.0001.
2.6. Third Round Validation on GENT2 Database
As a third-round validation, the expression levels of the four genes NOD2, BAX, IL-18 and ADRB2 were investigated in skin cancer samples from the GENT2 database (available at http://gent2.appex.kr/gent2/ accession on 28 May 2021), which collects data from 310 skin cancers samples and 201 controls. All four genes show a strongly significant differential expression in skin cancers as compared to the appropriate controls. Namely NOD2 shows a LOG2 FC (Fold change expressed in Log2) expression of −1.34, p < 0.001; BAX shows a LOG2FC expression of 1.3, p < 0.001; IL-18 shows a LOG2FC expression of 3.09, p < 0.001; ADRB2 shows a LOG2FC expression of 1.6, p < 0.001.
2.7. Principal Component Analysis (PCA) and Multiple Logistic Analysis
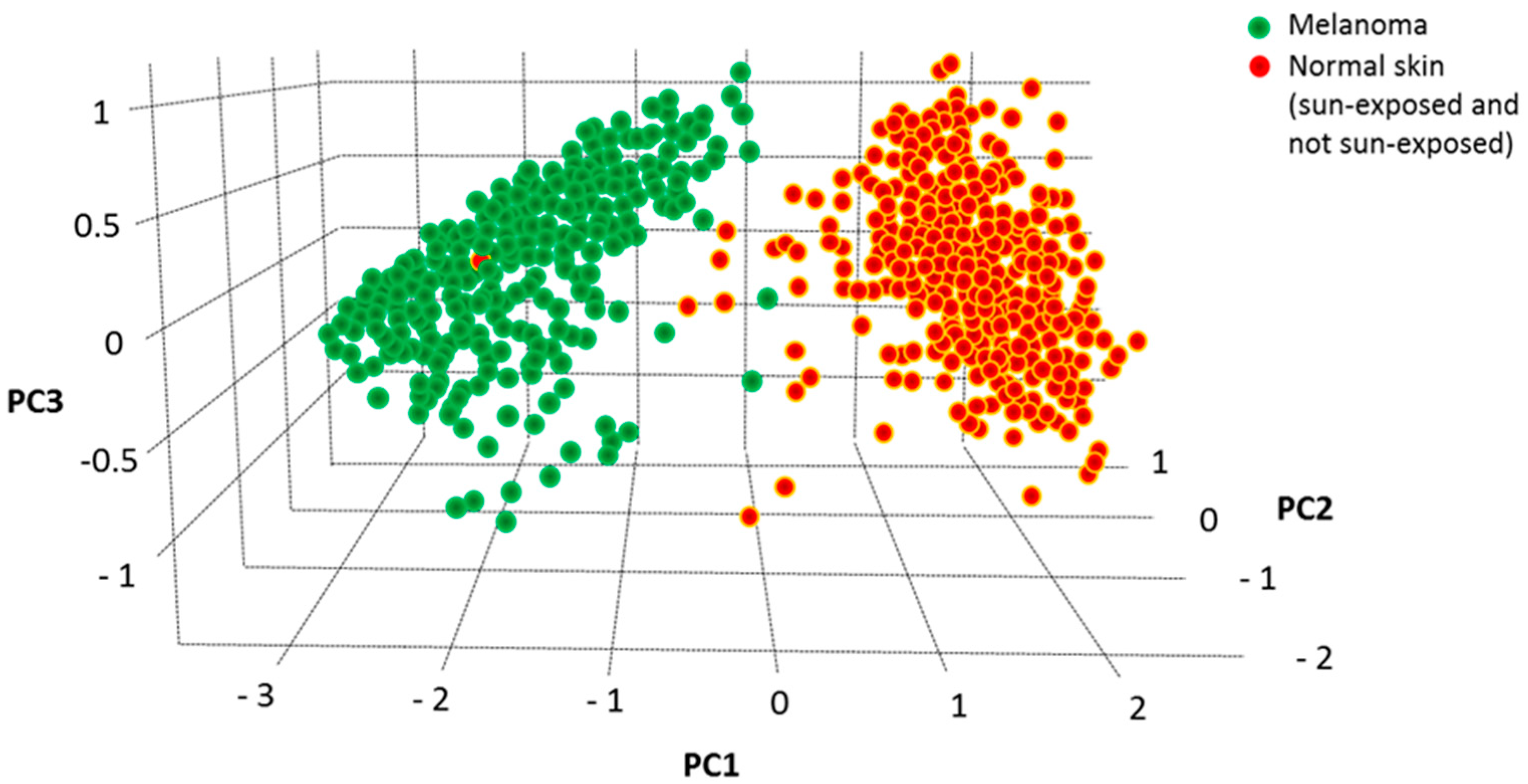
We have previously used Principal Component Analysis (PCA) to identify the best genes-combination able to discriminate melanoma from controls within a large selection of nicotinamide receptors [20] and cytokines/chemokines [21]. The combined expression level of the four validated genes (NOD2, BAX, IL-18 and ADRB2) was then analyzed according to the PCA dimensionality reduction tool in the GEPIA2 database. As reported in Figure 5, the three-dimensional space defined by the three variance components (PC1, PC2 and PC3) associated with the expression values of these four genes show a striking separation between 461 melanoma and 558 healthy controls, indicating that the expression profile of such four genes strongly differentiates melanoma from healthy controls.
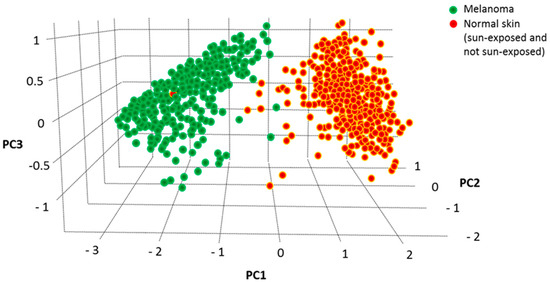
Figure 5.
PCA analysis of the combined values of NOD2, BAX, IL-18 and ADRB2 carried out according to GEPIA2 (http://gepia2.cancer-pku.cn/#dimension accession at 10 June 2021). Melanoma samples appear clearly separated from controls in the three-dimensional space, indicating that the combined expression values of these four genes shows distinct variance in melanoma vs. healthy controls.
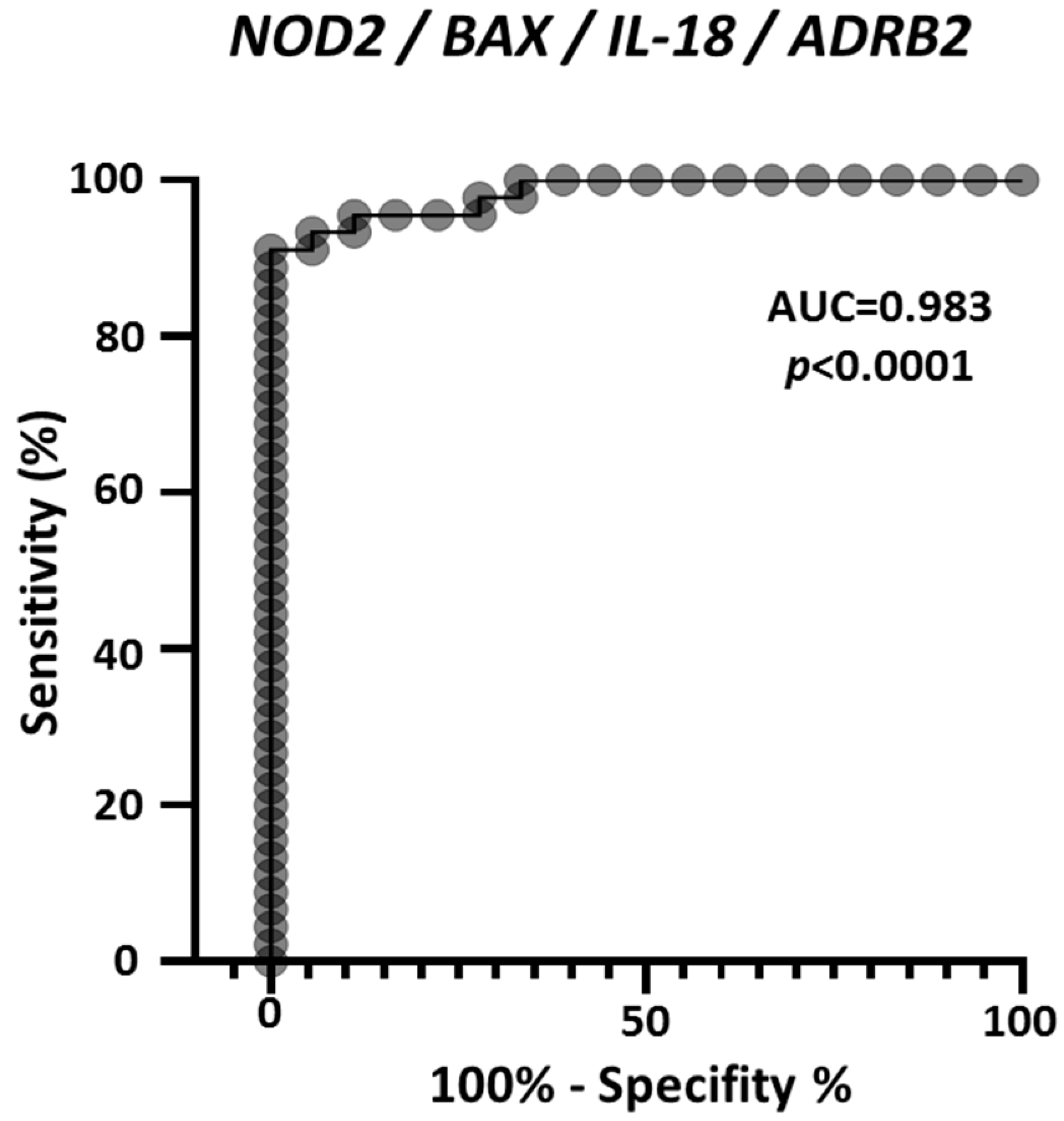
As an additional evaluation, multiple logistic analysis and ROC analysis were carried out on the four genes combined expression profile, in the 45 melanoma and 18 nevi samples derived from the GEO GDS1375 dataset. Figure 6 reports the ROC curve and demonstrates that combining the expression values of NOD2, BAX, IL-18 and ADRB2 achieves an extremely high efficacy to discriminate controls from melanoma samples, with AUC = 0.983. In addition, 88.9% of observed nevi were correctly classified and 93.3% of observed melanoma were correctly classified.
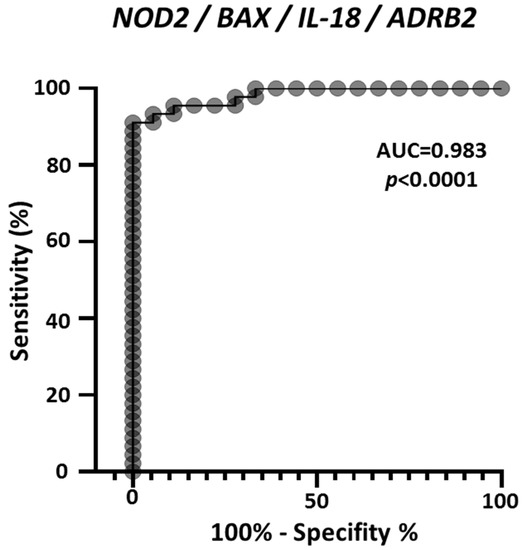
Figure 6.
ROC analysis on the combined expression values of NOD2, BAX, IL-18 and ADRB2, according to the values reported in GDS1375 from the GEO Database. The area under the curve (AUC) is plotted as sensitivity% vs. 100%-specificity%. The calculated AUC and p value are reported.
2.8. NOD2, BAX, IL-18 and ADRB2 Gene Expression in Different Cancer Types
We then further characterized the gene-expression of the four validated genes in 28 different cancer types. Table 3 reports the tumors sorted from the lowest to the highest “Knowledge-based Autoimmunity Score” (kb-AS) according to Table 1. As indicated by the asterisks, expression of NOD2, BAX, IL-18 and ADRB2 is significantly altered mostly in cancer types showing the highest autoimmunity scores, further supporting the possible role of such genes in the interplay between autoimmunity and cancer.

Table 3.
The expression of the four validated genes was analyzed in all cancer types, in the GEPIA2 database. Asterisks indicate whether that specific cancer shows a significantly altered expression of that specific gene, compared to the corresponding healthy controls. Most of the significant differential expressions are found in cancers showing the higher “knowledge-based Autoimmunity Score” (kb-AS).
2.9. Survival in Patients Expressing High- and Low- Levels of NOD2, BAX, IL-18 and ADRB2
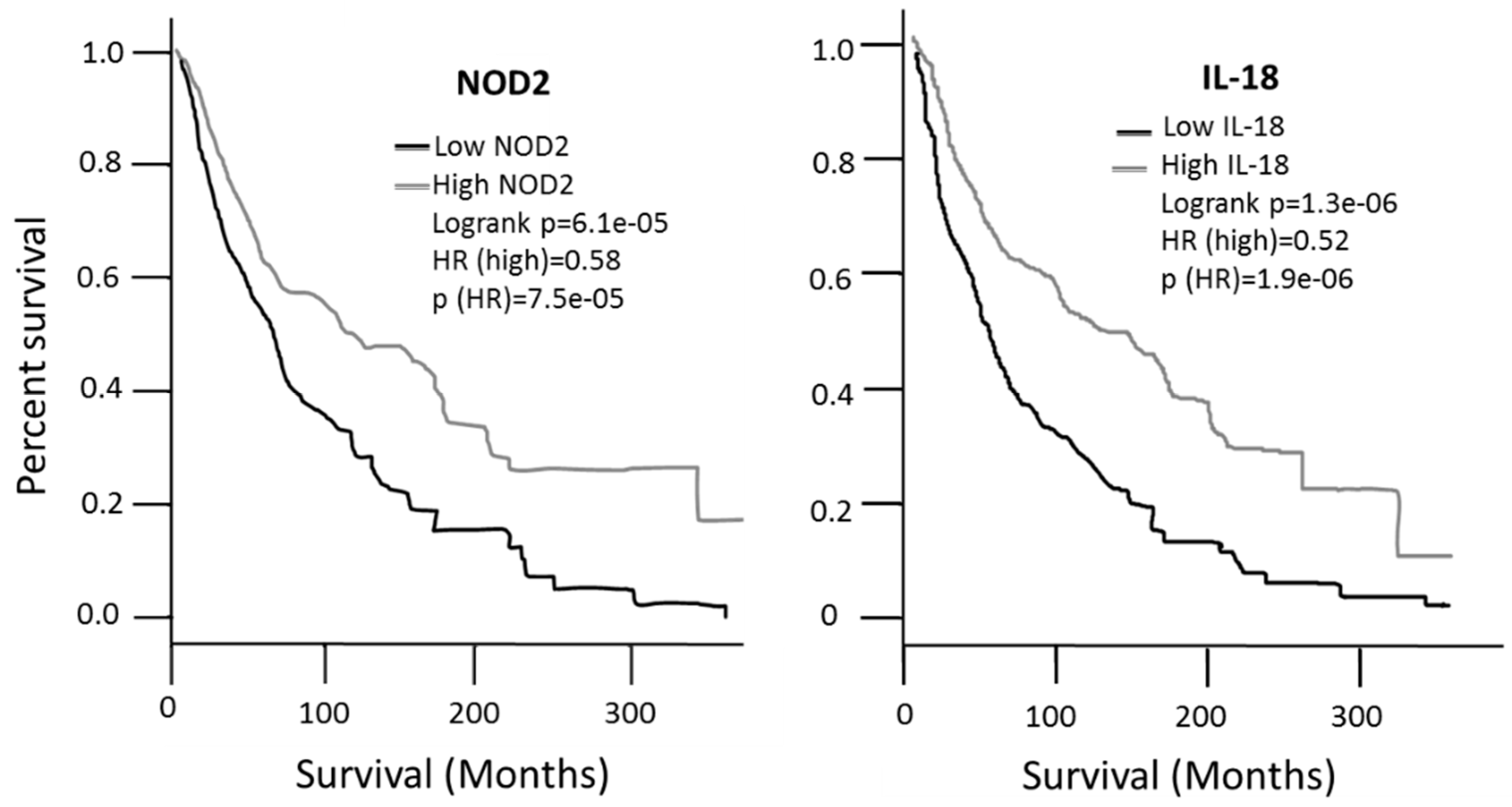
The survival associated with the expression values of each gene was then analyzed according to the GEPIA2 database. Patients expressing high levels of NOD2 or IL-18 showed a significantly better survival, with a Hazard Ratio (HR) of 0.58 and 0.52, respectively, compared to low expression patients (Figure 7); the analysis was performed on 229 high-expressing patients and 229 low-expressing patients. On the contrary, expression of BAX or ADRB2 has no significant impact on patients’ survival (data not shown). In addition to the possible diagnostic role reported in the previous figures, such data indicate that NOD2 and IL-18 expression levels may also have a prognostic value in melanoma.
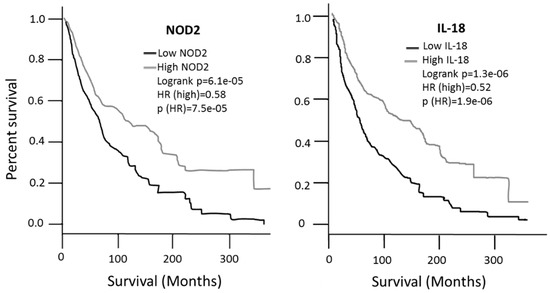
Figure 7.
Survival analysis carried out in the GEPIA2 database. NOD2 and IL-18 expression values show a significantly different survival in patients with high vs. low expression patients.
3. Discussion
A possible role of autoimmunity in melanoma is currently under intense investigation. Autoimmunity is generally recognized as a possible severe side effect of melanoma therapy, upon interferon- [22] or ipilimumab-treatment [23]. On the other hand, unchaining the immune system is the main goal of current immunotherapy protocols as well as previous approaches combining dacarbazine with IL-2 and GM-CSF [24]. Data published in 2018 indicate a high prevalence rate of pre-existing autoimmune comorbidities in melanoma patients as compared to the general population (28.3% and 19.8% in metastatic and not-metastatic melanoma, respectively, as compared to 9.2% in the general population) [25]. Other studies published in 2013 show that the co-occurrence of melanoma with other diseases, including autoimmune diseases, increases the melanoma-mortality rate [26], indicating a possible etiopathogenetic role of autoimmune response in the melanoma onset. Nevertheless, a significant prevalence of autoimmune diseases in melanoma was questioned in a previous study published in 2007 [10]. It is commonly reported that a dysregulation of the immune response, including autoimmune conditions, may predispose toward the cancer development, likely due to the chronic inflammation occurring in these patients [8].
On the other hand, autoimmunity may also have a protective role. In fact, patients undergoing an autoimmune reaction against melanocytes, such as vitiligo patients, show either a lower risk of developing melanoma [27,28] or a better prognosis [29,30], most likely due to a so-called “beneficial autoimmunity” [31]. The present study investigated the hypothesis that the expression of genes directly involved in autoimmunity may be altered in melanoma patients and may therefore represent suitable molecular targets for possible diagnostic, prognostic or therapeutic applications. To address this hypothesis, a non-exhaustive list of 98 genes related to autoimmunity [17,18,19] was investigated while the proper bioinformatic tools are being developed to address the complete list of 4186 genes contained in the GAAD autoimmunity-related genes database [19]. A sequential multi-step validation was carried out on four independent public databases (GEO, IST Online, GEPIA2 and GENT2) involving a total of 1024 melanoma patients and 924 controls. Four genes were identified and validated in all databases, showing a significantly altered expression in melanoma vs. controls, namely NOD2, BAX, IL-18 and ADRB2, NOD2 and IL-18 appear to be particularly interesting from a prognostic point of view since their expression was also found to be related to a significant survival improvement (Figure 7). NOD2 is a member of the Nod1/Apaf-1 family; the encoded protein has two caspase recruitment domains and six leucine-rich repeats giving to NOD2 a protein-binding activity. Leucin-rich repeats are functionally related to innate immunity activity, functioning as sensors of pathogen-associated molecular patterns [32] and triggering NF-kB activation upon recognition of bacterial lipopolysaccharides. NOD2 haplotypes have been associated with Crohn disease [33], and a gain-of-function mutation of NOD2 occurs in patients with Blau syndrome, a rare systemic granulomatous inflammatory disease [34]. Both Chron disease and Blau syndrome are reported to have a clear autoimmunity basis [35,36]. Several polymorphisms of NOD2 are known; some are recognized to be related to increased cancer risk, nevertheless melanoma appears to not be significantly related to NOD2 polymorphisms [37,38]. The present study suggests for the first time that NOD2 expression levels may be indeed related to melanoma onset. Interestingly, patients with Chron disease have an increased risk of developing melanoma and other cancers [39] and at least two cases of Blau Syndrome in melanoma patients have been described [40]. In all four databases investigated, NOD2 expression appears strongly and significantly reduced in melanoma patients, suggesting that the loss of its beneficial function in the immune and autoimmune surveillance may have a role in melanoma. Its protective role may be further recognized since high-expression patients show better survival, as compared to low-expression patients (Figure 7). NOD2 is involved in many processes such as embryogenesis [41] and in regeneration [42] and chronic inflammation [43], suggesting that the relationship between NOD2 and melanoma/cancer may not be limited to the immune system, but may involve also other mechanisms related to carcinogenesis and the evolution of malignancy [44].
IL-18 is one of the factors most related to autoimmunity [45,46,47,48] due to its potent and complex pro-inflammatory action. IL-18 is also known to play a key role in melanoma regulation [49] and is one of the AIM2 inflammasome components [50], recently recognized as a potential target for melanoma treatment [51]. Inducing IL-18 expression is known to exert an anti-melanoma effect in mice [52]; this is compatible with our data showing a significant reduction of IL-18 expression in melanoma patients, according to all four databases investigated, and is consistent with the better survival observed in patients showing high IL-18 expression (Figure 7). The role of IL-18 expression in melanoma patients has been previously reported [53], but the interplay with the autoimmunity is highlighted here for the first time, in the melanoma setting.
ADRB2 codes for the adrenoreceptor beta 2 G protein-coupled receptor. It has a central role in controlling immune and autoimmune processes, and in suppressing autoimmunity in the central nervous system [54]. As recently underlined [55], ADRB2 plays a dual role in inducing or inhibiting autoimmune disorders at both the systemic and local level, exerting opposite actions in different stages of autoimmune diseases such as Rheumatoid Arthritis [55]. Polymorphisms of this gene are related to susceptibility to autoimmune diseases such as Graves disease [56] and Rheumatoid Arthritis [57]. ADRB2 has been previously found associated with melanoma [58]; the present study investigates its expression in a melanoma onset, for the first time. The strong and significant reduced expression in melanoma patients suggests a loss-of-function and a reduced immune or autoimmune control toward melanoma-related antigens. ADRB2 is closely related to the C L type calcium channel Ca (V)1.2, further supporting the role we recently underlined ion channels play, as possible melanoma therapeutic targets [59,60].
BAX is a proapoptotic molecule, member of the BCL-2 family, with a recognized role to control immune tolerance and prevent autoimmune disorders in mice [61]. Its complete deletion in MCL-1 depleted cells has recently been found to completely rescue a lethal multiorgan autoimmunity [62], supporting a key role in the control of immune tolerance. We report here a significantly increased expression of BAX in melanoma patients, supporting it as a possible autoimmunity-related target in melanoma.
The link between autoimmunity and cancer is largely investigated but it is still controversial whether autoimmunity helps or blocks cancer set up or development [31,63,64]. Further investigations are needed to clarify also the potential therapeutic applications of drugs targeting autoimmunity.
Autoimmune manifestations occur in melanoma, which is known to express self-antigens such as B melanoma antigen 1 (BAGE) and melanoma-associated antigens (MAGEs). The co-occurrence of vitiligo, an autoimmunity-based disease, has been associated with better prognosis in melanoma patients [65] and a preexisting vitiligo is associated with a significantly reduced incidence of melanoma [66] and of other cancers [67]. Furthermore, vitiligo regression associates with melanoma progression [68]. All such data strongly support the hypothesis that autoimmunity may be beneficial for melanoma, as well as other cancer types, as reviewed by a recent study by Zitvogel et al. [31]. Data reported in the present study support the hypothesis of a beneficial autoimmunity mediated by NOD2, IL-18 and ADRB2, since their expression is shown to be almost abolished in melanoma patients. Interestingly, the multiple logistic analysis shows that the AUC = 0.98 computed for the four-genes combined profile (Figure 6), is much higher than the AUC computed for the single genes (AUC = 0.95 for NOD2, IL-18 and ADRB2; AUC = 0.93 for BAX) (Figure 2). This demonstrates that combining the expression values is much more effective to discriminate controls from melanoma, as compared to the values of the single genes.
Finally, when the expression analysis was extended to several cancer types other than melanoma, the four genes were found significantly altered especially in cancer types mostly related to autoimmunity, according to the knowledge-based Autoimmunity Score (kb-AS) computed in the present study. Some pitfalls may be recognized in the procedure followed to measure kb-AS. The first relates to the continuous growth of the PubMed database, which implies the possibility that the kb-AS may (or will) change over time. Further, less common cancer types were not investigated. We are aware that the searches carried out were not extensive, and we will address this issue in more details and larger completeness in a study focused on this specific aspect. However, we believe the current form of the kb-AS is sufficiently indicative to recognize at least the most common cancer-types showing the known highest relation to autoimmunity.
4. Materials and Methods
Ninety-Eight Autoimmunity-Related Genes Were Taken for the Literature [17,18,19]. The genes investigated are known to be related to allergies, ankylosing spondylitis, asthma celiac diseases, Crohn’s disease, type 1 and type 2 diabetes, Graves’ diseases, inflammatory bowel disease, multiple sclerosis, psoriasis, rheumatoid arthritis, Sjögren syndrome, systemic lupus erythematosus and ulcerative colitis. The expression of such genes was investigated in a total of 1948 human samples derived from four different public datasets, via one initial screening phase and three additional validation phases.
4.1. Calculating the “Knowledge-Based Autoimmunity Score” kb-AS
The known relation of 28 cancer types to autoimmunity was measured as follows: PubMed searches were carried out looking for the occurrence of the words relating to each cancer type in All-fields. Such a procedure was carried out as a stand-alone search or in the presence of autoimmunity-related words. Therefore, the co-occurrence of cancer-related words and autoimmunity-related words was measured. Words used to identify each cancer type and to identify autoimmunity are reported below.
The number of manuscripts presenting the co-occurrence, divided by the total number of manuscripts related to each cancer-type, expressed as percentage, was then reported as the “knowledge-based Autoimmunity Score” of cancers (kb-AS). The cancer types investigated were the 28 cancer types reported in the GEPIA2 database. The words related to each cancer type, searched in PubMed All-fields, are as follows:
Cervical = “Cervical squamous cell carcinoma” OR “Endocervical adenocarcinoma”;
Esophag = “Esophageal carcinoma” OR “Esophagus carcinoma”;
Pheoc-Para = “Pheochromocytoma” OR “Paraganglioma”;
Endometrial = “Uterine Corpus Endometrial carcinoma” OR “endometrial carcinoma” OR “endometrial cancer” OR “Endometrium cancer” OR “Uterine carcinosarcoma”;
Rectum = “Rectum adenocarcinoma” OR “rectum carcinoma” OR “rectum cancer”;
Adrenocortical = “Adrenocortical carcinoma”;
Prostate = “Prostate adenocarcinoma” OR “prostate carcinoma” OR “prostate cancer”;
Mesothelioma = “Mesothelioma”;
Ovary = “Ovarian serous cystadenocarcinoma” OR “Ovarian carcinoma” OR “Ovarian adenocarcinoma”;
Head and Neck = “Head and Neck squamous cell carcinoma” OR “Head and Neck carcinoma”;
Gliobl = “Glioblastoma”;
Sarcoma = “Sarcoma”;
Liver = “Liver hepatocellular carcinoma” OR “liver carcinoma”;
Bladder = “Bladder carcinoma”;
Testis = “testicular Germ cell tumors” OR “testicular cancer” OR “testis cancer “ OR “seminoma”; Colon = “Colon adenocarcinoma”;
Kidney = “Kidney carcinoma” OR “kidney cancer”;
Breast = “Breast carcinoma”;
Lung = “Lung adenocarcinoma” OR “Lung carcinoma”;
Glioma = “Glioma”;
Leukemia = “Leukemia”;
Gastric = “Gastric adenocarcinoma” OR “stomach adenocarcinoma”;
Pancreati c = “Pancreatic adenocarcinoma” OR “pancreatic carcinoma”;
Melanoma = “Melanoma”;
Chol = “Cholangio carcinoma”;
Thyroid = “Thyroid carcinoma”;
Lymphoma = “Lymphoma”;
Thymoma = “Thymoma”.
The autoimmunity related words with wild cards indicated by the asterisks were: “autoimmun*” OR “autoantigen*” OR “selfantigen*” OR “selftolerance”.
4.2. Expression of Autoimmunity-Related Genes, in GEO Database: Melanoma vs. Nevi Samples
The expression of 98 autoimmunity-related genes was evaluated in the melanoma GDS1375 dataset, from the GEO public database (https://www.ncbi.nlm.nih.gov/gds/), accessed on 8 April 2021 reporting the actual expression values in 63 samples (45 melanoma-patients vs. 18 nevi-patients). Calculations such as mean, Mann–Whitney test and ROC analysis were then carried out. ROC analysis is a binary test; it was used here to measure how effective the expression-level of any given gene to discriminate healthy from melanoma-biopsies is. The computed Area Under Curve (AUC) value ranges from 0.5 to 1, indicating 50% to 100% discrimination ability.
4.3. First Validation of Gene Expression Data in IST Online Database
The first validation step was carried out by analyzing expression levels of the selected genes in the independent IST Online database (http://ist.medisapiens.com/) accessed on 6 May 2021, having 208 melanoma samples and 147 healthy-skin samples. Different to the GEO database, IST Online does not show actual numbers; rather it expresses data as scatter plots, where each dot is one patient. Genes showing expression levels in melanoma significantly different to controls, according to the Fisher’s exact test, were then considered validated and were selected for the following validation step.
4.4. Second Validation of Gene Expression Data in GEPIA2 Database
During the finalization of the present study, the IST Online database, previously public for many years, became unavailable. In order to assure a full reproducibility of the results, we therefore decided to carry out additional validation steps. A second round validation step was carried out on the public database GEPIA2, available at http://gepia2.cancer-pku.cn/#index [69] accessed on 17 May 2021, having 461 melanoma and 558 healthy-skin samples. Genes matching stringent significance threshold parameters (namely, Log2 [FC] > 1.5 and p value < 0.0001) were considered fully validated.
4.5. Third Validation of Gene Expression Data in GENT2 Database
The four selected genes (NOD2, BAX, IL-18 and ADRB2) were investigated in control and in skin cancer samples (total 1042 samples, including 201 controls and 310 melanoma samples), taken from the GENT2 database at the link http://gent2.appex.kr/gent2/ accessed on 28 May 2021. The gene profile tools were used, indicating as types: “Tissue”, terms: “Gene symbol”, and as keyword the four genes symbol; data were then extracted from the table reporting T-test and Log2 fold changes results.
4.6. PCA Analysis and Multiple Logistic Analysis
The “dimensionality reduction” tool available on GEPIA2 at http://gepia2.cancer-pku.cn/#dimension accessed on 10 June 2021, was exploited to perform the PCA analysis [70] combining NOD2, BAX, IL-18 and ADRB2 expression levels, according to the methodology previously used [19,20]. Log scale was used; cancer type was SKCM tumor (Skin Cutaneous Melanoma); controls were SKCM normal, skin not sun exposed, skin sun exposed.
Multiple logistic analysis was carried out using the expression values of NOD2, BAX, IL-18 and ADRB2 in controls and melanoma patients, and ROC analysis on their combined expression values was computed.
4.7. Survival Analysis in Melanoma Patients
Survival analysis in melanoma patients was investigated according to GEPIA2 at http://gepia2.cancer-pku.cn/#survival accessed on 11 June 2021. The analysis was performed on 229 high-expressing patients and 229 low-expressing patients. Survival analyses were carried out on quartile distribution with the cutoff for high expression set at 70% and the cutoff for low expression set at 25%, using the skin cancer melanoma (SKCM) dataset, according to a methodology previously described [20].
4.8. Expression of NOD2, BAX, IL-18 and ADRB2 in 28 Cancer Types
The expression levels of NOD2, BAX, IL-18 and ADRB2 were analyzed in all cancer types present in GEPIA2 database, available at http://gepia2.cancer-pku.cn/#index [69] accessed on 1 July 2021. Log2 [FC] > 1.5 and p value < 0.0001 were selected as the significance threshold.
4.9. Statistics
The analyses were performed on 1948 human samples from four independent databases. The samples distribution was as follows: 45 melanoma and 18 nevi samples from GEO database; 208 melanoma and 147 healthy-skin samples from IST Online database; 461 melanoma and 558 healthy-skin samples from GEPIA2 database and 310 melanoma and 201 healthy skin samples from GENT2 database. Mean, two tails t Test, Mann–Whitney test and ROC analysis were carried out, with a significance-threshold set at p value < 0.0001. Fisher’s exact test was carried out to evaluate sample distribution above or below a given threshold, in data from Ist Online database. The statistical package GraphPad Prism version 9.3.0 was used (Graph Pad software Inc, 2365 Northside Dr., Suite 560, San Diego, CA 92108, USA).
5. Conclusions
In conclusion, the present study identifies four genes strongly related to autoimmunity. Their expression was found to be significantly altered in melanoma patients according to a multi-validation procedure carried out on 1948 patients from four independent databases. We believe such genes may represent suitable molecular targets to further investigate the role autoimmunity may play in melanoma setup and development. Such data suggest that autoimmunity may play a beneficial role in melanoma set up, at least to some extent.
Author Contributions
F.S.: Conceptualization, analysis, validation, writing. A.F.: Conceptualization: analysis, validation, writing, supervision, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ricerca Corrente 2021, RC 3.4, from Ministry of Health, Italy.
Data Availability Statement
Raw data regarding expression values from GEO and from Ist Online are deposited in Mendeley data at https://data.mendeley.com/GEO. Data were from https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS1375GEPIA2. Analysis was carried out on http://gepia2.cancer-pku.cn/ Ist Online data were from http://ist.medisapiens.com (currently not available). GENT2 analysis was carried out on http://gent2.appex.kr/gent2/.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goodnow, C.C.; Sprent, J.; Fazekas de St Groth, B.; Vinuesa, C.G. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 2005, 435, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, U.; Singh, S. Prevalence of autoantibodies in patients of psoriasis. J. Clin. Lab. Anal. 2010, 24, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Iturbe, B.; Pons HQuiroz, Y.; Lanaspa, M.A.; Richard, J.; Johnson, R.J. Autoimmunity in the pathogenesis of hypertension. Nat. Rev. Nephrol. 2014, 10, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.; Bastian, B.; Quiqueree, L.; Jones, C.; Morgan, R.; Reeves, G. Abnormal pulmonary vascular responses in patients registered with a systemic autoimmunity database: Pulmonary Hypertension Assessment and Screening Evaluation using stress echocardiography (PHASE-I). Eur. J. Echocardiogr. 2006, 7, 439–446. [Google Scholar] [CrossRef][Green Version]
- Bernatsky, S.; Ramsey-Goldman, R.; Clarke, A. Malignancy and auzhatoimmunity. Curr. Opin. Rheumatol. 2006, 18, 129–134. [Google Scholar] [CrossRef]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Felici, C.; Stucci, L.S.; Cives, M.; Silvestris, F. Immune System Evasion as Hallmark of Melanoma Progression: The Role of Dendritic Cells. Front. Oncol. 2019, 9, 1148. [Google Scholar] [CrossRef]
- Hurwitz, A.A.; Qingyong, J. Autoimmune Depigmentation Following Sensitization to Melanoma Antigens; (Methods in Molecular Medicine); Humana Press Inc.: Totowa, NJ, USA, 2004; Volume 102, pp. 421–427. [Google Scholar] [CrossRef]
- Logotheti, S.; Putzer, B.M. STAT3 and STAT5 Targeting for Simultaneous Management of Melanoma and Autoimmune Diseases. Cancers 2019, 11, 1448. [Google Scholar] [CrossRef]
- Ramirez-Montagut, T.; Turk MJo Wolchok, J.D.; Guevara-Patino, J.A.; Houghton, A.N. Immunity to melanoma: Unraveling the relation of tumorimmunity and autoimmunity. Oncogene 2003, 22, 3180–3187. [Google Scholar] [CrossRef]
- Kaae, J.; Wohlfahrt, J.; Boyd, H.A.; Wulf, H.C.; Biggar, R.J.; Melbye, M. The Impact of Autoimmune Diseases on the Incidence and Prognosis of Cutaneous Malignant Melanoma. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1840–1844. [Google Scholar] [CrossRef]
- Lu, Y.; Jia, L.; He, S.; Hurley, M.C.; Leys, M.J.; Jayasundera, T.; Heckenlively, J.R. Melanoma-associated retinopathy: A paraneoplastic autoimmune complication. Arch. Ophthalmol. 2009, 127, 1572–1580. [Google Scholar] [CrossRef]
- Gogas, H.; Ioannovich, J.; Dafni, U.; Stavropoulou-Giokas, C.; Frangia, K.; Tsoutsos, D.; Panagiotou, P.; Polyzos, A.; Papadopoulos, O.; Stratigos, A.; et al. Prognostic significance of autoimmunity during treatment of melanoma with interferon. N. Engl. J. Med. 2006, 354, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Maire, C.; Vercambre-Darras, S.; Devos, P.; D’Herbomez, M.; Dubucquoi, S.; Mortier, L. Metastatic melanoma: Spontaneous occurrence of auto antibodies is a good prognosis factor in a prospective cohort. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Bouwhuis, M.G.; Suciu, S.; Collette, S.; Aamdal, S.; Kruit, W.H.; Bastholt, L.; Stierner, U.; Salès, F.; Patel, P.; Punt, C.J.A.; et al. Autoimmune antibodies and recurrence-free interval in melanoma patients treated with adjuvant interferon. JNCI J. Natl. Cancer Inst. 2009, 101, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Li, Q.; Luo, H.; Holmdahl, R. Neutrophil-derived reactive oxygen species promote tumor colonization. Commun. Biol. 2021, 4, 865. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, S.J.; Takizawa, K.E.; Robertson, A.A.B.; Sester, D.P.; Stacey, K.J. Compromised NLRP3 and AIM2 inflammasome function in autoimmune NZB/W F1 mouse macrophages. Immunol. Cell Biol. 2019, 97, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.K.; Olsson, L.M. Recent Advances in the Genetics of Autoimmune Disease. Annu. Rev. Immunol. 2009, 27, 363–391. [Google Scholar] [CrossRef]
- Genes and Mutations Associated with Autoimmune Diseases. Available online: https://www.eupedia.com/genetics/autoimmune_diseases_snp.shtml (accessed on 8 April 2021).
- Lu, G.; Hao, X.; Chen, W.H.; Mu, S. GAAD: A Gene and Autoimmiune Disease Association Database. Genom. Proteom. Bioinform. 2018, 16, 252–261. [Google Scholar] [CrossRef]
- Scatozza, F.; Moschella, F.; D’Arcangelo, D.; Rossi, S.; Tabolacci, C.; Giampietri, C.; Proietti, E.; Facchiano, F.; Facchiano, A. Nicotinamide inhibits melanoma in vitro and in vivo. J. Exp. Clin. Cancer Res. 2020, 39, 211. [Google Scholar] [CrossRef]
- Cesati, M.; Scatozza, F.; D’Arcangelo, D.; Antonini-Cappellini, G.C.; Rossi, S.; Tabolacci, C.; Nudo, M.; Palese, E.; Lembo, L.; Di Lella, G.; et al. Investigatum Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as Highly Effective Melanoma Marker. Cancers 2020, 12, 3680. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Shin, D.; Lee, S.J.; Stuckert, J.; Sander, C.A.; Kirkwood, J.M. Serologic evidence of autoimmunity in E2696 and E1694 patients with high-risk melanoma treated with adjuvant interferon alfa. Melanoma Res. 2014, 24, 150–157. [Google Scholar] [CrossRef]
- Lang, N.; Dick, J.; Slynko, A.; Schulz, C.; Dimitrakopoulou-Strauss, A.; Sachpekidis, C.; Enk, A.H.; Hassel, J.C. Clinical significance of signs of autoimmune colitis in 18 F-fluorodeoxyglucose positron emission tomography-computed tomography of 100 stage-IV melanoma patients. Immunotherapy 2019, 11, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, Q.Y.; Kang, X.M.; Wang, J.X.; Sun, W.Z. Malignant melanoma therapy by chemotherapy and autoimmunity induced by cytokine. Cancer Biother. Radiopharm. 2009, 24, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Shilkrut, M.; Zhao, Z.; Li, M.; Batty, N.; Barber, B. Autoimmune comorbidities in patients with metastatic melanoma: A retrospective analysis of us claims data. BMC Cancer 2018, 18, 145. [Google Scholar] [CrossRef]
- Grann, A.F.; Frøslev, T.; Olesen, A.B.; Schmidt, H.; Lash, T.L. The impact of comorbidity and stage on prognosis of Danish melanoma patients, 1987–2009: A registry-based cohort study. Br. J. Cancer 2013, 109, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Teulings, H.E.; Overkamp, M.; Ceylan, E.; Nieuweboer-Krobotova, L.; Bos, J.D.; Nijsten, T.; Wolkerstorfer, A.W.; Luiten, R.M.; van der Veen, J.P. Decreased risk of melanoma and nonmelanoma skin cancer in patients with vitiligo: A survey among 1307 patients and their partners. Br. J. Dermatol. 2012, 168, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Failla, C.M.; Carbone, M.L.; Fortes, C.; Pagnanelli, G.; D’Atri, S. Melanoma and Vitiligo: In Good Company. Int. J. Mol. Sci. 2019, 20, 5731. [Google Scholar] [CrossRef] [PubMed]
- Piqué-Duran, E.; Palacios-Llopis, S.; Martínez-Martín, M.; Pérez-Cejudo, J.A. Complete regression of melanoma associated with vitiligo. Dermatol. Online J. 2011, 17, 4. [Google Scholar] [CrossRef]
- Lommerts, J.E.; Bekkenk, M.W.; Luiten, R.M. Vitiligo induced by immune checkpoint inhibitors in melanoma patients: An expert opinion. Expert Opin. Drug Saf. 2021, 20, 883–888. [Google Scholar] [CrossRef]
- Zitvogel, L.; Perreault, C.; Finn, O.J.; Guido Kroemer, G. Beneficial autoimmunity improves cancer prognosis. Nat. Rev. Clin. Oncol. 2021, 18, 591–602. [Google Scholar] [CrossRef]
- Aylwin Ng Ramnik, J.X. Leucine-rich repeat (LRR) proteins. Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef]
- Kaczmarek-Ryś, M.; Hryhorowicz, S.T.; Lis, E.; Banasiewicz, T.; Paszkowski, J.; Borejsza-Wysocki, M.; Walkowiak, J.; Cichy, W.; Krokowicz, P.; Czkwianianc, E.; et al. Crohn’s Disease Susceptibility and Onset Are Strongly Related to Three NOD2 Gene Haplotypes. J. Clin. Med. 2021, 10, 3777. [Google Scholar] [CrossRef] [PubMed]
- Caso, F.; Galozzi, P.; Costa, L.; Sfriso, P. Autoinflammatory granulomatous diseases: From Blau syndrome and early-onset sarcoidosis to NOD2-mediated disease and Crohn’s disease. RMD Open 2015, 1, e000097. [Google Scholar] [CrossRef] [PubMed]
- Bar Yehuda, S.; Axlerod, R.; Toker, O.; Zigman, N.; Goren, I.; Mourad, V.; Lederman, N.; Cohen, N.; Matz, E.; Dushnitzky, D.; et al. The Association of Inflammatory Bowel Diseases with Autoimmune Disorders: A Report from the epi-IIRN. Crohn’s Colitis 2018, 13, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Raphael, S.A.; Blau, E.B.; Zhang, W.H.; Hsu, S.H. Analysis of a large kindred with Blau syndrome for HLA, autoimmunity, and sarcoidosis. Am. J. Dis. Child. 1993, 147, 842–848. [Google Scholar] [CrossRef]
- Jingwei, L.; Caiyun, H.; Qian, X.; Chengzhong, X.; Yuan, Y. NOD2 polymorphisms associated with cancer risk: A meta-analysis. PLoS ONE 2014, 9, e89340. [Google Scholar] [CrossRef]
- Debniak, T.; Kurzawski, G.; Huzarski, T.; Byrski, T.; Gronwald, J.; Debniak, B.; ARozmiarek, A.; Dziuba, I.; Złowocka, E.; Suchy, J.; et al. NOD2 variants and the risk of malignant melanoma. Eur. J. Cancer Prev. 2005, 14, 143–146. [Google Scholar] [CrossRef]
- Garg, S.K.; Velayos, F.S.; Kisiel, J.B. Intestinal and Nonintestinal Cancer Risks for Patients with Crohn’s Disease. Gastroenterol. Clin. N. Am. 2017, 46, 515–529. [Google Scholar] [CrossRef]
- Murphy, K.P.; Kennedy, M.P.; Barry, J.E.; O’Regan, K.N.; Power, D.G. New-onset mediastinal and central nervous system sarcoidosis in a patient with metastatic melanoma undergoing CTLA4 monoclonal antibody treatment. Oncol. Res. Treat. 2014, 37, 351–353. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Fu, S.; Chen, X.; Zhang, S.; Li, Y.; Du, M.; Zhang, J. Different expression of NOD2 in decidual stromal cells between normal and unexplained recurrent spontaneous abortion women during first trimester gestation. Int. J. Clin. Exp. Pathol. 2014, 7, 8784–8790. [Google Scholar]
- Campbell, L.; Williams, H.; Crompton, R.A.; Cruickshank, S.M.; Hardman, M.J. Nod2 deficiency impairs inflammatory and epithelial aspects of the cutaneous wound-healing response. J. Pathol. 2013, 229, 121–131. [Google Scholar] [CrossRef]
- Orlando, R.C. Mechanisms of epithelial injury and inflammation in gastrointestinal diseases. Rev. Gastroenterol. Disord. 2002, 2 (Suppl. 2), S2–S8. [Google Scholar] [PubMed]
- Motofei, I.G. Biology of Cancer; from Cellular and Molecular Mechanisms to Developmental Processes and Adaptation. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Boraschi, D.; Dinarello, C.A. IL-18 in autoimmunity: Review. Eur. Cytokine Netw. 2006, 17, 224–252. [Google Scholar] [PubMed]
- Yang, C.A.; Chiang, B.L. Inflammasomes and human autoimmunity: A comprehensive review. J. Autoimmun. 2015, 61, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.I.; Lee, K.H.; Joo, Y.H.; Lee, J.M.; Jeon, J.; Jung, H.J.; Shin, M.; Cho, S.; Kim, T.H.; Park, S.; et al. Inflammasomes and autoimmune and rheumatic diseases: A comprehensive review. J. Autoimmun. 2019, 103, 102299. [Google Scholar] [CrossRef]
- Yang, C.A.; Chiang, B.L. Inflammasomes and Childhood Autoimmune Diseases: A Review of Current Knowledge. Clin. Rev. Allergy Immunol. 2020, 61, 156–170. [Google Scholar] [CrossRef]
- Song, H.; Kim, J.; Lee, H.K.; Park, H.J.; Nam, J.; Park, G.B.; Kim, Y.S.; Cho, D.; Hur, D.Y. Selenium inhibits migration of murine melanoma cells via down-modulation of IL-18 expression. Int. Immunopharmacol. 2011, 11, 2208–2213. [Google Scholar] [CrossRef]
- Ratsimandresy, R.A.; Indramohan, M.; Dorfleutner, A.; Stehlik, C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell. Mol. Immunol. 2017, 14, 127–142. [Google Scholar] [CrossRef]
- Fukuda, K.; Okamura, K.; Riding, R.L.; Fan, X.; Afshari, K.; Haddadi, N.S.; McCauley, S.M.; Guney, M.H.; Luban, J.; Funakoshi, T.; et al. AIM2 regulates anti-tumor immunity and is a viable therapeutic target for melanoma. J. Exp. Med. 2021, 218, e20200962. [Google Scholar] [CrossRef]
- Zheng, J.N.; Pei, D.S.; Mao, L.J.; Liu, X.Y.; Sun, F.H.; Zhang, B.F.; Liu, Y.Q.; Liu, J.J.; Li, W.; Han, D. Oncolytic adenovirus expressing interleukin-18 induces significant antitumor effects against melanoma in mice through inhibition of angiogenesis. Cancer Gene Ther. 2010, 17, 28–36. [Google Scholar] [CrossRef]
- Gil, M.; Kyung Eun Kim, K.E. Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. J. Clin. Med. 2019, 8, 1993. [Google Scholar] [CrossRef]
- Araujo, L.P.; Maricato, J.T.; Guereschi, M.G.; Takenaka, M.C.; Nascimento, V.N.; Menegatti de Melo, F.; Quintana, F.J.; Brum, P.C.; Basso, A.S. The Sympathetic Nervous System Mitigates CNS Autoimmunity via β2-Adrenergic Receptor Signaling in Immune Cells. Cell Rep. 2019, 28, 3120.e5–3130.e5. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Tai, Y.; Hu, S.; Zhang, M.; Wang, R.; Zhou, W.; Tao, J.; Han, Y.; Wang, Q.; Wei, W. Bidirectional Role of β2-Adrenergic Receptor in Autoimmune Diseases. Front. Pharmacol. 2018, 9, 1313. [Google Scholar] [CrossRef] [PubMed]
- Jazdzewski, K.; Bednarczuk, T.; Stepnowska, M.; Liyanarachchi, S.; Suchecka-Rachon, K.; Limon, J.; Narkiewicz, K. Beta-2-adrenergic receptor gene polymorphism confers susceptibility to Graves disease. Int. J. Mol. Med. 2007, 191, 181–186. [Google Scholar]
- Xu, B.; Ärlehag, L.; Rantapää-Dahlquist, B.S.; Lefvert, K.A. Beta2-adrenergic receptor gene single-nucleotide polymorphisms are associated with rheumatoid arthritis in northern Sweden. Scand. J. Rheumatol. 2004, 33, 395–398. [Google Scholar] [CrossRef]
- Györffy, B.; Lage, H. A Web-Based Data Warehouse on Gene Expression in Human Malignant Melanoma. J. Investig. Dermatol. 2007, 127, 394–399. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, D.; Scatozza, F.; Giampietri, C.; Marchetti, P.; Facchiano, F.; Facchiano, A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers 2019, 11, 446. [Google Scholar] [CrossRef] [PubMed]
- Biasiotta, A.; D’Arcangelo, D.; Passarelli, F.; Nicodemi, E.M.; Facchiano, A. Ion channels expression and function are strongly modified in solid tumors and vascular malformations. J. Transl. Med. 2016, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.D.; Lin, A.; Robb, L.; Josefsson, E.C.; Henley, K.J.; Gray, D.H.D.; Kile, B.T.; Roberts, A.W.; Strasser, A.; Huang, D.C.S.; et al. Proapoptotic Bak and Bax guard against fatal systemic and organ-specific autoimmune disease. Proc. Natl. Acad. Sci. USA 2013, 110, 2599–2604. [Google Scholar] [CrossRef] [PubMed]
- Teh, C.E.; Robbins, A.K.; Henstridge, D.C.; Dewson, G.; Diepstraten, S.T.; Kelly, G.; Febbraio, M.A.; Gabriel, S.S.; O’Reilly, L.A.; Strasser, A.; et al. MCL-1 is essential for survival but dispensable for metabolic fitness of FOXP3 + regulatory T cells. Cell Death Differ. 2020, 27, 3374–3385. [Google Scholar] [CrossRef]
- Chris ML Zhibin, C. Autoimmunity as an Etiological Factor of Cancer: The Transformative Potential of Chronic Type 2 Inflammation. Front. Cell Dev. Biol. 2021, 9, 664305. [Google Scholar] [CrossRef]
- Masetti, R.; Tiri, A.; Tignanelli, A.; Turrini, E.; Argentiero, A.; Pession, A.; Esposito, S. Autoimmunity and cancer. Autoimmun. Rev. 2021, 20, 102882. [Google Scholar] [CrossRef] [PubMed]
- Teulings, H.E.; Limpens, J.; Jansen, S.N.; Zwinderman, A.H.; Reitsma, J.B.; Phyllis ISpuls, P.I.; Luiten, R.M. M. Vitiligo-like depigmentation in patients with stage III-IV melanoma receiving immunotherapy and its association with survival: A systematic review and meta-analysis. J. Clin. Oncol. 2015, 33, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Paradisi, A.; Tabolli, S.; Didona, B.; Sobrino, L.; Russo, N.; Abeni, D. Markedly reduced incidence of melanoma and nonmelanoma skin cancer in a nonconcurrent cohort of 10,040 patients with vitiligo. J. Am. Acad. Dermatol. 2014, 71, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Chung KYSook Jung Yun SJHeesu Kim, H.; Byung Cheol Park, B.C.; Kim, J.S.; Seo, S.H.; Ahn, H.H.; Lee, D.; You Kim, Y.C.; Park, H.J.; Kim, M. Markedly reduced risk of internal malignancies in patients with vitiligo: A nationwide population-based cohort study. J. Clin. Oncol. 2019, 37, 903–911. [Google Scholar] [CrossRef]
- Nardin, C.; Pelletier, F.; Puzenat, E.; Aubin, F. Vitiligo repigmentation with melanoma progression during pembrolizumab treatment. Acta Derm. Venereol. 2019, 99, 913–914. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Chenwei Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Todorov, H.; Fournier, D.; Susanne Gerber, S. Principal Components Analysis: Theory and Application to Gene Expression Data Analysis. Genom. Comput. Biol. 2018, 4, e100041. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).