RAB20 Promotes Proliferation via G2/M Phase through the Chk1/cdc25c/cdc2-cyclinB1 Pathway in Penile Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Patient Cohort, Samples, and Research Ethics
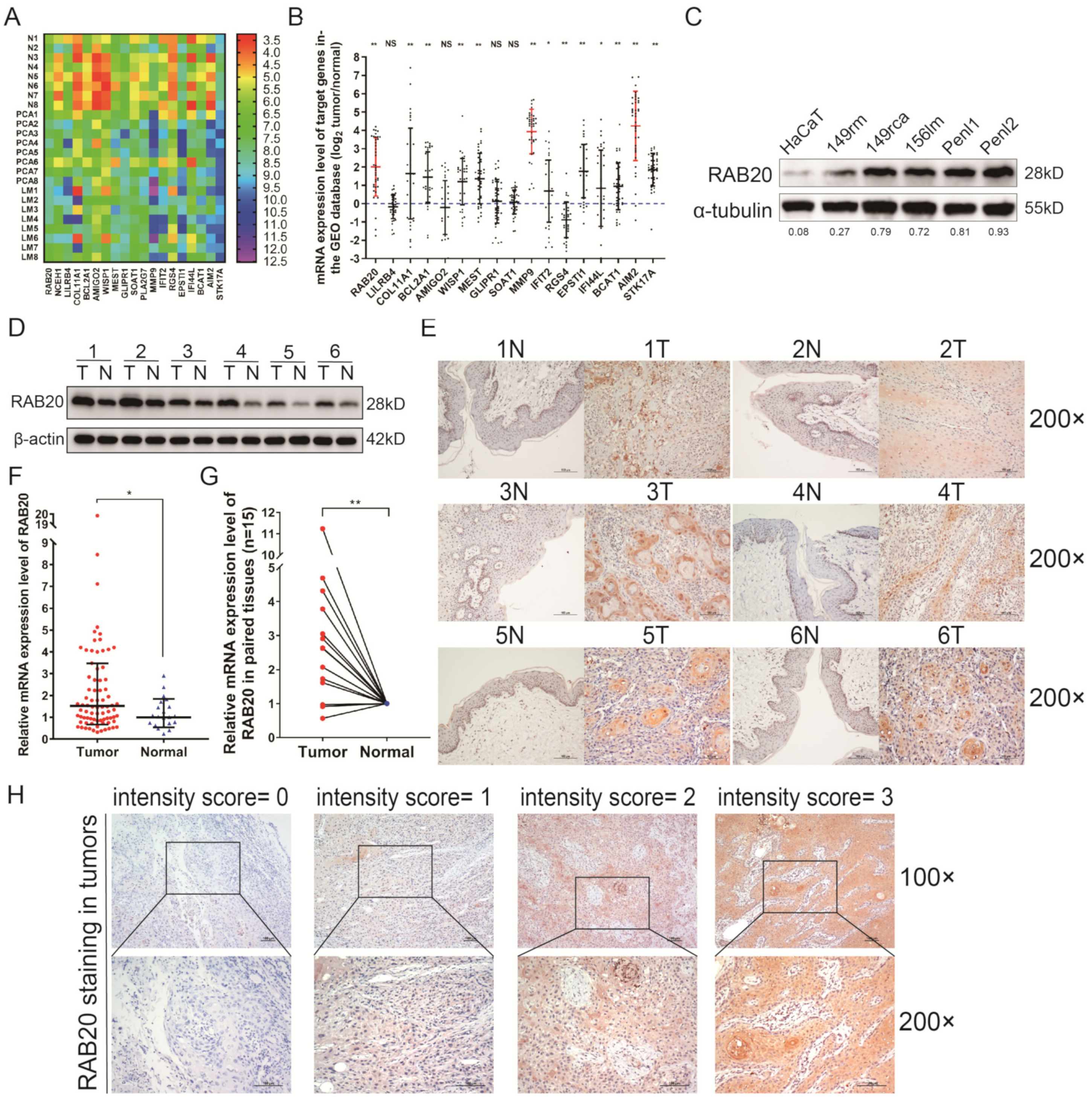
2.2. Target Gene Screening for the Progression of PSCC
2.3. Cell Lines, Culture Conditions, and Transfection Methods
2.4. Gene-Set Enrichment Analysis (GSEA)
2.5. Immunohistochemistry Assay
2.6. Western Blot (WB)
2.7. Quantitative Real-Time Polymerase Chain Reaction (qPCR) Assay
2.8. Cell Proliferation, Migration, Colony Formation, and Wound Healing Assays
2.9. Cell Cycle Assay
2.10. Xenograft Assay
3. Results
3.1. The Expression Profiles of mRNAs in PSCC Tissues
3.2. RAB20 Is Overexpressed in PSCC Cell Lines and Tissues
3.3. Overexpression of RAB20 Was Associated with Poor Clinical Features in PSCC
3.4. RAB20 Knockdown Inhibits Cell Proliferation and Cell Cycle Progression in PSCC
3.5. RAB20 Overexpression Promotes Cell Proliferation in PSCC
3.6. Knockdown of RAB20 Induced G2/M Cell Arrest in PSCC
3.7. RAB20 Regulates the Cell Cycle via the Chk1/cdc25c/cdc2-cyclinB1 Pathway in PSCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hakenberg, O.W.; Drager, D.L.; Erbersdobler, A.; Naumann, C.M.; Junemann, K.P.; Protzel, C. The Diagnosis and Treatment of Penile Cancer. Deutsches Ärzteblatt Int. 2018, 115, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ficarra, V.; Akduman, B.; Bouchot, O.; Palou, J.; Tobias-Machado, M. Prognostic factors in penile cancer. Urology 2010, 76, S66–S73. [Google Scholar] [CrossRef] [PubMed]
- Leone, A.; Diorio, G.J.; Pettaway, C.; Master, V.; Spiess, P.E. Contemporary management of patients with penile cancer and lymph node metastasis. Nat. Rev. Urol. 2017, 14, 335–347. [Google Scholar] [CrossRef]
- O’Brien, J.S.; Perera, M.; Manning, T.; Bozin, M.; Cabarkapa, S.; Chen, E.; Lawrentschuk, N. Penile Cancer: Contemporary Lymph Node Management. J. Urol. 2017, 197, 1387–1395. [Google Scholar] [CrossRef]
- Hu, J.; Cui, Y.; Liu, P.; Zhou, X.; Ren, W.; Chen, J.; Zu, X. Predictors of inguinal lymph node metastasis in penile cancer patients: A meta-analysis of retrospective studies. Cancer Manag. Res. 2019, 11, 6425–6441. [Google Scholar] [CrossRef] [Green Version]
- Busso-Lopes, A.F.; Marchi, F.A.; Kuasne, H.; Scapulatempo-Neto, C.; Trindade-Filho, J.C.; de Jesus, C.M.; Lopes, A.; Guimaraes, G.C.; Rogatto, S.R. Genomic profiling of human penile carcinoma predicts worse prognosis and survival. Cancer Prev. Res. 2015, 8, 149–156. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.M.; Pal, S.K.; Wang, K.; Palma, N.A.; Sanford, E.; Bailey, M.; He, J.; Elvin, J.A.; Chmielecki, J.; Squillace, R.; et al. Comprehensive Genomic Profiling of Advanced Penile Carcinoma Suggests a High Frequency of Clinically Relevant Genomic Alterations. Oncologist 2016, 21, 33–39. [Google Scholar] [CrossRef] [Green Version]
- McDaniel, A.S.; Hovelson, D.H.; Cani, A.K.; Liu, C.J.; Zhai, Y.; Zhang, Y.; Weizer, A.Z.; Mehra, R.; Feng, F.Y.; Alva, A.S.; et al. Genomic Profiling of Penile Squamous Cell Carcinoma Reveals New Opportunities for Targeted Therapy. Cancer Res. 2015, 75, 5219–5227. [Google Scholar] [CrossRef] [Green Version]
- Jacob, J.M.; Ferry, E.K.; Gay, L.M.; Elvin, J.A.; Vergilio, J.A.; Ramkissoon, S.; Severson, E.; Necchi, A.; Killian, J.K.; Ali, S.M.; et al. Comparative Genomic Profiling of Refractory and Metastatic Penile and Nonpenile Cutaneous Squamous Cell Carcinoma: Implications for Selection of Systemic Therapy. J. Urol. 2019, 201, 541–548. [Google Scholar] [CrossRef]
- Marchi, F.A.; Martins, D.C.; Barros-Filho, M.C.; Kuasne, H.; Busso Lopes, A.F.; Brentani, H.; Trindade Filho, J.C.S.; Guimaraes, G.C.; Faria, E.F.; Scapulatempo-Neto, C.; et al. Multidimensional integrative analysis uncovers driver candidates and biomarkers in penile carcinoma. Sci. Rep. 2017, 7, 6707. [Google Scholar] [CrossRef]
- Zhou, Q.H.; Han, H.; Lu, J.B.; Liu, T.Y.; Huang, K.B.; Deng, C.Z.; Li, Z.S.; Chen, J.P.; Yao, K.; Qin, Z.K.; et al. Up-regulation of indoleamine 2,3-dioxygenase 1 (IDO1) expression and catalytic activity is associated with immunosuppression and poor prognosis in penile squamous cell carcinoma patients. Cancer Commun. 2020, 40, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.H.; Deng, C.Z.; Chen, J.P.; Huang, K.B.; Liu, T.Y.; Yao, K.; Liu, Z.W.; Qin, Z.K.; Li, Y.H.; Guo, S.J.; et al. Elevated serum LAMC2 is associated with lymph node metastasis and predicts poor prognosis in penile squamous cell carcinoma. Cancer Manag. Res. 2018, 10, 2983–2995. [Google Scholar] [CrossRef] [Green Version]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Amillet, J.M.; Ferbus, D.; Real, F.X.; Antony, C.; Muleris, M.; Gress, T.M.; Goubin, G. Characterization of human Rab20 overexpressed in exocrine pancreatic carcinoma. Hum. Pathol. 2006, 37, 256–263. [Google Scholar] [CrossRef]
- Turner, N.; Lambros, M.B.; Horlings, H.M.; Pearson, A.; Sharpe, R.; Natrajan, R.; Geyer, F.C.; van Kouwenhove, M.; Kreike, B.; Mackay, A.; et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010, 29, 2013–2023. [Google Scholar] [CrossRef] [Green Version]
- Habermann, J.K.; Brucker, C.A.; Freitag-Wolf, S.; Heselmeyer-Haddad, K.; Kruger, S.; Barenboim, L.; Downing, T.; Bruch, H.P.; Auer, G.; Roblick, U.J.; et al. Genomic instability and oncogene amplifications in colorectal adenomas predict recurrence and synchronous carcinoma. Mod. Pathol. 2011, 24, 542–555. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.H.; Deng, C.Z.; Li, Z.S.; Chen, J.P.; Yao, K.; Huang, K.B.; Liu, T.Y.; Liu, Z.W.; Qin, Z.K.; Zhou, F.J.; et al. Molecular characterization and integrative genomic analysis of a panel of newly established penile cancer cell lines. Cell Death Dis. 2018, 9, 684. [Google Scholar] [CrossRef] [Green Version]
- Raman, T.; O’Connor, T.P.; Hackett, N.R.; Wang, W.; Harvey, B.G.; Attiyeh, M.A.; Dang, D.T.; Teater, M.; Crystal, R.G. Quality control in microarray assessment of gene expression in human airway epithelium. BMC Genom. 2009, 10, 493. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kuasne, H.; Colus, I.M.; Busso, A.F.; Hernandez-Vargas, H.; Barros-Filho, M.C.; Marchi, F.A.; Scapulatempo-Neto, C.; Faria, E.F.; Lopes, A.; Guimaraes, G.C.; et al. Genome-wide methylation and transcriptome analysis in penile carcinoma: Uncovering new molecular markers. Clin. Epigenet. 2015, 7, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, M.P.; Dong, J.; Wandinger-Ness, A. Rab proteins and endocytic trafficking: Potential targets for therapeutic intervention. Adv. Drug Deliv. Rev. 2003, 55, 1421–1437. [Google Scholar] [CrossRef] [PubMed]
- Camp, R.L.; Dolled-Filhart, M.; Rimm, D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Chen, D.; Guo, S.; Wang, Y.; Zou, Y.; Wu, Z.; Zhou, F.; Qin, Z.; Liu, Z.; Cao, Y.; et al. Molecular stratification by BCL2A1 and AIM2 provides additional prognostic value in penile squamous cell carcinoma. Theranostics 2021, 11, 1364–1376. [Google Scholar] [CrossRef]
- Vandooren, J.; Van den Steen, P.E.; Opdenakker, G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 222–272. [Google Scholar] [CrossRef]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef]
- Taylor, W.R.; Stark, G.R. Regulation of the G2/M transition by p53. Oncogene 2001, 20, 1803–1815. [Google Scholar] [CrossRef] [Green Version]
- Bartek, J.; Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Löbrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef]
- Bouldin, C.M.; Kimelman, D. Cdc25 and the importance of G2 control: Insights from developmental biology. Cell Cycle 2014, 13, 2165–2171. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.L.; Lin, J.Y.; Shieh, J.C.; Yeh, H.F.; Hsieh, Y.H.; Cheng, Y.C.; Lee, H.J.; Shen, C.Y.; Cheng, C.W. Induction of G2/M phase arrest by diosgenin via activation of Chk1 kinase and Cdc25C regulatory pathways to promote apoptosis in human breast cancer cells. Int. J. Mol. Sci. 2019, 21, 172. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef]
- Christodoulidou, M.; Sahdev, V.; Houssein, S.; Muneer, A. Epidemiology of penile cancer. Curr. Probl. Cancer 2015, 39, 126–136. [Google Scholar] [CrossRef]
- Pang, C.; Guan, Y.; Li, H.; Chen, W.; Zhu, G. Urologic cancer in China. Jpn. J. Clin. Oncol. 2016, 46, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Sali, A.P.; Menon, S.; Murthy, V.; Prakash, G.; Bakshi, G.; Joshi, A.; Desai, S.B. A Modified Histopathologic Staging in Penile Squamous Cell Carcinoma Predicts Nodal Metastasis and Outcome Better Than the Current AJCC Staging. Am. J. Surg. Pathol. 2020, 44, 1112–1117. [Google Scholar] [CrossRef]
- Liu, B.H.M.; Tey, S.K.; Mao, X.; Ma, A.P.Y.; Yeung, C.L.S.; Wong, S.W.K.; Ng, T.H.; Xu, Y.; Yao, Y.; Fung, E.Y.M.; et al. TPI1-reduced extracellular vesicles mediated by Rab20 downregulation promotes aerobic glycolysis to drive hepatocarcinogenesis. J. Extracell. Vesicles 2021, 10, e12135. [Google Scholar] [CrossRef]
- Oguchi, M.E.; Etoh, K.; Fukuda, M. Rab20, a novel Rab small GTPase that negatively regulates neurite outgrowth of PC12 cells. Neurosci. Lett. 2018, 662, 324–330. [Google Scholar] [CrossRef]




| RAB20 IHC Staining | |||||
|---|---|---|---|---|---|
| Variable | PSCC Cohort (n = 259), % | Low Expression (n = 112), % | High Expression (n = 147), % | χ2 | p-Value a |
| Age | 0.424 | 0.515 | |||
| <55 | 149 (57.5) | 67 (25.9) | 82 (31.7) | ||
| ≥55 | 110 (42.5) | 45 (17.4) | 65 (25.1) | ||
| pT status | 17.137 | 0.002 b | |||
| ≤pT1 c | 94 (36.3) | 56 (21.6) | 38 (14.7) | ||
| pT2 | 35 (13.5) | 13 (5.0) | 22 (8.5) | ||
| pT3 | 96 (37.1) | 34 (13.1) | 62 (23.9) | ||
| pT4 | 11 (4.2) | 2 (0.8) | 9 (3.5) | ||
| Tx | 23 (8.9) | 7 (2.7) | 16 (6.2) | ||
| pN status | 35.484 | 0.000 | |||
| N0 | 123 (47.5) | 75 (29.0) | 48 (18.5) | ||
| N1 | 32 (12.4) | 11 (4.2) | 21 (8.1) | ||
| N2 | 32 (12.4) | 13 (5.0) | 19 (7.3) | ||
| N3 | 72 (27.8) | 13 (5.0) | 59 (22.8) | ||
| Metastasis | 8.679 | 0.003 b | |||
| M0 | 244 (94.2) | 111 (42.9) | 133 (51.4) | ||
| M1 | 15 (5.8) | 1 (0.4) | 14 (5.4) | ||
| Clinical stage d | 39.060 | 0.000 | |||
| Stage I | 65 (25.1) | 46 (17.8) | 19 (7.3) | ||
| Stage II | 57 (22.0) | 29 (11.2) | 28 (10.8) | ||
| Stage III | 56 (21.6) | 20 (7.7) | 36 (13.9) | ||
| Stage IV | 81 (31.3) | 17 (6.6) | 64 (24.7) | ||
| Histology | 3.322 | 0.190 | |||
| G1 | 133 (51.4) | 63 (24.3) | 70 (27.0) | ||
| G2 | 90 (34.7) | 38 (14.7) | 52 (20.1) | ||
| G3 | 36 (13.9) | 11 (4.2) | 25 (9.7) | ||
| ENE | 19.743 | 0.000 | |||
| No | 199 (76.8) | 101 (39.0) | 98 (37.8) | ||
| Yes | 60 (23.2) | 11 (4.2) | 49 (18.9) | ||
| Univariate Analysis a | Multivariate Analysis b | |||||
|---|---|---|---|---|---|---|
| Variable | Total n | Events (%) | 5-Year CSS Rate (95% Cl) | p-Value | Hazard Ratio (95% Cl) | p-Value |
| Age | 0.037 | 0.271 | ||||
| <55 | 149 | 51 (34.2) | 0.665 (0.583–0.747) | Reference | ||
| ≥55 | 110 | 49 (44.5) | 0.542 (0.442–0.642) | 1.304 (0.813–2.091) | ||
| pT status c | 0.000 | 0.039 | ||||
| ≤pT1 | 94 | 18 (19.1) | 0.829 (0.747–0.911) | Reference | Reference | - |
| pT2 | 35 | 15 (42.9) | 0.539 (0.365–0.713) | 0.001 | 2.135 (1.027–4.438) | 0.042 |
| pT3 | 96 | 41 (42.7) | 0.589 (0.483–0.695) | 0.000 | 1.970 (1.101–3.526) | 0.022 |
| pT4 | 11 | 10 (90.9) | 0.000 | 0.000 | 3.307 (1.290–8.479) | 0.013 |
| Histology | 0.000 | 0.109 | ||||
| G1 | 133 | 37 (27.8) | 0.746 (0.670–0.822) | Reference | Reference | - |
| G2 | 90 | 35 (38.9) | 0.550 (0.428–0.672) | 0.010 | 1.005 (0.567–1.781) | 0.986 |
| G3 | 36 | 28 (77.8) | 0.248 (0.097–0.399) | 0.000 | 1.766 (0.936–3.332) | 0.079 |
| pN status | 0.000 | 0.000 | ||||
| N0 | 123 | 13 (10.6) | 0.910 (0.853–0.967) | Reference | Reference | - |
| N1 | 32 | 11 (34.4) | 0.664 (0.492–0.836) | 0.000 | 2.135 (1.027–4.438) | 0.005 |
| N2 | 32 | 16 (50.0) | 0.481 (0.289–0.673) | 0.000 | 1.970 (1.101–3.526) | 0.000 |
| N3 | 72 | 60 (83.3) | 0.112 (0.020–0.204) | 0.000 | 3.307 (1.290–8.479) | 0.000 |
| Metastasis | 0.000 | 0.008 | ||||
| M0 | 244 | 85 (34.8) | 0.655 (0.590–0.720) | Reference | ||
| M1 | 15 | 15 (100) | 0.000 | 2.686 (1.291–5.588) | ||
| Clinical stage d | 0.000 | |||||
| Stage I | 65 | 7 (10.8) | 0.900 (0.816–0.984) | Reference | Excluded e | |
| Stage II | 57 | 5 (8.8) | 0.942 (0.879–1.000) | 0.776 | ||
| Stage III | 56 | 21 (37.5) | 0.636 (0.499–0.773) | 0.000 | ||
| Stage IV | 81 | 67 (82.7) | 0.104 (0.020–0.188) | 0.000 | ||
| ENE | 0.000 | 0.109 | ||||
| No | 199 | 52 (26.1) | 0.754 (0.689–0.819) | Reference | ||
| Yes | 60 | 48 (80.0) | 0.116 (0.008–0.224) | 1.293 (0.600–2.786) | ||
| RAB20 | 0.000 | 0.011 | ||||
| Low expression | 112 | 21 (18.7) | 0.819 (0.739–0.899) | Reference | ||
| High expression | 147 | 79 (53.7) | 0.467 (0.383–0.551) | 2.090 (1.183–4.692) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, X.; Yuan, G.; Wang, Y.; Zou, Y.; Luo, S.; Han, H.; Qin, Z.; Liu, Z.; Zhou, F.; Liu, Y.; et al. RAB20 Promotes Proliferation via G2/M Phase through the Chk1/cdc25c/cdc2-cyclinB1 Pathway in Penile Squamous Cell Carcinoma. Cancers 2022, 14, 1106. https://doi.org/10.3390/cancers14051106
Tan X, Yuan G, Wang Y, Zou Y, Luo S, Han H, Qin Z, Liu Z, Zhou F, Liu Y, et al. RAB20 Promotes Proliferation via G2/M Phase through the Chk1/cdc25c/cdc2-cyclinB1 Pathway in Penile Squamous Cell Carcinoma. Cancers. 2022; 14(5):1106. https://doi.org/10.3390/cancers14051106
Chicago/Turabian StyleTan, Xingliang, Gangjun Yuan, Yanjun Wang, Yuantao Zou, Sihao Luo, Hui Han, Zike Qin, Zhuowei Liu, Fangjian Zhou, Yanling Liu, and et al. 2022. "RAB20 Promotes Proliferation via G2/M Phase through the Chk1/cdc25c/cdc2-cyclinB1 Pathway in Penile Squamous Cell Carcinoma" Cancers 14, no. 5: 1106. https://doi.org/10.3390/cancers14051106
APA StyleTan, X., Yuan, G., Wang, Y., Zou, Y., Luo, S., Han, H., Qin, Z., Liu, Z., Zhou, F., Liu, Y., & Yao, K. (2022). RAB20 Promotes Proliferation via G2/M Phase through the Chk1/cdc25c/cdc2-cyclinB1 Pathway in Penile Squamous Cell Carcinoma. Cancers, 14(5), 1106. https://doi.org/10.3390/cancers14051106






