NRG1/ERBB3/ERBB2 Axis Triggers Anchorage-Independent Growth of Basal-like/Triple-Negative Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of Breast Cancer Patients’ Data
2.2. Cell Cultures
2.3. Proliferation and Random/Directional Migration Analysis in Monolayer Conditions
2.4. Anchorage-Independent Growth Assay
2.5. RNA Isolation and Real Time PCR
2.6. Analysis of Cell Cycle Activity on Cultured Cells by Immunofluorescence Evaluation of KI67
2.7. Protein Extraction and Evaluation by Western Blot Analysis
2.8. Cell Survival Analysis by Flow Cytometry
2.9. Statistical Analysis
3. Results
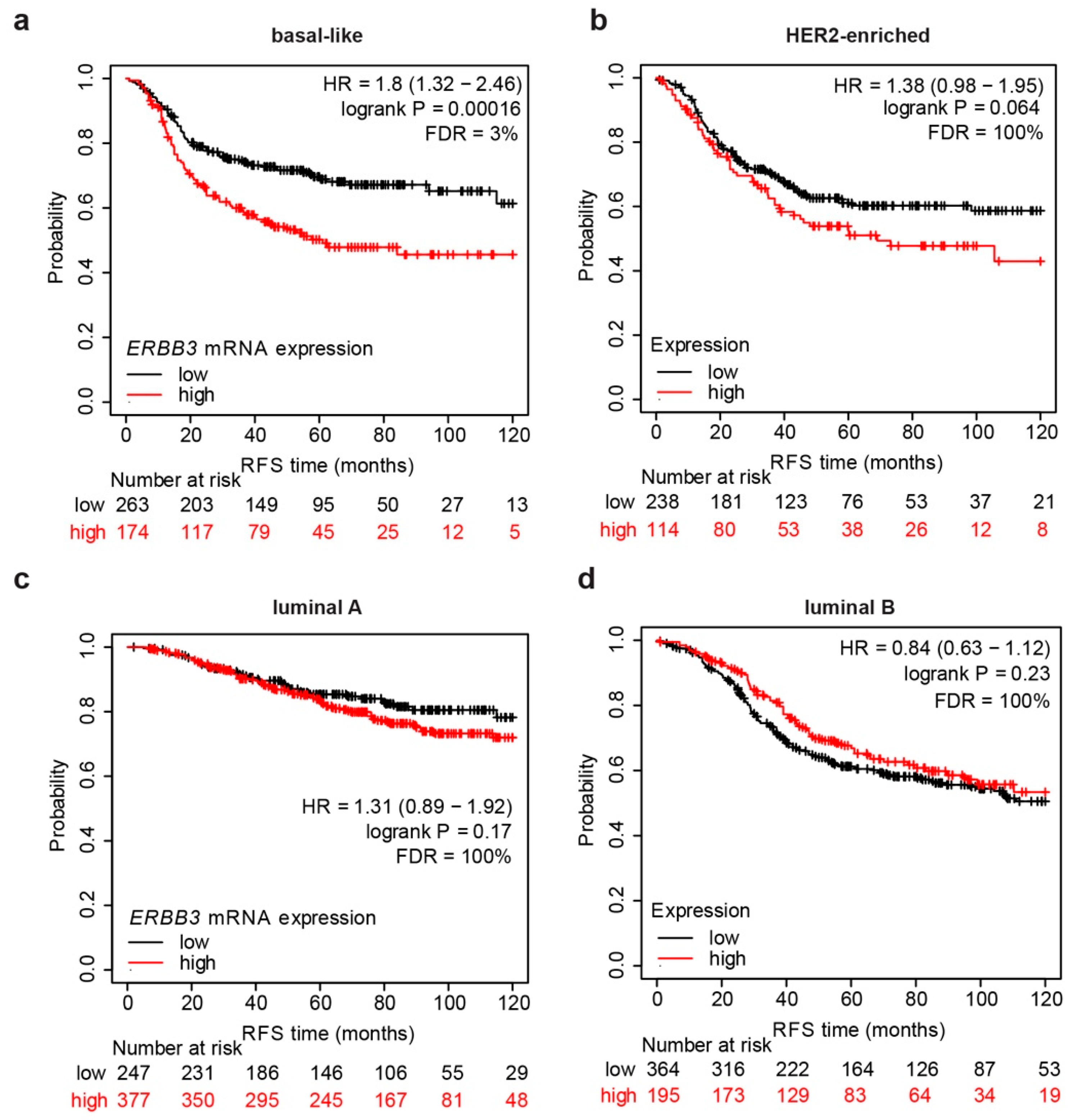
3.1. mRNA Expression Levels of ERBB3 Correlate with Shorter Relapse-Free Survival in Basal-like Breast Cancer Patients
3.2. Neuregulin 1β (NRG1β) Does Not Significantly Induce Proliferation or Motility in Basal-like/Triple-Negative Breast Cells Cultured in Monolayer Conditions
3.3. Administration of NRG1β Does Not Significantly Impact Cell Differentiation in Basal-like/Triple-Negative Breast Cells
3.4. NRG1β/ERBB3/ERBB2 Robustly Promotes Anchorage-Independent Cell Growth of Basal-like Breast Cells
3.5. Pertuzumab Efficiently Blocks NRG1β-Induced Cell Growth in Non-Adherent Conditions in HER2-Overexpressing Basal-like Breast Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yarden, Y.; Pines, G. The ERBB network: At last, cancer therapy meets systems biology. Nat. Rev. Cancer 2012, 12, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, C.L.; Engelman, J.A. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014, 25, 282–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Citri, A.; Skaria, K.B.; Yarden, Y. The deaf and the dumb: The biology of ErbB-2 and ErbB-3. Exp Cell Res 2003, 284, 54–65. [Google Scholar] [CrossRef]
- Baselga, J.; Swain, S.M. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 2009, 9, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, B.S.; Kljavin, N.M.; Stawiski, E.W.; Chan, E.; Parikh, C.; Durinck, S.; Chaudhuri, S.; Pujara, K.; Guillory, J.; Edgar, K.A.; et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 2013, 23, 603–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiavue, N.; Cabel, L.; Melaabi, S.; Bataillon, G.; Callens, C.; Lerebours, F.; Pierga, J.-Y.; Bidard, F.-C. ERBB3 mutations in cancer: Biological aspects, prevalence and therapeutics. Oncogene 2020, 39, 487–502. [Google Scholar] [CrossRef]
- Tzahar, E.; Waterman, H.; Chen, X.; Levkowitz, G.; Karunagaran, D.; Lavi, S.; Ratzkin, B.J.; Yarden, Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 1996, 16, 5276–5287. [Google Scholar] [CrossRef] [Green Version]
- Holbro, T.; Beerli, R.R.; Maurer, F.; Koziczak, M.; Barbas, C.F.; Hynes, N.E. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 2003, 100, 8933–8938. [Google Scholar] [CrossRef] [Green Version]
- Vaught, D.B.; Stanford, J.C.; Young, C.; Hicks, D.J.; Wheeler, F.; Rinehart, C.; Sanchez, V.; Koland, J.; Muller, W.J.; Arteaga, C.L.; et al. HER3 Is Required for HER2-Induced Preneoplastic Changes to the Breast Epithelium and Tumor Formation. Cancer Res. 2012, 72, 2672–2682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.Y.-F.; Lin, Y.; Bratman, S.V.; Feng, W.; Kuo, A.H.; Scheeren, F.A.; Engreitz, J.M.; Varma, S.; West, R.B.; Diehn, M. Neuregulin autocrine signaling promotes self-renewal of breast tumor-initiating cells by triggering HER2/HER3 activation. Cancer Res. 2014, 74, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Kim, J.; Lee, Y.; Seo, J.H.; Hong, S.R.; Kim, A. Neuregulin-1 induces cancer stem cell characteristics in breast cancer cell lines. Oncol. Rep. 2014, 32, 1218–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Núñez Abad, M.; Calabuig-Fariñas, S.; Lobo de Mena, M.; Torres-Martínez, S.; García González, C.; García García, J.Á.; Iranzo González-Cruz, V.; Camps Herrero, C. Programmed Death-Ligand 1 (PD-L1) as Immunotherapy Biomarker in Breast Cancer. Cancers 2022, 14, 307. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Bernard, P.S.; Perou, C.M. Molecular portraits and the family tree of cancer. Nat. Genet. 2002, 32, 533–540. [Google Scholar] [CrossRef] [PubMed]
- van ’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnitt, S.J. Classification and prognosis of invasive breast cancer: From morphology to molecular taxonomy. Mod. Pathol. 2010, 23 (Suppl. 2), S60–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheang, M.C.U.; Voduc, D.; Bajdik, C.; Leung, S.; McKinney, S.; Chia, S.K.; Perou, C.M.; Nielsen, T.O. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin. Cancer Res. 2008, 14, 1368–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: Intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020, 6, 54. [Google Scholar] [CrossRef]
- Badve, S.; Dabbs, D.J.; Schnitt, S.J.; Baehner, F.L.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011, 24, 157–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Jézéquel, P.; Gouraud, W.; Ben Azzouz, F.; Guérin-Charbonnel, C.; Juin, P.P.; Lasla, H.; Campone, M. bc-GenExMiner 4.5: New mining module computes breast cancer differential gene expression analyses. Database 2021, 2021, baab007. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Ringnér, M.; Fredlund, E.; Häkkinen, J.; Borg, Å.; Staaf, J. GOBO: Gene expression-based outcome for breast cancer online. PLoS ONE 2011, 6, e17911. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Pradeep, C.R.; Kostler, W.J.; Lauriola, M.; Granit, R.Z.; Zhang, F.; Jacob-Hirsch, J.; Rechavi, G.; Nair, H.B.; Hennessy, B.T.; Gonzalez-Angulo, A.M.; et al. Modeling ductal carcinoma in situ: A HER2-Notch3 collaboration enables luminal filling. Oncogene 2012, 31, 907–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradeep, C.R.; Zeisel, A.; Kostler, W.J.; Lauriola, M.; Jacob-Hirsch, J.; Haibe-Kains, B.; Amariglio, N.; Ben-Chetrit, N.; Emde, A.; Solomonov, I.; et al. Modeling invasive breast cancer: Growth factors propel progression of HER2-positive premalignant lesions. Oncogene 2012, 31, 3569–3583. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Han, B.; Yu, Y.; Yao, W.; Bose, S.; Karlan, B.Y.; Giuliano, A.E.; Cui, X. Evaluation of MCF10A as a Reliable Model for Normal Human Mammary Epithelial Cells. PLoS ONE 2015, 10, e0131285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauriola, M.; Enuka, Y.; Zeisel, A.; D’Uva, G.; Roth, L.; Sharon-Sevilla, M.; Lindzen, M.; Sharma, K.; Nevo, N.; Feldman, M.; et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 2014, 5, 5073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amit, I.; Citri, A.; Shay, T.; Lu, Y.; Katz, M.; Zhang, F.; Tarcic, G.; Siwak, D.; Lahad, J.; Jacob-Hirsch, J.; et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007, 39, 503–512. [Google Scholar] [CrossRef]
- Muellner, M.K.; Mair, B.; Ibrahim, Y.; Kerzendorfer, C.; Lechtermann, H.; Trefzer, C.; Klepsch, F.; Müller, A.C.; Leitner, E.; Macho-Maschler, S.; et al. Targeting a cell state common to triple-negative breast cancers. Mol. Syst. Biol. 2015, 11, 789. [Google Scholar] [CrossRef]
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010, 32, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Saeki, K.; Chang, G.; Kanaya, N.; Wu, X.; Wang, J.; Bernal, L.; Ha, D.; Neuhausen, S.L.; Chen, S. Mammary cell gene expression atlas links epithelial cell remodeling events to breast carcinogenesis. Commun. Biol. 2021, 4, 660. [Google Scholar] [CrossRef]
- Wuidart, A.; Sifrim, A.; Fioramonti, M.; Matsumura, S.; Brisebarre, A.; Brown, D.; Centonze, A.; Dannau, A.; Dubois, C.; Van Keymeulen, A.; et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 2018, 20, 666–676. [Google Scholar] [CrossRef]
- Cristea, S.; Polyak, K. Dissecting the mammary gland one cell at a time. Nat. Commun. 2018, 9, 2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020, 100, 489–523. [Google Scholar] [CrossRef]
- Seldin, L.; Le Guelte, A.; Macara, I.G. Epithelial plasticity in the mammary gland. Curr. Opin. Cell Biol. 2017, 49, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Hennighausen, L.; Robinson, G.W. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 2005, 6, 715–725. [Google Scholar] [CrossRef]
- Stingl, J.; Caldas, C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat. Rev. Cancer 2007, 7, 791–799. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U SA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef]
- Junttila, T.T.; Akita, R.W.; Parsons, K.; Fields, C.; Lewis Phillips, G.D.; Friedman, L.S.; Sampath, D.; Sliwkowski, M.X. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell 2009, 15, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.-S.; Mason, K.; Ramyar, K.X.; Stanley, A.M.; Gabelli, S.B.; Denney, D.W.; Leahy, D.J. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003, 421, 756–760. [Google Scholar] [CrossRef]
- Capelan, M.; Pugliano, L.; De Azambuja, E.; Bozovic, I.; Saini, K.S.; Sotiriou, C.; Loi, S.; Piccart-Gebhart, M.J. Pertuzumab: New hope for patients with HER2-positive breast cancer. Ann. Oncol. 2013, 24, 273–282. [Google Scholar] [CrossRef]
- Agus, D.B.; Akita, R.W.; Fox, W.D.; Lewis, G.D.; Higgins, B.; Pisacane, P.I.; Lofgren, J.A.; Tindell, C.; Evans, D.P.; Maiese, K.; et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell 2002, 2, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Miglietta, F.; Griguolo, G.; Bottosso, M.; Giarratano, T.; Lo Mele, M.; Fassan, M.; Cacciatore, M.; Genovesi, E.; De Bartolo, D.; Vernaci, G.; et al. Evolution of HER2-low expression from primary to recurrent breast cancer. NPJ Breast Cancer 2021, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Shu, L.; Chen, A.; Li, L.; Yao, L.; He, Y.; Xu, J.; Gu, W.; Li, Q.; Wang, K.; Zhang, T.; et al. NRG1 regulates Fra-1 transcription and metastasis of triple-negative breast cancer cells via the c-Myc ubiquitination as manipulated by ERK1/2-mediated Fbxw7 phosphorylation. Oncogene 2022, 41, 907–919. [Google Scholar] [CrossRef]
- Aceto, N.; Duss, S.; MacDonald, G.; Meyer, D.S.; Roloff, T.-C.; Hynes, N.E.; Bentires-Alj, M. Co-expression of HER2 and HER3 receptor tyrosine kinases enhances invasion of breast cells via stimulation of interleukin-8 autocrine secretion. Breast Cancer Res. 2012, 14, R131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerji, U.; van Herpen, C.M.L.; Saura, C.; Thistlethwaite, F.; Lord, S.; Moreno, V.; Macpherson, I.R.; Boni, V.; Rolfo, C.; de Vries, E.G.E.; et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: A phase 1 dose-escalation and dose-expansion study. Lancet. Oncol. 2019, 20, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Modi, S.; Park, H.; Murthy, R.K.; Iwata, H.; Tamura, K.; Tsurutani, J.; Moreno-Aspitia, A.; Doi, T.; Sagara, Y.; Redfern, C.; et al. Antitumor Activity and Safety of Trastuzumab Deruxtecan in Patients with HER2-Low-Expressing Advanced Breast Cancer: Results from a Phase Ib Study. J. Clin. Oncol. 2020, 38, 1887–1896. [Google Scholar] [CrossRef]
- Eiger, D.; Agostinetto, E.; Saúde-Conde, R.; de Azambuja, E. The Exciting New Field of HER2-Low Breast Cancer Treatment. Cancers 2021, 13, 1015. [Google Scholar] [CrossRef]
- Gianni, L.; Lladó, A.; Bianchi, G.; Cortes, J.; Kellokumpu-Lehtinen, P.-L.; Cameron, D.A.; Miles, D.; Salvagni, S.; Wardley, A.; Goeminne, J.-C.; et al. Open-label, phase II, multicenter, randomized study of the efficacy and safety of two dose levels of Pertuzumab, a human epidermal growth factor receptor 2 dimerization inhibitor, in patients with human epidermal growth factor receptor 2-negative metastat. J. Clin. Oncol. 2010, 28, 1131–1137. [Google Scholar] [CrossRef] [Green Version]
- Chervo, M.F.; Cordo Russo, R.I.; Petrillo, E.; Izzo, F.; De Martino, M.; Bellora, N.; Cenciarini, M.E.; Chiauzzi, V.A.; Santa María de la Parra, L.; Pereyra, M.G.; et al. Canonical ErbB-2 isoform and ErbB-2 variant c located in the nucleus drive triple negative breast cancer growth. Oncogene 2020, 39, 6245–6262. [Google Scholar] [CrossRef]
- Phillips, G.D.L.; Fields, C.T.; Li, G.; Dowbenko, D.; Schaefer, G.; Miller, K.; Andre, F.; Burris, H.A.; Albain, K.S.; Harbeck, N.; et al. Dual targeting of HER2-positive cancer with trastuzumab emtansine and pertuzumab: Critical role for neuregulin blockade in antitumor response to combination therapy. Clin. Cancer Res. 2014, 20, 456–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, A.W.; Habbous, S.; Hoey, C.; Jerzak, K.J.; Das, S. Dual- versus single-agent HER2 inhibition and incidence of intracranial metastatic disease: A systematic review and meta-analysis. NPJ Breast Cancer 2021, 7, 17. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miano, C.; Morselli, A.; Pontis, F.; Bongiovanni, C.; Sacchi, F.; Da Pra, S.; Romaniello, D.; Tassinari, R.; Sgarzi, M.; Pantano, E.; et al. NRG1/ERBB3/ERBB2 Axis Triggers Anchorage-Independent Growth of Basal-like/Triple-Negative Breast Cancer Cells. Cancers 2022, 14, 1603. https://doi.org/10.3390/cancers14071603
Miano C, Morselli A, Pontis F, Bongiovanni C, Sacchi F, Da Pra S, Romaniello D, Tassinari R, Sgarzi M, Pantano E, et al. NRG1/ERBB3/ERBB2 Axis Triggers Anchorage-Independent Growth of Basal-like/Triple-Negative Breast Cancer Cells. Cancers. 2022; 14(7):1603. https://doi.org/10.3390/cancers14071603
Chicago/Turabian StyleMiano, Carmen, Alessandra Morselli, Francesca Pontis, Chiara Bongiovanni, Francesca Sacchi, Silvia Da Pra, Donatella Romaniello, Riccardo Tassinari, Michela Sgarzi, Elvira Pantano, and et al. 2022. "NRG1/ERBB3/ERBB2 Axis Triggers Anchorage-Independent Growth of Basal-like/Triple-Negative Breast Cancer Cells" Cancers 14, no. 7: 1603. https://doi.org/10.3390/cancers14071603





