Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection
Abstract
:Simple Summary
Abstract
1. Introduction
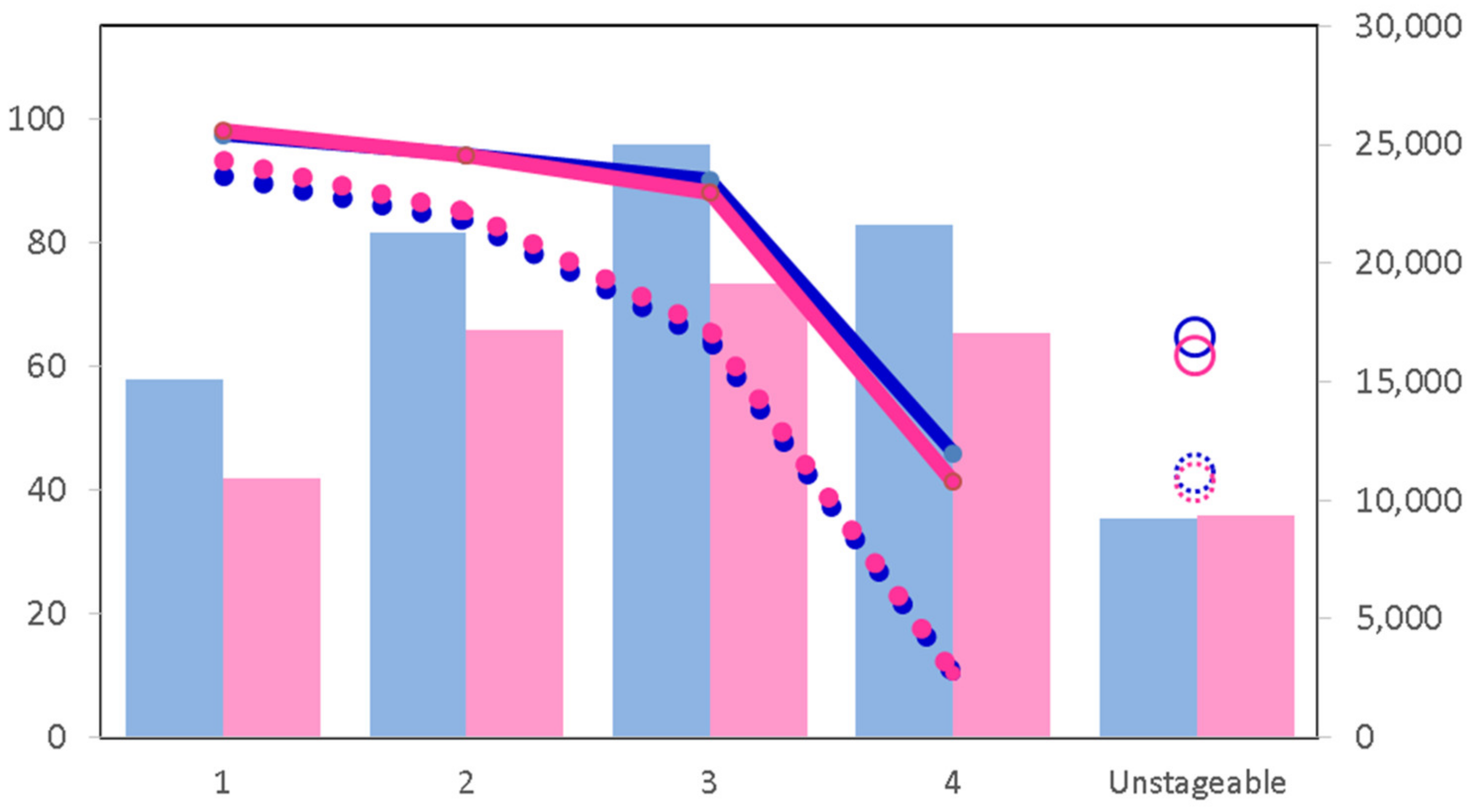
2. Detection and Screening in Colorectal Cancer
3. Pathogenesis of Colorectal Cancer
4. Obstacles and Limitations to the Use of Biomarkers as a Screening Tool
5. Current Advances in CRC Biomarker Detection
5.1. DNA-Based Molecular Markers and Tests
5.2. Circulating Tumor Cells
5.3. microRNAs and Other Non-Coding RNAs
5.4. Differential Gene and Protein Expression in CRC
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Abd Algfoor, Z.; Shahrizal Sunar, M.; Abdullah, A.; Kolivand, H. Identification of metabolic pathways using pathfinding approaches: A systematic review. Brief. Funct. Genom. 2017, 16, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Noori, R.; Marolin, I.; Franken, P.; Brandao, J.; Biermann, K.; Collini, P.; Grigorian, M.; Lukanidin, E.; Ambartsumian, N.; et al. The role of S100a4 (Mts1) in Apc- and Smad4-driven tumour onset and progression. Eur. J. Cancer 2016, 68, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, Y. Alternative RNA splicing and gastric cancer. Mutat. Res. 2017, 773, 263–273. [Google Scholar] [CrossRef]
- Muller, M.F.; Ibrahim, A.E.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. Int. J. Pathol. 2016, 469, 125–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, B.D.; James, T.; East, J.E.; Grimshaw, D.; Paddon, M.; Justice, S.; Oke, J.L.; Shine, B. Experience of adopting faecal immunochemical testing to meet the NICE colorectal cancer referral criteria for low-risk symptomatic primary care patients in Oxfordshire, UK. Frontline Gastroenterol. 2019, 10, 347–355. [Google Scholar] [CrossRef]
- O’Connor, L.; Gilmour, J.; Bonifer, C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J. Biol. Med. 2016, 89, 513–525. [Google Scholar]
- Cancer Research, UK. Bowel Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer (accessed on 12 October 2021).
- Huang, Q.; Li, S.; Cheng, P.; Deng, M.; He, X.; Wang, Z.; Yang, C.-H.; Zhao, X.-Y.; Huang, J. High expression of anti-apoptotic protein Bcl-2 is a good prognostic factor in colorectal cancer: Result of a meta-analysis. World J. Gastroenterol. 2017, 23, 5018–5033. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef]
- Whiffin, N.; Hosking, F.J.; Farrington, S.M.; Palles, C.; Dobbins, S.E.; Zgaga, L.; Lloyd, A.; Kinnersley, B.; Gorman, M.; Tenesa, A.; et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 2014, 23, 4729–4737. [Google Scholar] [CrossRef] [Green Version]
- Nandakumar, G.; Morgan, J.A.; Silverberg, D.; Steinhagen, R.M. Familial polyposis coli: Clinical manifestations, evaluation, management and treatment. Mt. Sinai. J. Med. 2004, 71, 384–391. [Google Scholar]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef] [Green Version]
- Shussman, N.; Wexner, S.D. Colorectal polyps and polyposis syndromes. Gastroenterol. Rep. 2014, 2, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ahnen, D.J. The American College of Gastroenterology Emily Couric Lecture—The adenoma-carcinoma sequence revisited: Has the era of genetic tailoring finally arrived? Am. J. Gastroenterol. 2011, 106, 190–198. [Google Scholar] [CrossRef]
- Wieszczy, P.; Kaminski, M.; Franczyk, R.; Loberg, M.; Kobiela, J.; Rupinska, M.; Kocot, B.; Rupinski, M.; Holme, O.; Wojciechowska, U.; et al. Colorectal Cancer Incidence and Mortality After Removal of Adenomas During Screening Colonoscopies. Gastroenterology 2020, 158, 875–883.e5. [Google Scholar] [CrossRef]
- Morgan, J.; Thomas, K.; Lee-Robichaud, H.; Nelson, R.L.; Braungart, S. Transparent cap colonoscopy versus standard colonoscopy to improve caecal intubation. Cochrane Database Syst. Rev. 2012, 12, Cd008211. [Google Scholar] [CrossRef]
- Nayor, J.; Saltzman, J.R. Colonoscopy quality: Measuring the patient experience. Endoscopy 2018, 50, 4–5. [Google Scholar]
- National Cancer Research Institute. National Cancer Registry. Available online: http://www.ncin.org.uk/cancer_type_and_topic_specific_work/topic_specific_work/tww_conversion_and_detection (accessed on 14 October 2021).
- NHS England. National Tariff Payment System. Available online: https://www.england.nhs.uk/publication/national-tariff-payment-system-documents-annexes-and-supporting-documents/ (accessed on 14 October 2021).
- Cancer Research, UK. Bowel Cancer Survival. Available online: https://www.cancerresearchuk.org/about-cancer/bowel-cancer/survival (accessed on 24 October 2021).
- Cancer Research, UK. Bowel Cancer Survial Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer/survival#heading-Three (accessed on 10 February 2022).
- Lansdorp-Vogelaar, I.; von Karsa, L. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First Edition—Introduction. Endoscopy 2012, 44 (Suppl. S3), 15–30. [Google Scholar] [CrossRef] [Green Version]
- Triantafillidis, J.K.; Vagianos, C.; Malgarinos, G. Colonoscopy in Colorectal Cancer Screening: Current Aspects. Indian J. Surg. Oncol. 2015, 6, 237–250. [Google Scholar] [CrossRef] [Green Version]
- Atkin, W.S.; Edwards, R.; Kralj-Hans, I.; Wooldrage, K.; Hart, A.R.; Northover, J.M.; Parkin, D.M.; Wardle, J.; Duffy, S.W.; Cuzick, J. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: A multicentre randomised controlled trial. Lancet 2010, 375, 1624–1633. [Google Scholar] [CrossRef] [Green Version]
- NHS, UK. NHS UK Bowel Screening Guidelines. Available online: https://www.nhs.uk/conditions/bowel-cancer-screening/ (accessed on 14 October 2021).
- D’Souza, N.; Georgiou Delisle, T.; Chen, M.; Benton, S.; Abulafi, M. Faecal immunochemical test is superior to symptoms in predicting pathology in patients with suspected colorectal cancer symptoms referred on a 2WW pathway: A diagnostic accuracy study. Gut 2021, 70, 1130–1138. [Google Scholar] [CrossRef]
- FIT Screening. OC Sensor. Available online: http://www.fit-screening.co.uk/ (accessed on 25 January 2022).
- NICE, UK. Quantitative Faecal Immunochemical Tests to Guide Referral for Colorectal Cancer in Primary Care. Available online: https://www.nice.org.uk/guidance/dg30/chapter/3-The-diagnostic-tests (accessed on 26 January 2022).
- Alpha, L. The HM-JACKarc System. Available online: https://www.faecal-immunochemical-test.co.uk/products/ (accessed on 1 February 2022).
- R-Biopharm AG. RIDASCREEN® Haemoglobin. Available online: https://clinical.r-biopharm.com/products/ridascreen-haemoglobin-2/ (accessed on 1 February 2022).
- Sentinel Diagnostics. FOB Gold. Available online: https://www.sentineldiagnostics.com/fit-line/fob-gold-line/fob-gold/ (accessed on 1 February 2022).
- Cardoso, R.; Guo, F.; Heisser, T.; Hackl, M.; Ihle, P.; De Schutter, H.; Van Damme, N.; Valerianova, Z.; Atanasov, T.; Májek, O.; et al. Colorectal cancer incidence, mortality, and stage distribution in European countries in the colorectal cancer screening era: An international population-based study. Lancet Oncol. 2021, 22, 1002–1013. [Google Scholar] [CrossRef]
- CDC. Colorectal Cancer Screening Tests. Available online: https://www.cdc.gov/cancer/colorectal/basic_info/screening/tests.htm (accessed on 14 October 2021).
- Ladabaum, U.; Mannalithara, A. Comparative Effectiveness and Cost Effectiveness of a Multitarget Stool DNA Test to Screen for Colorectal Neoplasia. Gastroenterology 2016, 151, 427–439.e6. [Google Scholar] [CrossRef] [Green Version]
- Das, V.; Kalita, J.; Pal, M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed. Pharm. 2017, 87, 8–19. [Google Scholar] [CrossRef]
- Au, F.C.; Stein, B.; Tang, C.K. Carcinoembryonic antigen levels in colonic lesions. Am. J. Surg. 1986, 151, 61–64. [Google Scholar] [CrossRef]
- Chang, J.C.; Kundranda, M. Novel Diagnostic and Predictive Biomarkers in Pancreatic Adenocarcinoma. Int. J. Mol. Sci. 2017, 18, 667. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, B.D.; Shinkins, B.; Pathiraja, I.; Roberts, N.W.; James, T.J.; Mallett, S.; Perera, R.; Primrose, J.N.; Mant, D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015, CD011134. [Google Scholar] [CrossRef] [Green Version]
- Tejpar, S.; Celik, I.; Schlichting, M.; Sartorius, U.; Bokemeyer, C.; Van Cutsem, E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 3570–3577. [Google Scholar] [CrossRef]
- Erstad, D.J.; Tumusiime, G.; Cusack, J.C., Jr. Prognostic and Predictive Biomarkers in Colorectal Cancer: Implications for the Clinical Surgeon. Ann. Surg. Oncol. 2015, 22, 3433–3450. [Google Scholar] [CrossRef] [PubMed]
- KEGG Disease. Wnt map04310 Pathway. Available online: https://www.genome.jp/dbget-bin/www_bget?pathway:map04310 (accessed on 24 January 2022).
- AmiGO-2. GO_0016055 Pathway. Available online: http://amigo.geneontology.org/amigo/term/GO:0016055 (accessed on 24 January 2022).
- Schatoff, E.M.; Leach, B.I.; Dow, L.E. Wnt Signaling and Colorectal Cancer. Curr. Colorectal Cancer Rep. 2017, 13, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koni, M.; Pinnarò, V.; Brizzi, M.F. The Wnt Signalling Pathway: A Tailored Target in Cancer. Int. J. Mol. Sci. 2020, 21, 7697. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, J.; Prenen, H. Molecular genetics of colorectal cancer. Ann. Gastroenterol. 2014, 27, 9–14. [Google Scholar]
- KEGG Disease. Colorectal Cancer map05210 Pathway. Available online: https://www.genome.jp/dbget-bin/www_bget?pathway:map05210 (accessed on 21 January 2022).
- Marmol, I.; Sanchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [Green Version]
- Grady, W.M.; Carethers, J.M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008, 135, 1079–1099. [Google Scholar] [CrossRef] [Green Version]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef] [Green Version]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef] [Green Version]
- Bujanda, L.; Cosme, A.; Gil, I.; Arenas-Mirave, J.I. Malignant colorectal polyps. World J. Gastroenterol. 2010, 16, 3103–3111. [Google Scholar] [CrossRef]
- Williams, J.G.; Pullan, R.D.; Hill, J.; Horgan, P.G.; Salmo, E.; Buchanan, G.N.; Rasheed, S.; McGee, S.G.; Haboubi, N. Management of the malignant colorectal polyp: ACPGBI position statement. Colorectal Dis. Off. J. Assoc. Coloproctol. Great Br. Irel. 2013, 15 (Suppl. S2), 1–38. [Google Scholar] [CrossRef] [Green Version]
- Wang, H. MicroRNAs and Apoptosis in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 5353. [Google Scholar] [CrossRef]
- Zhou, K.; Liu, M.; Cao, Y. New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef] [Green Version]
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Dutkiewicz, A.; Giefing, M.; Lewandowska, M.A.; Kozlowski, P. Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants. Cancers 2019, 11, 793. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.M.D. Adiponectin: Role in Physiology and Pathophysiology. Int. J. Prev. Med. 2020, 11, 136. [Google Scholar] [CrossRef]
- Chyra, M.; Roczniak, W.; Świętochowska, E.; Dudzińska, M.; Oświęcimska, J. The Effect of the Ketogenic Diet on Adiponectin, Omentin and Vaspin in Children with Drug-Resistant Epilepsy. Nutrients 2022, 14, 479. [Google Scholar] [CrossRef]
- Vetvik, K.K.; Sonerud, T.; Lindeberg, M.; Lüders, T.; Størkson, R.H.; Jonsdottir, K.; Frengen, E.; Pietiläinen, K.H.; Bukholm, I. Globular adiponectin and its downstream target genes are up-regulated locally in human colorectal tumors: Ex vivo and in vitro studies. Metabolism 2014, 63, 672–681. [Google Scholar] [CrossRef]
- Tae, C.H.; Kim, S.-E.; Jung, S.-A.; Joo, Y.-H.; Shim, K.-N.; Jung, H.-K.; Kim, T.H.; Cho, M.-S.; Kim, K.H.; Kim, J.S. Involvement of adiponectin in early stage of colorectal carcinogenesis. BMC Cancer 2014, 14, 811. [Google Scholar] [CrossRef] [Green Version]
- Medina, E.A.; Oberheu, K.; Polusani, S.R.; Ortega, V.; Velagaleti, G.V.; Oyajobi, B.O. PKA/AMPK signaling in relation to adiponectin’s antiproliferative effect on multiple myeloma cells. Leukemia 2014, 28, 2080–2089. [Google Scholar] [CrossRef]
- Li, W.; Saud, S.M.; Young, M.R.; Chen, G.; Hua, B. Targeting AMPK for cancer prevention and treatment. Oncotarget 2015, 6, 7365–7378. [Google Scholar] [CrossRef] [Green Version]
- Wei, E.K.; Ma, J.; Pollak, M.N.; Rifai, N.; Fuchs, C.S.; Hankinson, S.E.; Giovannucci, E. A prospective study of C-peptide, insulin-like growth factor-I, insulin-like growth factor binding protein-1, and the risk of colorectal cancer in women. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2005, 14, 850–855. [Google Scholar] [CrossRef] [Green Version]
- Świerczyński, M.; Szymaszkiewicz, A.; Fichna, J.; Zielińska, M. New insights into molecular pathways in colorectal cancer: Adiponectin, interleukin-6 and opioid signaling. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188460. [Google Scholar] [CrossRef]
- Cox, A.J.; West, N.P.; Cripps, A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015, 3, 207–215. [Google Scholar] [CrossRef]
- Zeng, J.; Tang, Z.H.; Liu, S.; Guo, S.S. Clinicopathological significance of overexpression of interleukin-6 in colorectal cancer. World J. Gastroenterol. 2017, 23, 1780–1786. [Google Scholar] [CrossRef]
- Waldner, M.J.; Foersch, S.; Neurath, M.F. Interleukin-6—A key regulator of colorectal cancer development. Int. J. Biol. Sci. 2012, 8, 1248–1253. [Google Scholar] [CrossRef]
- Scarà, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 247–260. [Google Scholar]
- Pavai, S.; Yap, S.F. The clinical significance of elevated levels of serum CA 19-9. Med. J. Malays. 2003, 58, 667–672. [Google Scholar]
- Tsen, A.; Barbara, M.; Rosenkranz, L. Dilemma of elevated CA 19-9 in biliary pathology. Pancreatology 2018, 18, 862–867. [Google Scholar] [CrossRef]
- Bottoni, P.; Scatena, R. The Role of CA 125 as Tumor Marker: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 229–244. [Google Scholar]
- Fiala, L.; Bob, P.; Raboch, J. Oncological markers CA-125, CA 19-9 and endometriosis. Medicine 2018, 97, e13759. [Google Scholar] [CrossRef]
- Presti, J., Jr.; Alexeeff, S.; Horton, B.; Prausnitz, S.; Avins, A.L. Changes in Prostate Cancer Presentation Following the 2012 USPSTF Screening Statement: Observational Study in a Multispecialty Group Practice. J. Gen. Intern. Med. 2019, 35, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, S.; Gillatt, D. Managing patients with acute urinary retention. Practitioner 2011, 255, 21–24. [Google Scholar] [PubMed]
- American Cancer Society. Colorectal Cancer Staging. Available online: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/acs-recommendations.html (accessed on 24 October 2021).
- Heisser, T.; Weigl, K.; Hoffmeister, M.; Brenner, H. Age-specific sequence of colorectal cancer screening options in Germany: A model-based critical evaluation. PLoS Med. 2020, 17, e1003194. [Google Scholar] [CrossRef] [PubMed]
- Pellat, A.; Deyra, J.; Coriat, R.; Chaussade, S. Results of the national organised colorectal cancer screening program with FIT in Paris. Sci. Rep. 2018, 8, 4162. [Google Scholar] [CrossRef]
- Danish Cancer Society. Colon Cancer Screening Guidelines. Available online: https://www.cancer.dk/international/english/screening-colon-cancer-english (accessed on 21 October 2021).
- NHS. The NICE FIT Study. Available online: https://www.nicefitstudy.com/ (accessed on 10 October 2021).
- Cunningham, C.; Leong, K.; Clark, S.; Plumb, A.; Taylor, S.; Geh, I.; Karandikar, S.; Moran, B. Association of Coloproctology of Great Britain & Ireland (ACPGBI): Guidelines for the Management of Cancer of the Colon, Rectum and Anus (2017)—Diagnosis, Investigations and Screening. Colorectal Dis. Off. J. Assoc. Coloproctol. Great Br. Irel. 2017, 19 (Suppl. S1), 9–17. [Google Scholar]
- Imperiale, T.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef] [Green Version]
- Vakil, N.; Ciezki, K.; Huq, N.; Singh, M. Multitarget stool DNA testing for the prevention of colon cancer: Outcomes in a large integrated healthcare system. Gastrointest. Endosc. 2020, 92, 334–341. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Wu, P.H.; Chen, Y.J.; Yang, C.H.; Huang, J.L.; Chou, Y.C.; Chang, P.K.; Wen, C.C.; Jao, S.W.; Huang, H.H.; et al. Using Comorbidity Pattern Analysis to Detect Reliable Methylated Genes in Colorectal Cancer Verified by Stool DNA Test. Genes 2021, 12, 1539. [Google Scholar] [CrossRef]
- Liu, C.; Xu, L.; Li, W.; Jie, M.; Xue, W.; Yu, W. Multiple Biomarker-Combined Screening for Colorectal Cancer Based on Bisulfate Conversion-Free Detection of Fecal DNA Methylation. BioMed Res. Int. 2021, 2021, 1479748. [Google Scholar] [CrossRef]
- Exact Sciences. Cologuard Was Superior to a Leading FIT* in Detecting Colorectal Cancer (CRC) and Precancer. Available online: https://www.cologuardhcp.com/about/cologuard-vs-fit (accessed on 3 February 2022).
- Shirley, M. Epi proColon® for Colorectal Cancer Screening: A Profile of Its Use in the USA. Mol. Diagn. Ther. 2020, 24, 497–503. [Google Scholar] [CrossRef]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef] [Green Version]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [Green Version]
- U.S. FDA. Epi proColon® Screening Approval. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf13/P130001C.pdf (accessed on 24 October 2021).
- Amir, S.; Golan, M.; Mabjeesh, N.J. Targeted knockdown of SEPT9_v1 inhibits tumor growth and angiogenesis of human prostate cancer cells concomitant with disruption of hypoxia-inducible factor-1 pathway. Mol. Cancer Res. 2010, 8, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Hubbard, S.L.; Peraud, A.; Salhia, B.; Sakai, K.; Rutka, J.T. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia 2004, 6, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Burrows, J.F.; Chanduloy, S.; McIlhatton, M.A.; Nagar, H.; Yeates, K.; Donaghy, P.; Price, J.; Godwin, A.K.; Johnston, P.G.; Russell, S.H. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. J. Pathol. 2003, 201, 581–588. [Google Scholar] [CrossRef]
- Montagna, C.; Lyu, M.-S.; Hunter, K.; Lukes, L.; Lowther, W.; Reppert, T.; Hissong, B.; Weaver, Z.; Ried, T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003, 63, 2179–2187. [Google Scholar]
- Kojima, K.; Sakai, I.; Hasegawa, A.; Niiya, H.; Azuma, T.; Matsuo, Y.; Fujii, N.; Tanimoto, M.; Fujita, S. FLJ10849, a septin family gene, fuses MLL in a novel leukemia cell line CNLBC1 derived from chronic neutrophilic leukemia in transformation with t(4;11)(q21;q23). Leukemia 2004, 18, 998–1005. [Google Scholar] [CrossRef]
- Oh, T.; Kim, N.; Moon, Y.; Kim, M.S.; Hoehn, B.D.; Park, C.H.; Kim, T.S.; Kim, N.K.; Chung, H.C.; An, S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J. Mol. Diagn. 2013, 15, 498–507. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, L.; Lu, C.; Huang, W.; Yang, C.; Wang, Q.; Wang, Q.; Lei, R.; Sun, R.; Wan, K.; et al. Methylation of SDC2/TFPI2 and Its Diagnostic Value in Colorectal Tumorous Lesions. Front. Mol. Biosci. 2021, 8, 706754. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, G.; Wang, K.; Wang, X.; Ma, Y.; Xiong, S.; Zheng, M.; Fei, S. Blood leukocytes methylation levels analysis indicate methylated plasma test is a promising tool for colorectal cancer early detection. J. Cancer 2021, 12, 3678–3685. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, C.; Wang, S.; Xiang, Z.; Dou, R.; Lin, Z.; Zheng, J.; Xiong, B. SDC2 and TFPI2 Methylation in Stool Samples as an Integrated Biomarker for Early Detection of Colorectal Cancer. Cancer Manag. Res. 2021, 13, 3601–3617. [Google Scholar] [CrossRef]
- Müller, H.M.; Oberwalder, M.; Fiegl, H.; Morandell, M.; Goebel, G.; Zitt, M.; Mühlthaler, M.; Öfner, D.; Margreiter, R.; Widschwendter, M. Methylation changes in faecal DNA: A marker for colorectal cancer screening? Lancet 2004, 363, 1283–1285. [Google Scholar] [CrossRef]
- Shirahata, A.; Hibi, K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer. Res. 2014, 34, 4121–4125. [Google Scholar]
- Shirahata, A.; Sakuraba, K.; Goto, T.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; Hibi, K. Detection of vimentin (VIM) methylation in the serum of colorectal cancer patients. Anticancer. Res. 2010, 30, 5015–5018. [Google Scholar]
- Yi, J.M.; Dhir, M.; Guzzetta, A.A.; Iacobuzio-Donahue, C.A.; Heo, K.; Yang, K.M.; Suzuki, H.; Toyota, M.; Kim, H.M.; Ahuja, N. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2012, 33, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Nassar, F.J.; Msheik, Z.S.; Nasr, R.R.; Temraz, S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin. Epigenet. 2021, 13, 111. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Balic, M.; El-Ashry, D.; Cote, R.J. Circulating Tumor Cells: Strategies for Capture, Analyses, and Propagation. Cancer J. 2018, 24, 70–77. [Google Scholar] [CrossRef]
- Li, H.; Courtois, E.T.; Sengupta, D.; Tan, Y.; Chen, K.H.; Goh, J.J.L.; Kong, S.L.; Chua, C.; Hon, L.K.; Tan, W.S.; et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017, 49, 708–718. [Google Scholar] [CrossRef]
- Bian, S.; Hou, Y.; Zhou, X.; Li, X.; Yong, J.; Wang, Y.; Wang, W.; Yan, J.; Hu, B.; Guo, H.; et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 2018, 362, 1060–1063. [Google Scholar] [CrossRef] [Green Version]
- Denis, J.A.; Patroni, A.; Guillerm, E.; Pépin, D.; Benali-Furet, N.; Wechsler, J.; Manceau, G.; Bernard, M.; Coulet, F.; Larsen, A.K.; et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol. Oncol. 2016, 10, 1221–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Therkildsen, C.; Bergmann, T.K.; Henrichsen-Schnack, T.; Ladelund, S.; Nilbert, M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014, 53, 852–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.T.; Ung, T.T.; Li, S.; Sah, D.K.; Park, S.Y.; Lian, S.; Jung, Y.D. Lithocholic Acid Induces miR21, Promoting PTEN Inhibition via STAT3 and ERK-1/2 Signaling in Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 10209. [Google Scholar] [CrossRef] [PubMed]
- Durán-Vinet, B.; Araya-Castro, K.; Calderón, J.; Vergara, L.; Weber, H.; Retamales, J.; Araya-Castro, P.; Leal-Rojas, P. CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers 2021, 13, 4640. [Google Scholar] [CrossRef]
- Gungormez, C.; Gumushan Aktas, H.; Dilsiz, N.; Borazan, E. Novel miRNAs as potential biomarkers in stage II colon cancer: Microarray analysis. Mol. Biol. Rep. 2019, 46, 4175–4183. [Google Scholar] [CrossRef]
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Vrielink, J.A.O.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802. [Google Scholar] [CrossRef] [Green Version]
- Yang, I.-P.; Tsai, H.-L.; Miao, Z.-F.; Huang, C.-W.; Kuo, C.-H.; Wu, J.-Y.; Wang, W.-M.; Juo, S.-H.H.; Wang, J.-Y. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J. Transl. Med. 2016, 14, 108. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Liu, Z. Serum miR-92a-1 is a novel diagnostic biomarker for colorectal cancer. J. Cell. Mol. Med. 2020, 24, 8363–8367. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, B.; Lin, M.; Yu, H.; Chen, H.; Zhang, Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019, 24, 299–305. [Google Scholar] [CrossRef]
- Shen, X.; Xue, Y.; Cong, H.; Wang, X.; Fan, Z.; Cui, X.; Ju, S. Circulating lncRNA DANCR as a potential auxillary biomarker for the diagnosis and prognostic prediction of colorectal cancer. Biosci. Rep. 2020, 40, BSR20191481. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, D.; Zhang, C.; Sun, Y. The Diagnostic and Prognostic Value of Serum lncRNA NEAT1 in Colorectal Cancer. Cancer Manag. Res. 2020, 12, 10985–10992. [Google Scholar] [CrossRef]
- Xie, Y.; Li, J.; Li, P.; Li, N.; Zhang, Y.; Binang, H.; Zhao, Y.; Duan, W.; Chen, Y.; Wang, Y.; et al. RNA-Seq Profiling of Serum Exosomal Circular RNAs Reveals Circ-PNN as a Potential Biomarker for Human Colorectal Cancer. Front. Oncol. 2020, 10, 982. [Google Scholar] [CrossRef]
- Chen, W.; Gao, C.; Liu, Y.; Wen, Y.; Hong, X.; Huang, Z. Bioinformatics Analysis of Prognostic miRNA Signature and Potential Critical Genes in Colon Cancer. Front. Genet. 2020, 11, 478. [Google Scholar] [CrossRef]
- Hauptman, N.; Jevšinek Skok, D.; Spasovska, E.; Boštjančič, E.; Glavač, D. Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4, and KCNA5 as potential diagnostic biomarkers in colorectal cancer. BMC Med. Genom. 2019, 12, 54. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.; Lee, S.; Choi, H.-S.; Kim, S.-N.; Lee, E.; Shin, Y.; Seo, J.; Kim, B.; Jung, Y.; Kim, W.; et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. 2011, 17, 700–709. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Zhu, X.; Luo, C.; Bu, F.; Zhu, J.; Zhu, Z. Data mining combined with experiments to validate CEP55 as a prognostic biomarker in colorectal cancer. Immun. Inflamm. Dis. 2021, 9, 167–182. [Google Scholar] [CrossRef]
- Piran, M.; Sepahi, N.; Moattari, A.; Rahimi, A.; Ghanbariasad, A. Systems Biomedicine of Primary and Metastatic Colorectal Cancer Reveals Potential Therapeutic Targets. Front. Oncol. 2021, 11, 597536. [Google Scholar] [CrossRef]
- Song, G.; Xu, S.; Zhang, H.; Wang, Y.; Xiao, C.; Jiang, T.; Wu, L.; Zhang, T.; Sun, X.; Zhong, L.; et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 148. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Li, Y.; Peng, Y.; Lu, D.; Zhang, F.; Cui, X.; Zhang, Q.; Li, Z. Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J. Cell. Physiol. 2019, 234, 15215–15224. [Google Scholar] [CrossRef]
- Yu, M.H.; Luo, Y.; Qin, S.L.; Wang, Z.S.; Mu, Y.F.; Zhong, M. Up-regulated CKS2 promotes tumor progression and predicts a poor prognosis in human colorectal cancer. Am. J. Cancer Res. 2015, 5, 2708–2718. [Google Scholar]
- Liu, J.; Li, H.; Sun, L.; Shen, S.; Zhou, Q.; Yuan, Y.; Xing, C. Epigenetic Alternations of MicroRNAs and DNA Methylation Contribute to Liver Metastasis of Colorectal Cancer. Dig. Dis. Sci. 2019, 64, 1523–1534. [Google Scholar] [CrossRef]
- Liu, C.; Pan, Z.; Chen, Q.; Chen, Z.; Liu, W.; Wu, L.; Jiang, M.; Lin, W.; Zhang, Y.; Lin, W.; et al. Pharmacological targeting PTK6 inhibits the JAK2/STAT3 sustained stemness and reverses chemoresistance of colorectal cancer. J. Exp. Clin. Cancer Res. 2021, 40, 297. [Google Scholar] [CrossRef]
- Li, R.; Hao, Y.; Wang, Q.; Meng, Y.; Wu, K.; Liu, C.; Xu, L.; Liu, Z.; Zhao, L. ECHS1, an interacting protein of LASP1, induces sphingolipid-metabolism imbalance to promote colorectal cancer progression by regulating ceramide glycosylation. Cell Death Dis. 2021, 12, 911. [Google Scholar] [CrossRef]
- Yang, G.; Huang, L.; Jia, H.; Aikemu, B.; Zhang, S.; Shao, Y.; Hong, H.; Yesseyeva, G.; Wang, C.; Li, S.; et al. NDRG1 enhances the sensitivity of cetuximab by modulating EGFR trafficking in colorectal cancer. Oncogene 2021, 40, 5993–6006. [Google Scholar] [CrossRef]
- Coss, A.; Tosetto, M.; Fox, E.J.; Sapetto-Rebow, B.; Gorman, S.; Kennedy, B.N.; Lloyd, A.T.; Hyland, J.M.; O’Donoghue, D.P.; Sheahan, K.; et al. Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett. 2009, 276, 228–238. [Google Scholar] [CrossRef]
- Qu, X.; Sandmann, T.; Frierson, H.; Fu, L.; Fuentes, E.; Walter, K.; Okrah, K.; Rumpel, C.; Moskaluk, C.; Lu, S.; et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene 2016, 35, 6403–6415. [Google Scholar] [CrossRef] [Green Version]
- Dong, B.; Chai, M.; Chen, H.; Feng, Q.; Jin, R.; Hu, S. Screening and verifying key genes with poor prognosis in colon cancer through bioinformatics analysis. Transl. Cancer Res. 2020, 9, 6720–6732. [Google Scholar] [CrossRef]
- Yu, D.; Sun, J.; Weng, Y.; Luo, L.; Sheng, J.; Xu, Z. Serum angiogenin as a potential biomarker for early detection of colorectal adenomas and colorectal cancer. Anti-Cancer Drugs 2021, 32, 703–708. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel, F.; Eltarhoni, K.; Nisar, P.; Soloviev, M. Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection. Cancers 2022, 14, 1889. https://doi.org/10.3390/cancers14081889
Kamel F, Eltarhoni K, Nisar P, Soloviev M. Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection. Cancers. 2022; 14(8):1889. https://doi.org/10.3390/cancers14081889
Chicago/Turabian StyleKamel, Faddy, Khadiga Eltarhoni, Pasha Nisar, and Mikhail Soloviev. 2022. "Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection" Cancers 14, no. 8: 1889. https://doi.org/10.3390/cancers14081889
APA StyleKamel, F., Eltarhoni, K., Nisar, P., & Soloviev, M. (2022). Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection. Cancers, 14(8), 1889. https://doi.org/10.3390/cancers14081889






