SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Extracellular Vesicle (EV) Isolation
2.3. Nanoparticle Tracking Analysis (NTA)
2.4. Protein Isolation from EVs
2.5. Mass Spectrometry
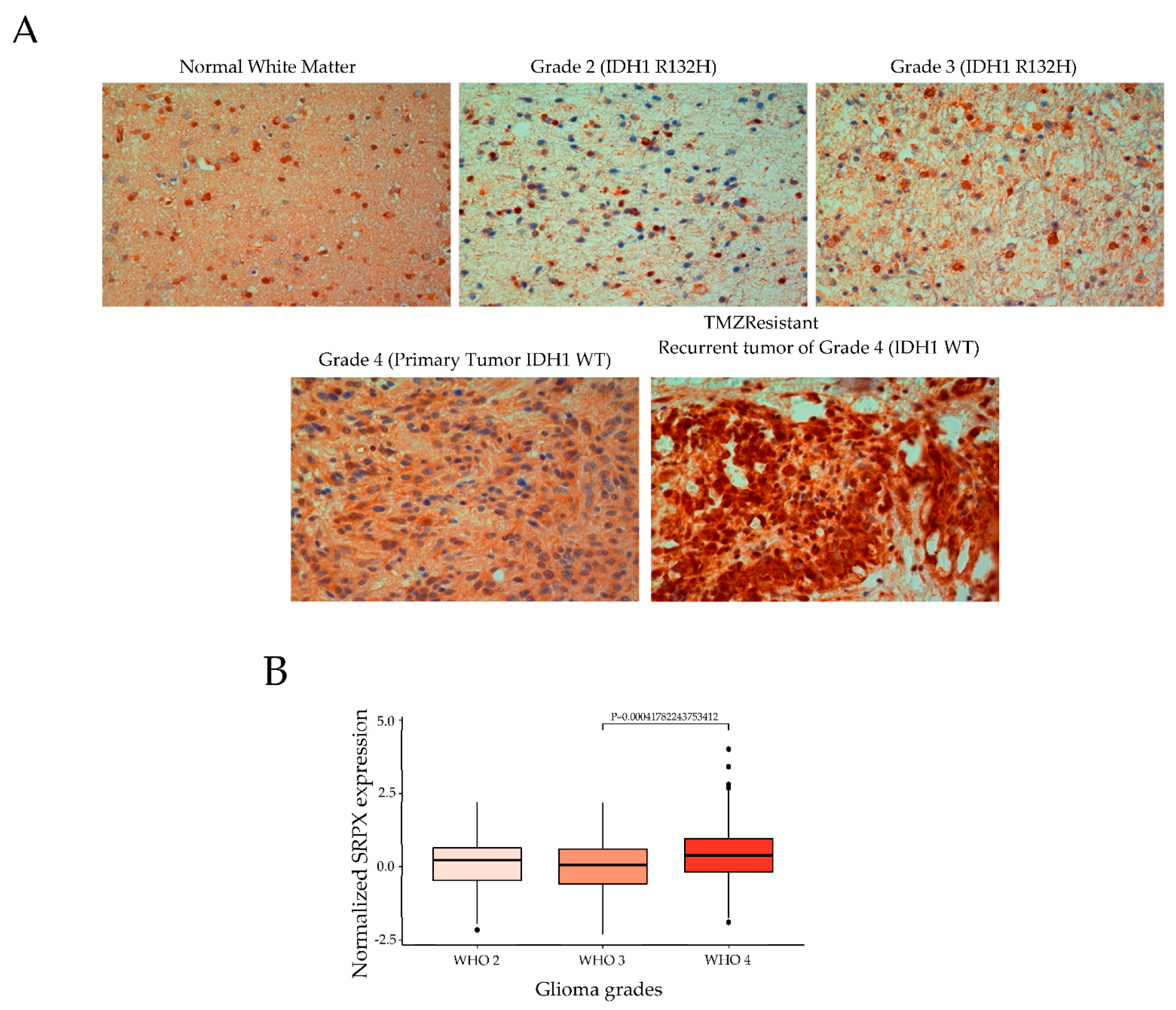
2.6. Immunohistochemical Staining
2.7. RNA isolation and RT-qPCR
2.8. Analysis of Chinese Glioblastoma Genome Atlas (CGGA) Datasets
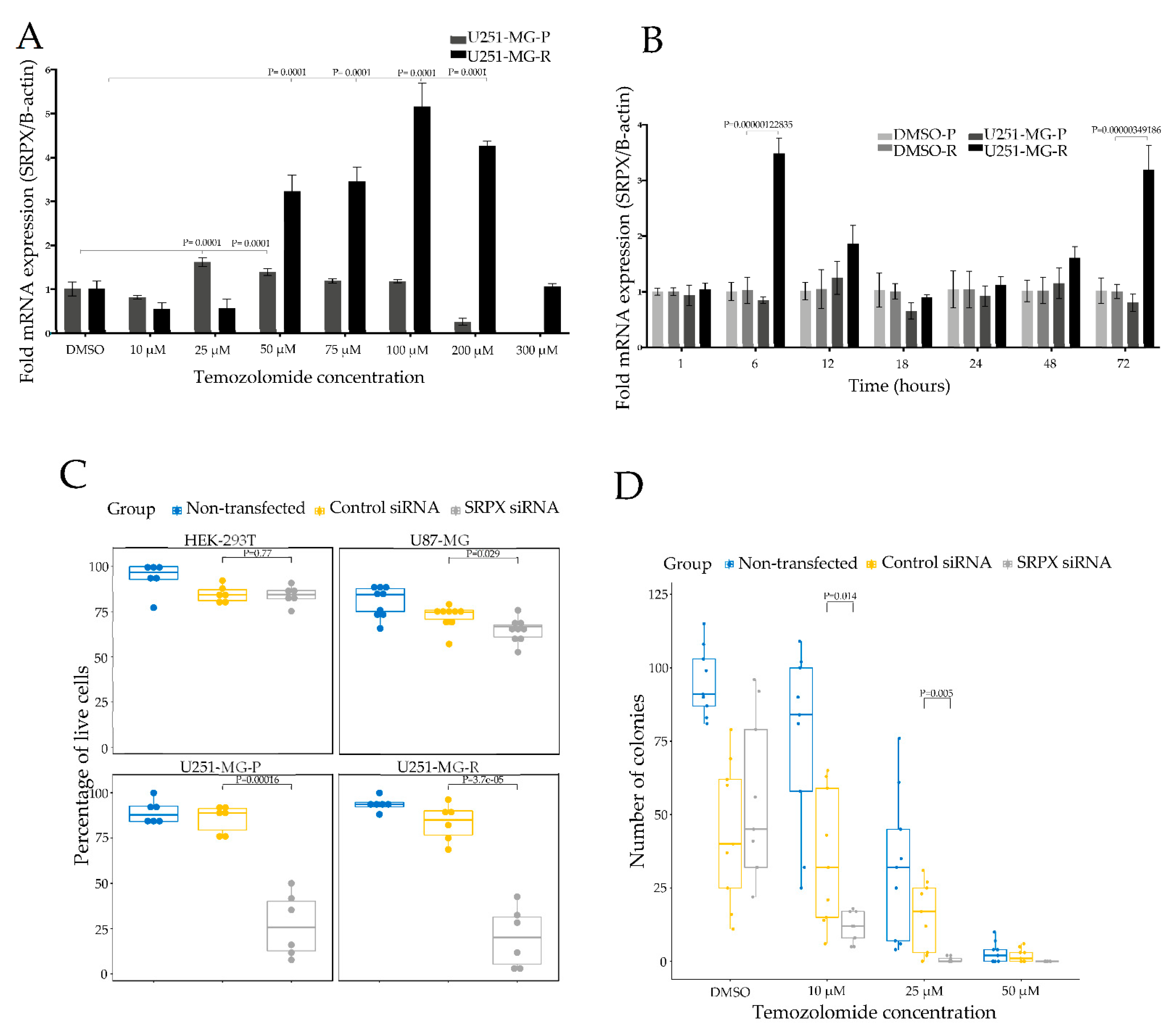
2.9. Generation of Temozolomide Resistant U251-MG Cells
2.10. Transfection
2.10.1. Cell Viability Assay
2.10.2. Cell Viability Assay for U251-MG-R Cells Treated with TMZ
2.11. Clonogenic Survival Assay
2.12. TMZ Sensitivity Assay
2.13. Statistical Analysis
3. Results
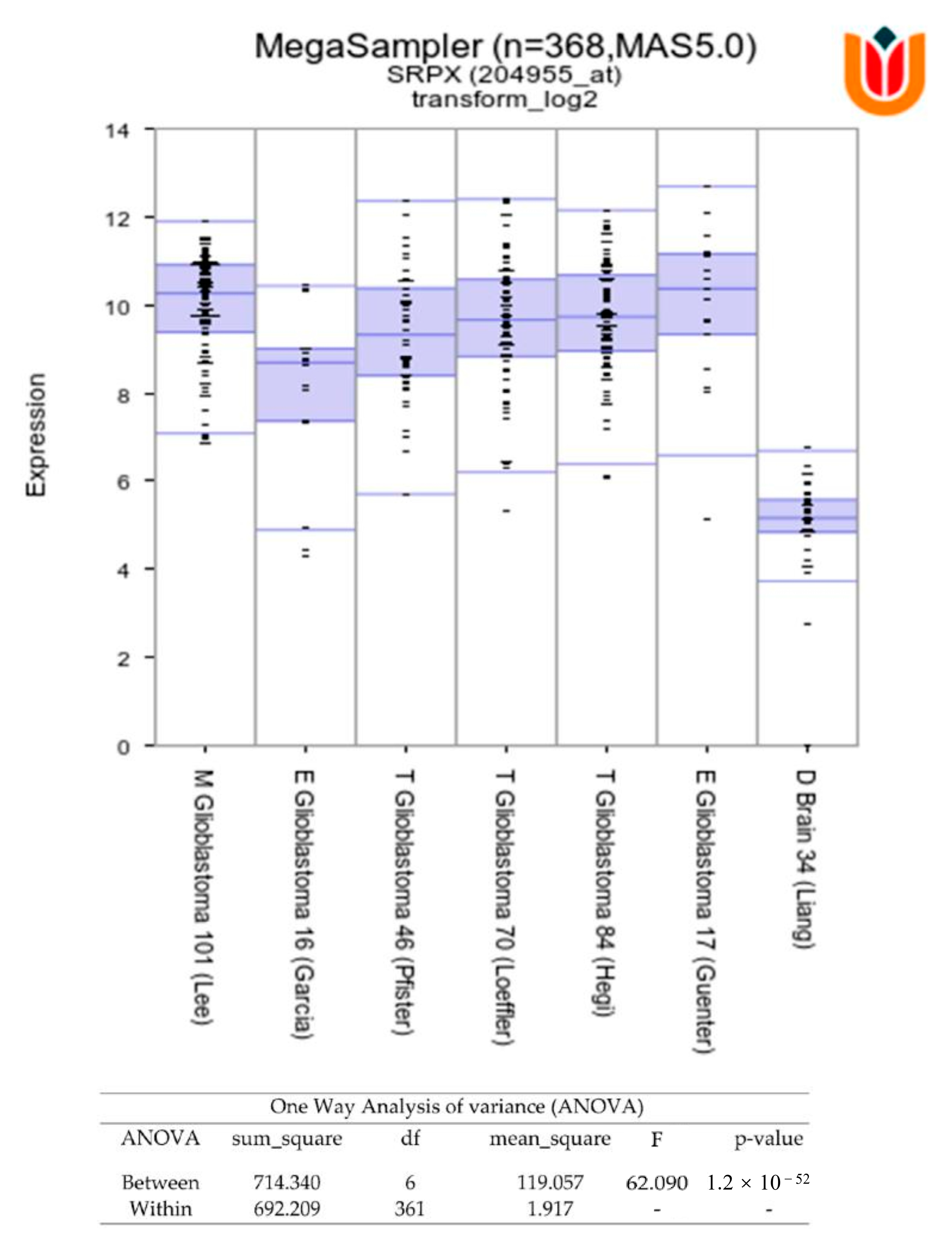
3.1. Proteomics Analysis of Glioblastoma- and HPA-Derived EVs
3.2. Association of SRPX with TMZ Resistance in Glioblastomas
3.3. SRPX Is Involved in Cell Viability of Glioblastoma Cells
3.4. SRPX Depletion via siRNA Sensitizes Glioblastoma Cells to TMZ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Saadatpour, L.; Fadaee, E.; Fadaei, S.; Mansour, R.N.; Mohammadi, M.; Mousavi, S.M.; Goodarzi, M.; Verdi, J.; Mirzaei, H. Glioblastoma: Exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 2016, 23, 415–418. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.A.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Erkan, E.P.; Saydam, O. Extracellular Vesicles as Novel Delivery Tools for Cancer Treatment. Curr. Cancer Drug Targets 2015, 16, 34–42. [Google Scholar] [CrossRef]
- Erkan, E.P.; Senfter, D.; Madlener, S.; Jungwirth, G.; Ströbel, T.; Saydam, N.; Saydam, O. Extracellular vesicle-mediated suicide mRNA/protein delivery inhibits glioblastoma tumor growth in vivo. Cancer Gene Ther. 2016, 24, 38–44. [Google Scholar] [CrossRef]
- Westphal, M.; Lamszus, K. Circulating biomarkers for gliomas. Nat. Rev. Neurol. 2015, 11, 556–566. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Klekner, A.; Szivos, L.; Virga, J.; Árkosy, P.; Bognár, L.; Birkó, Z.; Nagy, B. Significance of liquid biopsy in glioblastoma—A review. J. Biotechnol. 2019, 298, 82–87. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, P.; Sowers, J.L.; Machuca, D.F.; Mirfattah, B.; Herring, J.; Tang, H.; Chen, Y.; Tian, B.; Brasier, A.R.; et al. Proteome Analysis of Hypoxic Glioblastoma Cells Reveals Sequential Metabolic Adaptation of One-Carbon Metabolic Pathways. Mol. Cell. Proteom. 2017, 16, 1906–1921. [Google Scholar] [CrossRef]
- Sangar, V.; Funk, C.C.; Kusebauch, U.; Campbell, D.S.; Moritz, R.; Price, N.D. Quantitative Proteomic Analysis Reveals Effects of Epidermal Growth Factor Receptor (EGFR) on Invasion-promoting Proteins Secreted by Glioblastoma Cells. Mol. Cell. Proteom. 2014, 13, 2618–2631. [Google Scholar] [CrossRef]
- Miyauchi, E.; Furuta, T.; Ohtsuki, S.; Tachikawa, M.; Uchida, Y.; Sabit, H.; Obuchi, W.; Baba, T.; Watanabe, M.; Terasaki, T.; et al. Identification of blood biomarkers in glioblastoma by SWATH mass spectrometry and quantitative targeted absolute proteomics. PLoS ONE 2018, 13, e0193799. [Google Scholar] [CrossRef]
- Khwaja, F.W.; Reed, M.S.; Olson, J.J.; Schmotzer, B.J.; Gillespie, G.Y.; Guha, A.; Groves, M.D.; Kesari, S.; Pohl, J.; Van Meir, E.G. Proteomic Identification of Biomarkers in the Cerebrospinal Fluid (CSF) of Astrocytoma Patients. J. Proteome Res. 2007, 6, 559–570. [Google Scholar] [CrossRef]
- Choi, D.; Montermini, L.; Kim, D.-K.; Meehan, B.; Roth, F.; Rak, J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell. Proteom. 2018, 17, 1948–1964. [Google Scholar] [CrossRef]
- Ströbel, T.; Madlener, S.; Tuna, S.; Vose, S.; Lagerweij, T.; Wurdinger, T.; Vierlinger, K.; Wöhrer, A.; Price, B.D.; Demple, B.; et al. Ape1 guides DNA repair pathway choice that is associated with drug tolerance in glioblastoma. Sci. Rep. 2017, 7, 9674. [Google Scholar] [CrossRef]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Ströbel, T.; Erkan, E.P.; Fan, J.-B.; O Breakefield, X.; Saydam, O. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Mitulović, G.; Stingl, C.; Steinmacher, I.; Hudecz, O.; Hutchins, J.R.A.; Peters, J.-M.; Mechtler, K. Preventing Carryover of Peptides and Proteins in Nano LC-MS Separations. Anal. Chem. 2009, 81, 5955–5960. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Hurwitz, S.N.; Rider, M.A.; Bundy, J.L.; Liu, X.; Singh, R.K.; Meckes, D.G., Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 2016, 7, 86999–87015. [Google Scholar] [CrossRef]
- Zhao, Z.; Meng, F.; Wang, W.; Wang, Z.; Zhang, C.; Jiang, T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data 2017, 4, 170024. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: Identification of novel molecular targets in matrix and immune modulation. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef]
- Bai, K.; He, S.; Shu, L.; Wang, W.; Lin, S.; Zhang, Q.; Li, L.; Cheng, L.; Dai, Y. Identification of cancer stem cell characteristics in liver hepatocellular carcinoma by WGCNA analysis of transcriptome stemness index. Cancer Med. 2020, 9, 4290–4298. [Google Scholar] [CrossRef]
- Bhat, K.P.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal Differentiation Mediated by NF-κB Promotes Radiation Resistance in Glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef]
- Behnan, J.; Stangeland, B.; Hosainey, S.A.M.; Joel, M.; Olsen, T.K.; Micci, F.; Glover, J.C.; Isakson, P.; E Brinchmann, J. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene 2016, 36, 570–584. [Google Scholar] [CrossRef]
- Pan, Y.-B.; Zhang, C.-H.; Wang, S.-Q.; Ai, P.-H.; Chen, K.; Zhu, L.; Sun, Z.-L.; Feng, D.-F. Transforming growth factor beta induced (TGFBI) is a potential signature gene for mesenchymal subtype high-grade glioma. J. Neuro-Oncol. 2018, 137, 395–407. [Google Scholar] [CrossRef]
- Okawa, S.; Gagrica, S.; Blin, C.; Ender, C.; Pollard, S.M.; Krijgsveld, J. Proteome and Secretome Characterization of Glioblastoma-Derived Neural Stem Cells. Stem Cells 2016, 35, 967–980. [Google Scholar] [CrossRef]
- Sallinen, S.L.; Sallinen, P.K.; Haapasalo, H.K.; Helin, H.J.; Helén, P.T.; Schraml, P.; Kallioniemi, O.; Kononen, J. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000, 60, 6617–6622. [Google Scholar]
- Daubon, T.; Léon, C.; Clarke, K.; Andrique, L.; Salabert, L.; Darbo, E.; Pineau, R.; Guérit, S.; Maitre, M.; Dedieu, S.; et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Inoue, Y.; Ueda, M.; Tasaki, M.; Takeshima, A.; Nagatoshi, A.; Masuda, T.; Misumi, Y.; Kosaka, T.; Nomura, T.; Mizukami, M.; et al. Sushi repeat-containing protein 1: A novel disease-associated molecule in cerebral amyloid angiopathy. Acta Neuropathol. 2017, 134, 605–617. [Google Scholar] [CrossRef]
- Iragavarapu, S.; Algeciras, M.E.; Lee, R.K.; Bhattacharya, S.K. ETX1 is over-expressed in the glaucomatous trabecular meshwork. Mol. Vis. 2009, 15, 2061–2067. [Google Scholar]
- Tambe, Y.; Isono, T.; Haraguchi, S.; Yoshioka-Yamashita, A.; Yutsudo, M.; Inoue, H. A novel apoptotic pathway induced by the drs tumor suppressor gene. Oncogene 2004, 23, 2977–2987. [Google Scholar] [CrossRef]
- Meindl, A.; Carvalho, M.R.S.; Herrmann, K.; Achatz, H.; Lorenz, B.; Apfelstedt-Sylla, E.; Wittwer, B.; Ross, M.; Meitinger, T.; Lorenz, B.; et al. A gene (SRPX) encoding a sushi-repeat-containing protein is deleted in patients with X-linked retinitis pigmentosa. Hum. Mol. Genet. 1995, 4, 2339–2346. [Google Scholar] [CrossRef]
- Dry, K.L.; Aldred, M.A.; Edgar, A.J.; Brown, J.; Manson, F.D.; Ho, M.-F.; Prosser, J.; Hardwick, L.J.; Lennon, A.A.; Thomson, K.; et al. Identification of a novel gene, ETX1, from Xp21.1, a candidate gene for X-linked retinitis pigmentosa (RP3). Hum. Mol. Genet. 1995, 4, 2347–2353. [Google Scholar] [CrossRef]
- Inoue, H.; Pan, J.; Hakura, A. Suppression of v-src transformation by the drs gene. J. Virol. 1998, 72, 2532–2537. [Google Scholar] [CrossRef]
- Jing, P.; Kazuyoshi, N.; Masuo, Y.; Hirokazu, I.; Qin, L.; Kiyomasa, O.; Naohisa, Y.; Akira, H. Isolation of a novel gene down-regulated byv-src. FEBS Lett. 1996, 383, 21–25. [Google Scholar] [CrossRef][Green Version]
- Kim, C.J.; Shimakage, M.; Kushima, R.; Mukaisho, K.-I.; Shinka, T.; Okada, Y.; Inoue, H. Down-regulation of drs mRNA in human prostate carcinomas. Hum. Pathol. 2003, 34, 654–657. [Google Scholar] [CrossRef]
- Yamashita, A.; Hakura, A.; Inoue, H. Suppression of anchorage-independent growth of human cancer cell lines by the drs gene. Oncogene 1999, 18, 4777–4787. [Google Scholar] [CrossRef][Green Version]
- Shimakage, M.; Takami, K.; Kodama, K.; Mano, M.; Yutsudo, M.; Inoue, H. Expression of drs mRNA in human lung adenocarcinomas. Hum. Pathol. 2002, 33, 615–619. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019, 47, 7294–7305. [Google Scholar] [CrossRef]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Li, G.; Guo, J.; Shen, B.-Q.; Yadav, D.B.; Sliwkowski, M.X.; Crocker, L.M.; Lacap, J.A.; Phillips, G.D.L. Mechanisms of Acquired Resistance to Trastuzumab Emtansine in Breast Cancer Cells. Mol. Cancer Ther. 2018, 17, 1441–1453. [Google Scholar] [CrossRef]
- Shao, F.; Liu, C. Revisit the Candidacy of Brain Cell Types as the Cell(s) of Origin for Human High-Grade Glioma. Front. Mol. Neurosci. 2018, 11, 48. [Google Scholar] [CrossRef]
- Zong, H.; Parada, L.F.; Baker, S.J. Cell of Origin for Malignant Gliomas and Its Implication in Therapeutic Development. Cold Spring Harb. Perspect. Biol. 2015, 7, a020610. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.E.; Kahng, J.Y.; Kim, S.H.; Park, J.S.; Yoon, S.J.; Um, J.-Y.; Kim, W.K.; Lee, J.-K.; Park, J.; et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018, 560, 243–247. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ampudia-Mesias, E.; El-Hadad, S.; Cameron, C.S.; Wöhrer, A.; Ströbel, T.; Saydam, N.; Saydam, O. SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma. Cancers 2022, 14, 1984. https://doi.org/10.3390/cancers14081984
Ampudia-Mesias E, El-Hadad S, Cameron CS, Wöhrer A, Ströbel T, Saydam N, Saydam O. SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma. Cancers. 2022; 14(8):1984. https://doi.org/10.3390/cancers14081984
Chicago/Turabian StyleAmpudia-Mesias, Elisabet, Samia El-Hadad, Charles Scott Cameron, Adelheid Wöhrer, Thomas Ströbel, Nurten Saydam, and Okay Saydam. 2022. "SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma" Cancers 14, no. 8: 1984. https://doi.org/10.3390/cancers14081984
APA StyleAmpudia-Mesias, E., El-Hadad, S., Cameron, C. S., Wöhrer, A., Ströbel, T., Saydam, N., & Saydam, O. (2022). SRPX Emerges as a Potential Tumor Marker in the Extracellular Vesicles of Glioblastoma. Cancers, 14(8), 1984. https://doi.org/10.3390/cancers14081984





