Low Frequency of Cancer-Predisposition Gene Mutations in Liver Transplant Candidates with Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Controls
2.3. Library Preparation, Sequencing and Bioinformatics
2.4. Variant and Gene Prioritization
2.5. Statistical Analyses
3. Results
3.1. Germline Variants in Established and Candidate CPG
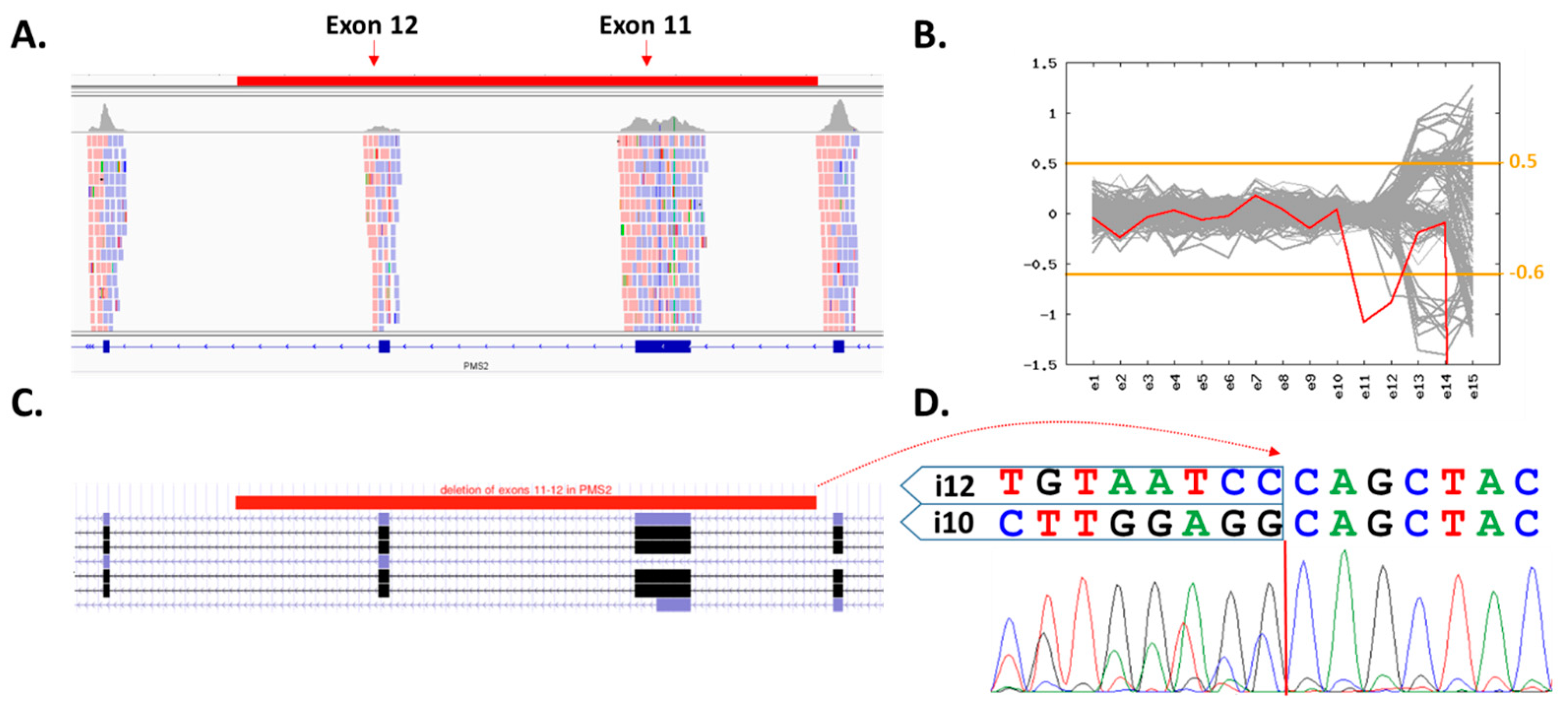
3.2. Clinical Characterization of PV Carriers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CI | confidence interval |
| CNV | copy number variants |
| CPG | cancer-predisposition genes |
| DDR | DNA damage repair |
| HBV | hepatitis B virus |
| HCC | hepatocellular carcinoma |
| HCV | hepatitis C virus |
| MAF | minor allele frequency |
| MRN | MRE11-RAD50-NBN |
| NASH | non-alcoholic steatohepatitis |
| OR | odds ratio |
| PARPi | PARP1 inhibitors |
| PV | pathogenic/likely pathogenic germline variant |
| PMC | population-matched controls |
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar]
- European Association For The Study of The Liver; European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef]
- Sangiovanni, A.; Prati, G.M.; Fasani, P.; Ronchi, G.; Romeo, R.; Manini, M.; Del Ninno, E.; Morabito, A.; Colombo, M. The natural history of compensated cirrhosis due to hepatitis C virus: A 17-year cohort study of 214 patients. Hepatology 2006, 43, 1303–1310. [Google Scholar] [CrossRef]
- van der Meer, A.J.; Veldt, B.J.; Feld, J.J.; Wedemeyer, H.; Dufour, J.F.; Lammert, F.; Duarte-Rojo, A.; Heathcote, E.J.; Manns, M.P.; Kuske, L.; et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA 2012, 308, 2584–2593. [Google Scholar] [CrossRef]
- Colmenero, J.; Tabrizian, P.; Bhangui, P.; Pinato, D.J.; Rodriguez-Peralvarez, M.L.; Sapisochin, G.; Bhoori, S.; Pascual, S.; Senzolo, M.; Al-Adra, D.; et al. De Novo Malignancy after Liver Transplantation: Risk Assessment, Prevention, and Management-Guidelines From the ILTS-SETH Consensus Conference. Transplantation 2022, 106, e30–e45. [Google Scholar] [CrossRef]
- Daniel, K.E.; Eickhoff, J.; Lucey, M.R. Why do patients die after a liver transplantation? Clin. Transpl. 2017, 31, e12906. [Google Scholar] [CrossRef]
- Ozturk, M. Genetic aspects of hepatocellular carcinogenesis. Semin. Liver Dis. 1999, 19, 235–242. [Google Scholar] [CrossRef]
- Uson Junior, P.L.; Kunze, K.L.; Golafshar, M.A.; Riegert-Johnson, D.; Boardman, L.; Borad, M.J.; Ahn, D.; Sonbol, M.B.; Faigel, D.O.; Fukami, N.; et al. Germline Cancer Susceptibility Gene Testing in Unselected Patients with Hepatobiliary Cancers: A Multi-Center Prospective Study. Cancer Prev. Res. 2022, 15, 121–128. [Google Scholar] [CrossRef]
- Mezina, A.; Philips, N.; Bogus, Z.; Erez, N.; Xiao, R.; Fan, R.; Olthoff, K.M.; Reddy, K.R.; Samadder, N.J.; Nielsen, S.M.; et al. Multigene Panel Testing in Individuals With Hepatocellular Carcinoma Identifies Pathogenic Germline Variants. JCO Precis. Oncol. 2021, 5, 988–1000. [Google Scholar] [CrossRef]
- Lhotova, K.; Stolarova, L.; Zemankova, P.; Vocka, M.; Janatova, M.; Borecka, M.; Cerna, M.; Jelinkova, S.; Kral, J.; Volkova, Z.; et al. Multigene Panel Germline Testing of 1333 Czech Patients with Ovarian Cancer. Cancers 2020, 12, 956. [Google Scholar] [CrossRef] [PubMed]
- Soukupova, J.; Zemankova, P.; Lhotova, K.; Janatova, M.; Borecka, M.; Stolarova, L.; Lhota, F.; Foretova, L.; Machackova, E.; Stranecky, V.; et al. Validation of CZECANCA (CZEch CAncer paNel for Clinical Application) for targeted NGS-based analysis of hereditary cancer syndromes. PLoS ONE 2018, 13, e0195761. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang Le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 29 November 2022).
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Weisburd, B.; Thomas, B.; Solomonson, M.; Ruderfer, D.M.; Kavanagh, D.; Hamamsy, T.; Lek, M.; Samocha, K.E.; Cummings, B.B.; et al. The ExAC browser: Displaying reference data information from over 60,000 exomes. Nucleic Acids Res. 2017, 45, D840–D845. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Exome Variant Server. Available online: https://evs.gs.washington.edu/EVS/ (accessed on 1 April 2022).
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Varon, R.; Seemanova, E.; Chrzanowska, K.; Hnateyko, O.; Piekutowska-Abramczuk, D.; Krajewska-Walasek, M.; Sykut-Cegielska, J.; Sperling, K.; Reis, A. Clinical ascertainment of Nijmegen breakage syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur. J. Hum. Genet. 2000, 8, 900–902. [Google Scholar] [CrossRef]
- Wieme, G.; Kral, J.; Rosseel, T.; Zemankova, P.; Parton, B.; Vocka, M.; Van Heetvelde, M.; Kleiblova, P.; Blaumeiser, B.; Soukupova, J.; et al. Prevalence of Germline Pathogenic Variants in Cancer Predisposing Genes in Czech and Belgian Pancreatic Cancer Patients. Cancers 2021, 13, 4430. [Google Scholar] [CrossRef] [PubMed]
- Elkholi, I.E.; Foulkes, W.D.; Rivera, B. MRN Complex and Cancer Risk: Old Bottles, New Wine. Clin. Cancer Res. 2021, 27, 5465–5471. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Rybicka, M.; Woziwodzka, A.; Sznarkowska, A.; Romanowski, T.; Stalke, P.; Dreczewski, M.; Verrier, E.R.; Baumert, T.F.; Bielawski, K.P. Liver Cirrhosis in Chronic Hepatitis B Patients Is Associated with Genetic Variations in DNA Repair Pathway Genes. Cancers 2020, 12, 3295. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Xiao, R.; Chen, X.; Yuan, C.; Sun, Y.; Li, J. A non-synonymous polymorphism in NBS1 is associated with progression from chronic hepatitis B virus infection to hepatocellular carcinoma in a Chinese population. Onco Targets Ther. 2018, 11, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Dumon Jones, V.; Frappart, P.-O.; Tong, W.-M.; Sajithlal, G.; Hulla, W.; Schmid, G.; Herceg, Z.; Digweed, M.; Wang, Z.-Q. Nbn Heterozygosity Renders Mice Susceptible to Tumor Formation and Ionizing Radiation-Induced Tumorigenesis. Cancer Res. 2003, 63, 7263–7269. [Google Scholar]
- Soukupova, J.; Zemankova, P.; Kleiblova, P.; Janatova, M.; Kleibl, Z. CZECANCA: CZEch CAncer paNel for Clinical Application—Design and Optimization of the Targeted Sequencing Panel for the Identification of Cancer Susceptibility in High-risk Individuals from the Czech Republic. Klin. Onkol. 2016, 29 (Suppl. 1), S46–S54. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Suo, P.; Liu, G.; Li, L.; Zhang, X.; Chen, S.; Xu, M.; Song, L. Identification of a novel germline frameshift mutation p.D300fs of PMS1 in a patient with hepatocellular carcinoma: A case report and literature review. Medicine 2020, 99, e19076. [Google Scholar] [CrossRef]
- Chau, C.; van Doorn, R.; van Poppelen, N.M.; van der Stoep, N.; Mensenkamp, A.R.; Sijmons, R.H.; van Paassen, B.W.; van den Ouweland, A.M.W.; Naus, N.C.; van der Hout, A.H.; et al. Families with BAP1-Tumor Predisposition Syndrome in The Netherlands: Path to Identification and a Proposal for Genetic Screening Guidelines. Cancers 2019, 11, 1114. [Google Scholar] [CrossRef]
- Caruso, S.; Calderaro, J.; Letouze, E.; Nault, J.C.; Couchy, G.; Boulai, A.; Luciani, A.; Zafrani, E.S.; Bioulac-Sage, P.; Seror, O.; et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J. Hepatol. 2017, 66, 734–742. [Google Scholar] [CrossRef]
- Fu, J.; Wang, T.; Zhai, X.; Xiao, X. Primary hepatocellular adenoma due to biallelic HNF1A mutations and its co-occurrence with MODY 3: Case-report and review of the literature. Endocrine 2020, 67, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Tovar, E.A.; Graveel, C.R. MET in human cancer: Germline and somatic mutations. Ann. Transl. Med. 2017, 5, 205. [Google Scholar] [CrossRef] [PubMed]
- Donati, B.; Pietrelli, A.; Pingitore, P.; Dongiovanni, P.; Caddeo, A.; Walker, L.; Baselli, G.; Pelusi, S.; Rosso, C.; Vanni, E.; et al. Telomerase reverse transcriptase germline mutations and hepatocellular carcinoma in patients with nonalcoholic fatty liver disease. Cancer Med. 2017, 6, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, J.J.; Frier, Q.J.; Sumarriva, D.; Oberley, M.; Bolton, D.; Deveras, R.A. Germline VHL Mutation Discovered in Association with EGFR-Positive Lung Cancer and Metachronous Hepatocellular Carcinoma: A Case Report. Case Rep. Oncol. 2021, 14, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.M.; Mahan, K.; Mettler, T.; Dunitz, J.M.; Khoruts, A. Case report of synchronous post-lung transplant colon cancers in the era of colorectal cancer screening recommendations in cystic fibrosis: Screening “too early” before it’s too late. BMC Gastroenterol. 2019, 19, 137. [Google Scholar] [CrossRef]
- Gozdowska, J.; Bieniasz, M.; Wszoła, M.; Kieszek, R.; Domagała, P.; Drozdowski, J.; Tomaszek, A.; Kwiatkowski, A.; Chmura, A.; Durlik, M. Determining eligibility for and preparation to kidney transplantation of a patient with Lynch syndrome—A case report and literature review. Ann. Transplant. 2014, 19, 124–128. [Google Scholar]
- Qudaih, A.T.; Al Ashour, B.H.; Naim, A.K.; Joudeh, A.A. Kidney Transplant Recipient With Multiple Contemporaneous Malignancies Secondary to Muir-Torre Syndrome. Cureus 2021, 13, e16642. [Google Scholar] [CrossRef]
- Wassano, N.S.; Sergi, F.; Ferro, G.; Genzini, T.; D’Alpino Peixoto, R. Rapid Disease Progression of Liver Metastases following Resection in a Liver-Transplanted Patient with Probable Lynch Syndrome—A Case Report and Review of the Literature. Case Rep. Oncol. 2017, 10, 244–251. [Google Scholar] [CrossRef]
- Yang, R.L.; Kurian, A.W.; Winton, L.M.; Weill, D.; Patel, K.; Kingham, K.; Wapnir, I.L. Addressing inherited predisposition for breast cancer in transplant recipients. J. Surg. Oncol. 2016, 113, 605–608. [Google Scholar] [CrossRef]
- Trepo, E.; Nahon, P.; Bontempi, G.; Valenti, L.; Falleti, E.; Nischalke, H.D.; Hamza, S.; Corradini, S.G.; Burza, M.A.; Guyot, E.; et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014, 59, 2170–2177. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Manjunath, H.; Yopp, A.C.; Beg, M.S.; Marrero, J.A.; Gopal, P.; Waljee, A.K. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: A meta-analysis. Am. J. Gastroenterol. 2014, 109, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Pelusi, S.; Baselli, G.; Pietrelli, A.; Dongiovanni, P.; Donati, B.; McCain, M.V.; Meroni, M.; Fracanzani, A.L.; Romagnoli, R.; Petta, S.; et al. Rare Pathogenic Variants Predispose to Hepatocellular Carcinoma in Nonalcoholic Fatty Liver Disease. Sci. Rep. 2019, 9, 3682. [Google Scholar] [CrossRef] [PubMed]
- Pirisi, M.; Toniutto, P.; Uzzau, A.; Fabris, C.; Avellini, C.; Scott, C.; Apollonio, L.; Beltrami, C.A.; Beltrami, C.; Bresadola, F. Carriage of HFE mutations and outcome of surgical resection for hepatocellular carcinoma in cirrhotic patients. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2000, 89, 297–302. [Google Scholar]
- Strnad, P.; Buch, S.; Hamesch, K.; Fischer, J.; Rosendahl, J.; Schmelz, R.; Brueckner, S.; Brosch, M.; Heimes, C.V.; Woditsch, V.; et al. Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut 2019, 68, 1099. [Google Scholar] [CrossRef]
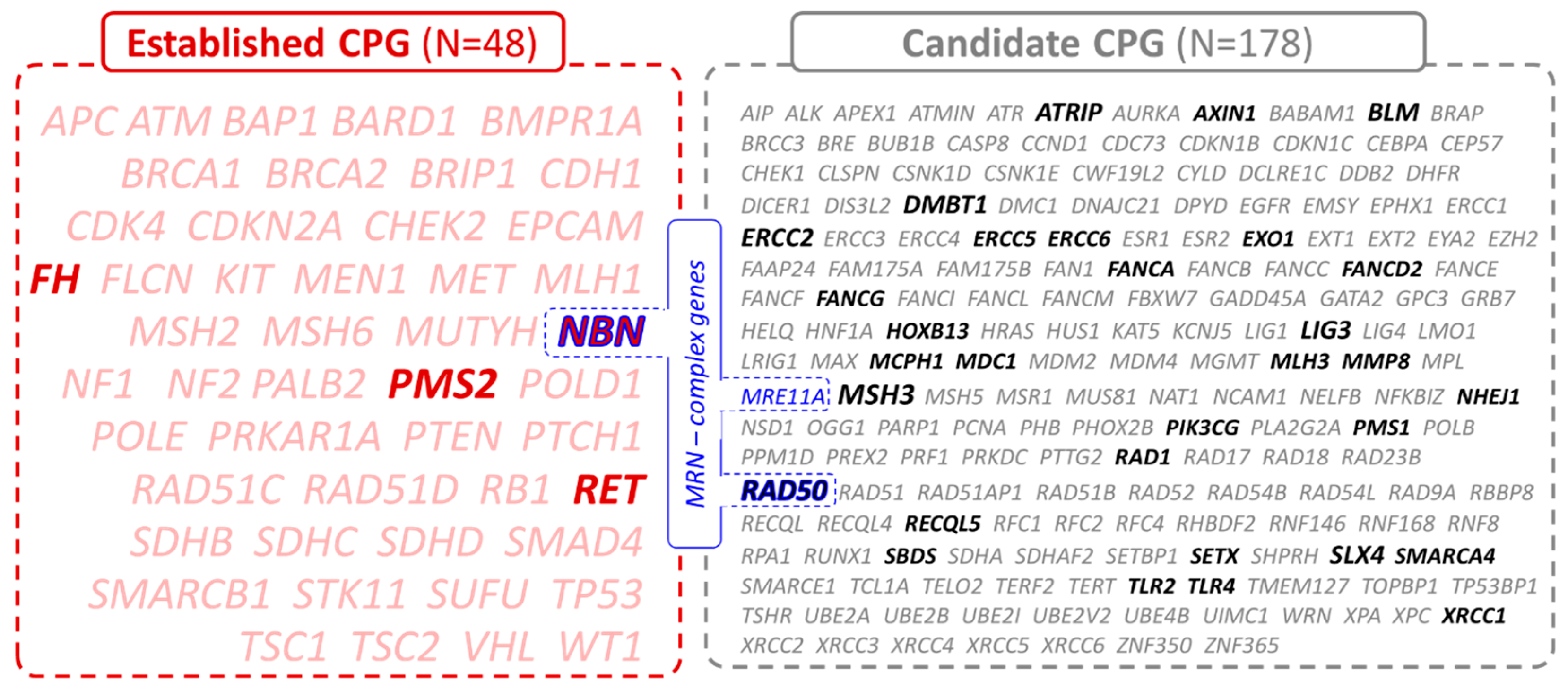
| Patients’ Characteristics | All Patients N = 334 | Males N = 258 | Females N = 76 |
|---|---|---|---|
| Age [years]; median (range) | 63 (26–77) | 63 (36–75) | 65 (26–77) |
| Cirrhosis; N (%) | 329 (98.5) | 254 (98.4) | 75 (98.7) |
| Alcoholic | 129 (38.6) | 115 (44.6) | 14 (18.4) |
| Viral | 120 (35.9) | 77 (29.8) | 43 (56.6) |
| Cholestatic and autoimmune | 48 (14.4) | 37 (14.3) | 11 (14.5) |
| NASH (non-alcoholic steatohepatitis) | 29 (8.7) | 23 (8.9) | 6 (7.9) |
| Metabolic | 3 (0.9) | 2 (0.8) | 1 (1.3) |
| Not present | 5 (1.5) | 4 (1.6) | 1 (1.3) |
| HCC treatment; N (%) | |||
| Liver transplantation | 299 (89.5) | 225 (87.2) | 74 (97.4) |
| Other | 35 (10.5) | 33 (12.8) | 2 (2.6) |
| HCC characteristics | |||
| AFP [ng/mL]; median (range) | 8.3 (0.9–5784) | 7.4 (0.9–5784) | 15.0 (1.9–1210) |
| Milan criteria; N (% of known) | 220 (65.9) | 168 (65.1) | 52 (68.4) |
| Microangioinvasion; N (% of known) | 128 (45.6) | 91 (43.8) | 37 (50.7) |
| Cholangiocarc. differentiation; N (% of known) | 18 (5.4) | 10 (4.6) | 8 (10.8) |
| Grade 1; N (% of known) | 37 (13.0) | 22 (10.3) | 15 (21.1) |
| Grade 2; N (% of known) | 154 (54.0) | 123 (57.5) | 31 (43.7) |
| Grade 3; N (% of known) | 94 (33.0) | 69 (32.2) | 25 (35.2) |
| Multiple primary tumor; N (%) | 57 (17.1) | 48 (18.7) | 9 (11.9) |
| Malignancy in 1st/2nd degree relatives; N (%) | 131 (39.2) | 99 (38.4) | 32 (42.1) |
| Diabetes; N (%) | 138 (41.3) | 117 (45.3) | 21 (27.6) |
| Obesity (BMI>30); N (%) | 94 (28.1) | 80 (31.0) | 14 (18.4) |
| Smoking; N (%) | 192 (57.5) | 161 (62.4) | 31 (40.8) |
| Age at HCC Diagnosis (Years), Sex. | Variant | Personal ca History 1 | Family ca History | Cirrhosis and HCC Features |
|---|---|---|---|---|
| A. Established high-to-moderate cancer-predisposition genes | ||||
| 71, F | PMS2: c.1144+250_2175-1948del8907 | 0 | 0 | Viral |
| 72, M | NBN: c.657del5 (p.Lys219fs) | PrC (post) | PrC (father) | Alcoholic |
| 68, M | NBN: c.657del5 (p.Lys219fs) | 0 | TC (father) | Alcoholic |
| 68, M | NBN: c.657del5 (p.Lys219fs) | PrC (post) | 0 | Autoimmune |
| 58, F | NBN: c.657del5 (p.Lys219fs) | 0 | 0 | Viral, HCC recurrence post-Tx |
| 53, F | FH: c.1127A>C (p.Gln376Pro) | 0 | 0 | Viral |
| 71, M | RET: c.2304G>C (p.Glu768Asp) | CRC (pre) | BC (mother); sarcoma (brother) | Alcoholic |
| B. Candidate cancer-predisposition genes | ||||
| 69, F | DMBT1: c.2177-2A>C | BC | 0 | Autoimmune |
| 66, M | DMBT1: c.4828+1G>A | 0 | Leu (brother); HNC (brother) | Alcoholic |
| 69, M | DMBT1: c.4611C>G (p.Tyr1537Ter) | 0 | H&N (brother) | Alcoholic |
| 71, M | RAD50: c.1875C>G (p.Tyr625Ter) | PrC (post) | BC (mother) | Viral |
| 65, M | RAD50: c.2043delC (p.Val683fs) | 0 | 0 | Autoimmune |
| 65, M | RAD50: c.2521del9 (p.Thr841fs) * | 0 | 0 | Viral, HCC recurrence post-Tx |
| 65, M | ATRIP: c.1870del2 (p.Cys624fs) | 0 | 0 | Viral, HCC recurrence post-Tx |
| 61, F | ATRIP: c.1152del4 (p.Gly385fs) * | 0 | 0 | Autoimmune |
| 65, M | BLM: c.1642C>T (p.Gln548Ter) | 0 | LC (father) | Non-cirrhotic |
| 64, M | BLM: c.1642C>T (p.Gln548Ter) | 0 | 0 | Autoimmune |
| 68, M | ERCC2: c.2150C>G (r.2144_2190del45) | 0 | 0 | Autoimmune |
| 53, M | ERCC2: c.2150C>G (r.2144_2190del45) | 0 | 0 | Viral, HCC recurrence post-Tx |
| 71, F | LIG3: c.1283delT (p.His428fs) | 0 | 0 | Viral, HCC recurrence post-Tx |
| 77, F | LIG3: c.799C>T (p.Arg267Ter) | 0 | LC (father) | Non-cirrhotic |
| 68, M | MSH3: c.2686G>T (p.Gly896Ter) | 0 | Mel (mother) | Alcoholic |
| 66, M | MSH3: c.1480delA (p.Asn494fs) | 0 | 0 | Alcoholic |
| 66, F | SLX4: c.4207G>T (p.Glu1403Ter) | SkC (post) | 0 | Viral |
| 58, M | SLX4: c.4024delA (p.Ser1342fs) | 0 | GaC (mother) | Autoimmune |
| 57, F | AXIN1: c.64C>T (p.Arg22Ter) | 0 | 0 | Viral |
| 62, M | ERCC5: c.3285del10 (p.Ser1096fs) | 0 | 0 | Viral, HCC recurrence post-Tx |
| 71, M | ERCC6: c.537T>A (p.Tyr179Ter) | 0 | 0 | Viral |
| 61, M | EXO1: c.1578del2 (p.Asp526fs) | CRC (pre) | 0 | NASH |
| 60, F | FANCA: del16-17 | 0 | 0 | NASH |
| 59, M | FANCD2: c.990-1G>A | Leu (post) | GaC (mother) | NASH |
| 60, F | FANCG: c.313G>T (p.Glu105Ter) | 0 | H&N (father); HCC (mother) | NASH |
| 64, M | HOXB13: c.251G>A (p.Gly84Glu) | 0 | 0 | Autoimmune |
| 59, M | MCPH1: c.126del2 (p.Phe43fs) | 0 | BC (mother) | Autoimmune, HCC recurrence post-Tx |
| 57, M | MDC1: c.6081delC (p.Ser2028fs) | 0 | 0 | Alcoholic |
| 40, M | MMP8: c.460G>T (p.Gly154Ter) | 0 | 0 | Viral |
| 61, M | MLH3: c.3393dup2 (p.Thr1132fs) | 0 | BC (mother) | Autoimmune, HCC recurrence post-Tx |
| 59, F | NHEJ1: c.169C>T (p.Arg57Ter) | 0 | HCC (mother) | Alcoholic |
| 65, M | PIK3CG: c.2519del2 (p.Gln840fs) | H&N; SkC; PrC (all pre) | LC (father) | Alcoholic |
| 54, M | PMS1: c.1009insA (p.Tyr337fs) | 0 | 0 | Viral |
| 60, M | RAD1: c.168del5 (p.Lys57fs) | 0 | 0 | Alcoholic |
| 41, F | RECQL5: c.2308C>T (p.Arg770Ter) | 0 | RCC (mother) | Viral |
| 73, F | SBDS: c.258+2T>C | 0 | 0 | Viral |
| 69, M | SETX: c.5074dup2 (p.Leu1692fs) | 0 | LC (father) | NASH |
| 59, M | SMARCA4: c.859+1G>A | 0 | 0 | Viral |
| 69, M | TLR2: c.1339C>T (p.Arg447Ter) | Lym (pre) | HCC (father) | Viral |
| 74, M | TLR4: c.261-1G>C | 0 | BC (mother) | NASH |
| 45, F | XRCC1: c.406dupT (p.Tyr136fs) | 0 | BC (mother) | Alcoholic |
| Gene | Carriers in 334 Patients; N (%) | Carriers in 1662 PMC; N (%) | Odds Ratio (95% Confidence Interval) | p-Value |
|---|---|---|---|---|
| A. Established high-to-moderate cancer-predisposition genes | ||||
| PMS2 | 1 (0.3) | 4 (0.3) | 1.2 (0.14–11.11) | 0.8 |
| NBN | 4 (1.2) | 4 (0.3) | 5.0 (1.25–20.17) | 0.012 |
| FH | 1 (0.3) | 0 | n.d. | n.d. |
| RET | 1 (0.3) | 2 (0.1) | 2.5 (0.23–27.49) | 0.4 |
| All carriers. | 7 (2.1) | 10 (0.6) 1 | ||
| B. Candidate cancer-predisposition genes | ||||
| DMBT1 | 3 (0.9) | 2 (0.1) | 7.5 (1.25–45.13) | 0.010 |
| RAD502 | 3 (0.9) | 3 (0.2) | 5.0 (1.01–24.90) | 0.029 |
| ATRIP2 | 2 (0.6) | 3 (0.2) | 3.3 (0.56–19.98) | 0.2 |
| BLM | 2 (0.6) | 7 (0.4) | 1.4 (0.30–6.87) | 0.7 |
| ERCC2 | 2 (0.6) | 8 (0.5) | 1.2 (0.26–5.88) | 0.8 |
| LIG3 | 2 (0.6) | 1 (0.1) | 10.0 (0.91–110.48) | 0.021 |
| MSH3 | 2 (0.6) | 6 (0.4) | 1.7 (0.33–8.26) | 0.5 |
| SLX4 | 2 (0.6) | 2 (0.1) | 5.0 (0.70–35.56) | 0.1 |
| AXIN1 | 1 (0.3) | 0 | n.d. | n.d. |
| ERCC5 | 1 (0.3) | 0 | n.d. | n.d. |
| ERCC6 | 1 (0.3) | 0 | n.d. | n.d. |
| EXO1 | 1 (0.3) | 2 (0.1) | 2.5 (0.23–27.49) | 0.4 |
| FANCA | 1 (0.3) | 7 (0.4) | 0.7 (0.09–5.77) | 0.7 |
| FANCD2 | 1 (0.3) | 0 | n.d. | n.d. |
| FANCG | 1 (0.3) | 2 (0.1) | 2.5 (0.23–27.49) | 0.4 |
| HOXB13 | 1 (0.3) | 4 (0.3) | 1.2 (0.14–11.14) | 0.8 |
| MCPH1 | 1 (0.3) | 10 (0.6) | 0.5 (0.06–3.88) | 0.5 |
| MDC1 | 1 (0.3) | 0 | n.d. | n.d. |
| MLH3 | 1 (0.3) | 1 (0.1) | 5.0 (0.31–79.75) | 0.2 |
| MMP8 | 1 (0.3) | 5 (0.3) | 1.0 (0.12–8.52) | 0.9 |
| NHEJ1 | 1 (0.3) | 0 | n.d. | n.d. |
| PIK3CG | 1 (0.3) | 0 | n.d. | n.d. |
| PMS1 | 1 (0.3) | 2 (0.1) | 2.5 (0.23–27.49) | 0.4 |
| RAD1 | 1 (0.3) | 0 | n.d. | n.d. |
| RECQL5 | 1 (0.3) | 6 (0.4) | 0.8 (0.10–6.89) | 0.9 |
| SBDS | 1 (0.3) | 13 (0.8) | 0.4 (0.05–2.91) | 0.3 |
| SETX | 1 (0.3) | 10 (0.6) | 0.5 (0.06–3.88) | 0.5 |
| SMARCA4 | 1 (0.3) | 0 | n.d. | n.d. |
| TLR2 | 1 (0.3) | 1 (0.1) | 5.0 (0.31–79.75) | 0.2 |
| TLR4 | 1 (0.3) | 2 (0.1) | 2.5 (0.23–27.49) | 0.4 |
| XRCC1 | 1 (0.3) | 7 (0.4) | 0.7 (0.09–5.77) | 0.7 |
| All carriers | 40 (12.0) | 104 (6.3) 1 | - | - |
| This Study; N (%) | Uson Junior et al. 2022 (Ref. [9]); N (%) | Mezina et al. 2021 (Prospective) (Ref. [10]); N (%) | Mezina et al. 2021 (Retrospective) (Ref. [10]); N (%) | |
|---|---|---|---|---|
| HCC patients analyzed (N) | 334 | 44 | 217 | 219 |
| Genes analyzed (N) | 226 | 83 | 134 | 1–154 |
| APC | 0 | 0 | 0 | 2 (0.91) |
| ATM | 0 | 0 | 0 | 1 (0.46) |
| BARD1 | 0 | 1 (2.27) | 0 | 1 (0.46) |
| BLM | 2 (0.59) | 0 | 0 | 0 |
| BRCA1 | 0 | 0 | 0 | 1 (0.46) |
| BRCA2 | 0 | 0 | 2 (0.92) | 6 (2.74) |
| BRIP1 | 0 | 0 | 4 * (1.84) | 1 (0.46) |
| CDKN2A | 0 | 1 (2.27) | 0 | 0 |
| CHEK2 | 0 | 0 | 3 * (1.38) | 2 (0.91) |
| FANCA | 1 (0.29) | n.a. | 5 (2.30) | 1 (0.46) |
| FANCD2 | 1 (0.29) | n.a. | 2 (0.92) | 0 |
| FANCG | 1 (0.29) | n.a. | 0 | 0 |
| FANCM | 0 | n.a. | 1 * (0.46) | 0 |
| FH | 1 (0.29) | 0 | 0 | 2 (0.91) |
| HOXB13 | 1 (0.29) | 0 | 0 | 0 |
| MITF | n.a. | 1 (2.27) | 1 * (0.46) | 1 (0.46) |
| MLH3 | 1 (0.29) | n.a. | 0 | 0 |
| MSH2 | 0 | 0 | 0 | 2 (0.91) |
| MSH3 | 2 (0.59) | n.a. | 1 * (0.46) | 0 |
| MSH6 | 0 | 0 | 1 (0.46) | 0 |
| MUTYH | 0 | 0 | 3 (1.38) | 2 (0.91) |
| NBN | 4 (1.19) | 2 (4.54) | 0 | 2 (0.91) |
| NF1 | 0 | 0 | 1 (0.46) | 0 |
| NTHL1 | n.a. | 0 | 0 | 1 (0.46) |
| PALB2 | 0 | 0 | 0 | 3 (1.37) |
| PMS2 | 1 (0.29) | 0 | 1 (0.46) | 0 |
| RAD50 | 3 * (0.89) | 1 (2.27) | 1 (0.46) | 0 |
| RAD51D | 0 | 1 (2.27) | 0 | 0 |
| RET | 1 (0.29) | 0 | 0 | 0 |
| SLX4 | 2 (0.59) | n.a. | 0 | 0 |
| SMARCA4 | 1 (0.29) | 0 | 0 | 0 |
| TMEM127 | 0 | 0 | 1 * (0.46) | 0 |
| TP53 | 0 | 0 | 0 | 2 (0.91) |
| Established PV carriers in CPG* | 7 (2.1) | 5 (11.4) | 12 (5.5) | 25 (11.4) |
| All carriers (referred in the study; N) | 47 (14.1) | 7 (15.9) | 25 (11.5) | 30 (13.7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horackova, K.; Frankova, S.; Zemankova, P.; Nehasil, P.; Cerna, M.; Neroldova, M.; Otahalova, B.; Kral, J.; Hovhannisyan, M.; Stranecky, V.; et al. Low Frequency of Cancer-Predisposition Gene Mutations in Liver Transplant Candidates with Hepatocellular Carcinoma. Cancers 2023, 15, 201. https://doi.org/10.3390/cancers15010201
Horackova K, Frankova S, Zemankova P, Nehasil P, Cerna M, Neroldova M, Otahalova B, Kral J, Hovhannisyan M, Stranecky V, et al. Low Frequency of Cancer-Predisposition Gene Mutations in Liver Transplant Candidates with Hepatocellular Carcinoma. Cancers. 2023; 15(1):201. https://doi.org/10.3390/cancers15010201
Chicago/Turabian StyleHorackova, Klara, Sona Frankova, Petra Zemankova, Petr Nehasil, Marta Cerna, Magdalena Neroldova, Barbora Otahalova, Jan Kral, Milena Hovhannisyan, Viktor Stranecky, and et al. 2023. "Low Frequency of Cancer-Predisposition Gene Mutations in Liver Transplant Candidates with Hepatocellular Carcinoma" Cancers 15, no. 1: 201. https://doi.org/10.3390/cancers15010201
APA StyleHorackova, K., Frankova, S., Zemankova, P., Nehasil, P., Cerna, M., Neroldova, M., Otahalova, B., Kral, J., Hovhannisyan, M., Stranecky, V., Zima, T., Safarikova, M., Kalousova, M., Consortium, C., Novotny, J., Sperl, J., Borecka, M., Jelinkova, S., Vocka, M., ... Soukupova, J. (2023). Low Frequency of Cancer-Predisposition Gene Mutations in Liver Transplant Candidates with Hepatocellular Carcinoma. Cancers, 15(1), 201. https://doi.org/10.3390/cancers15010201






