LASP1, CERS6, and Actin Form a Ternary Complex That Promotes Cancer Cell Migration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cultures
2.2. Antibodies and Chemical Reagents
2.3. siRNA Transfection
2.4. Plasmid Construction and Transfection
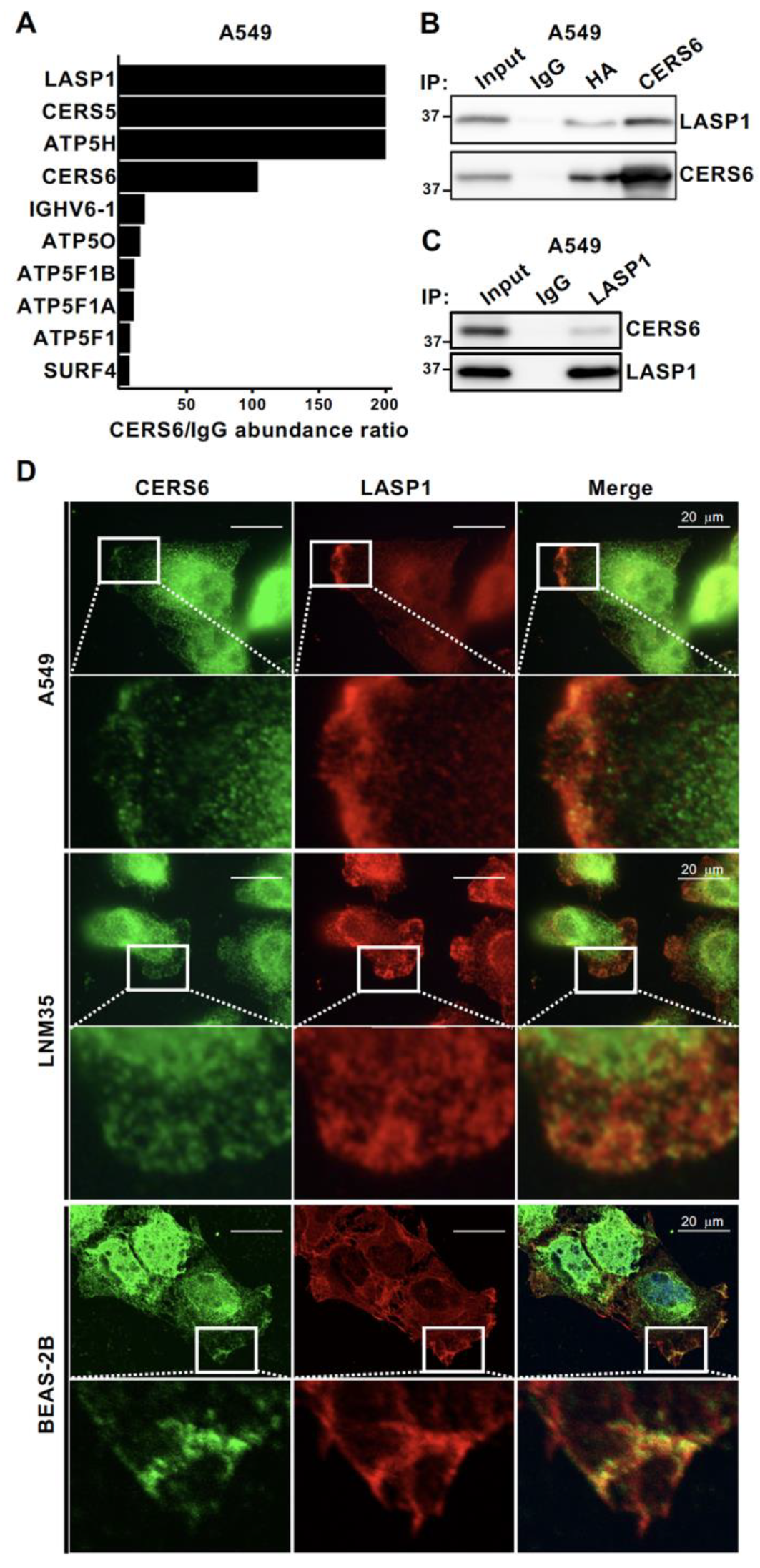
2.5. Co-immunoprecipitation, Liquid Chromatography, and Tandem Mass Spectrometry (IP and LC-MS/MS)
2.6. In Vitro Motility and Complementation Assays
2.7. Quantitative RT-PCR
2.8. Ceramide Quantitation
2.9. Co-Immunoprecipitation and Western Blotting (IP-WB)
2.10. Immunofluorescent Staining
2.11. Statistics
3. Results
3.1. LASP1 Is a CERS6 Binding Partner
3.2. LIM Domain of LASP1 Required for CERS6 Interaction
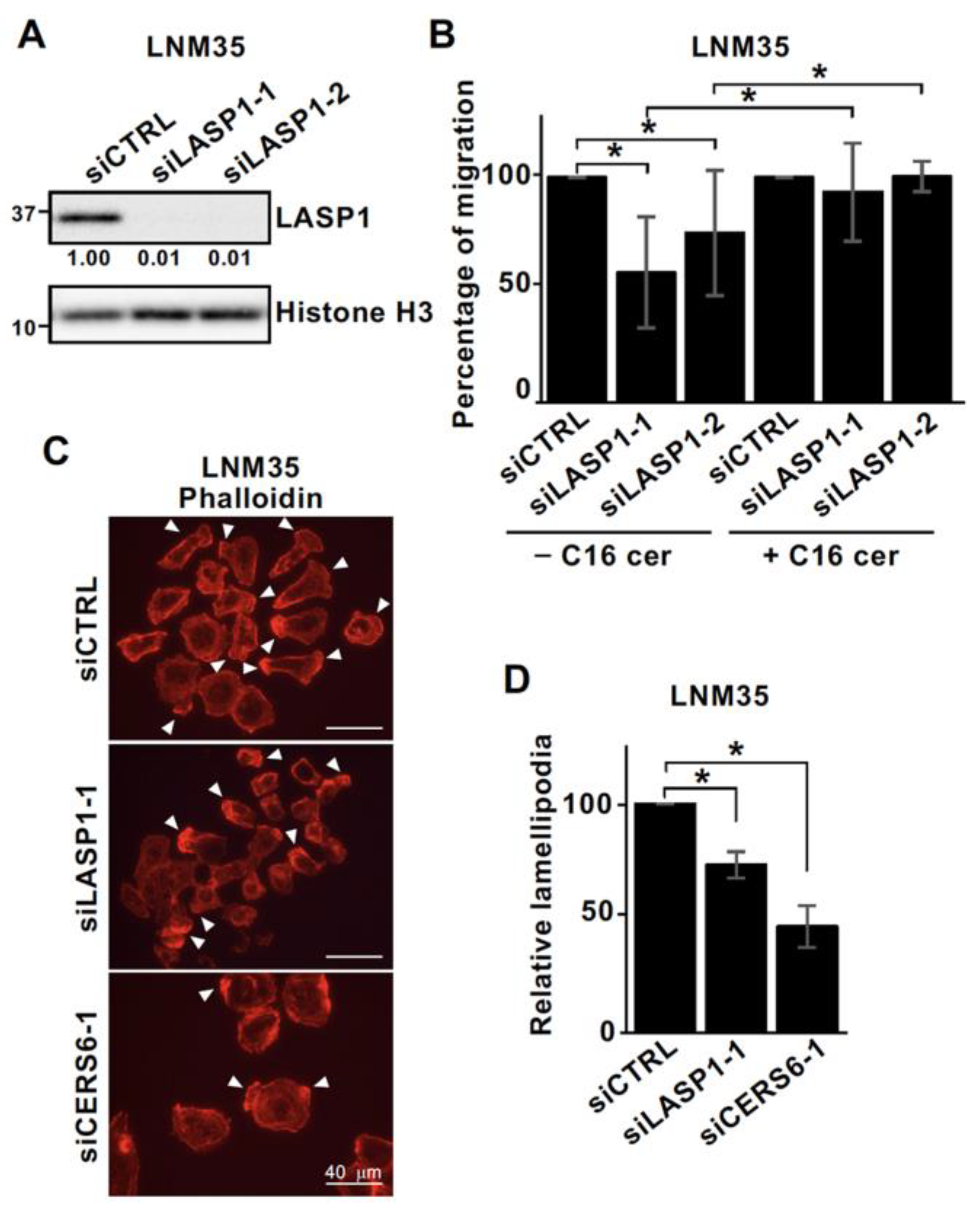
3.3. LASP1 and CERS6 Coordinate to Promote Cell Migration
3.4. Complex Formation May Alter LASP1 and CERS6 Levels in LNM35 Cells
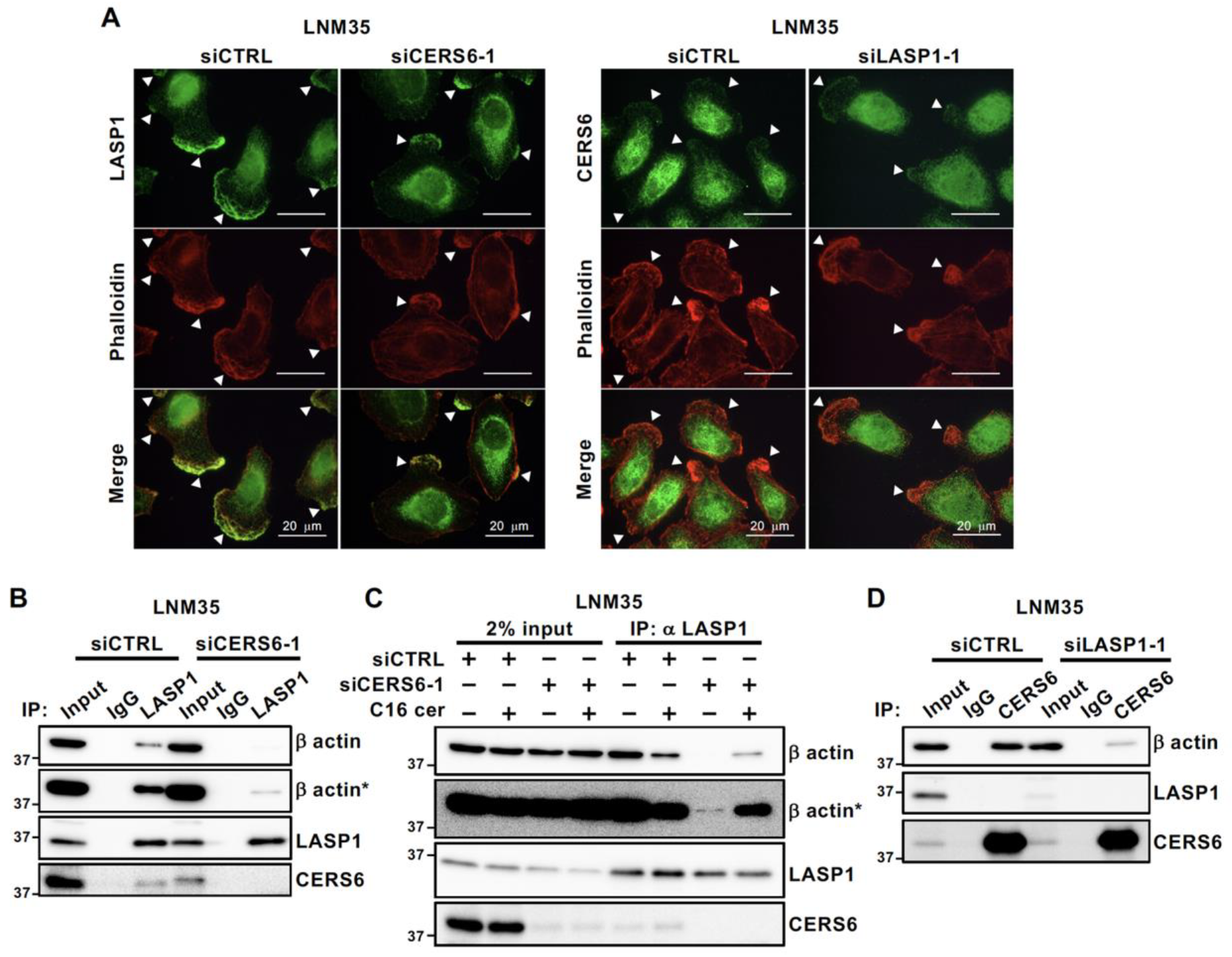
3.5. Complex Formation Facilitates LASP1– and CERS6–Actin Interaction
4. Discussion
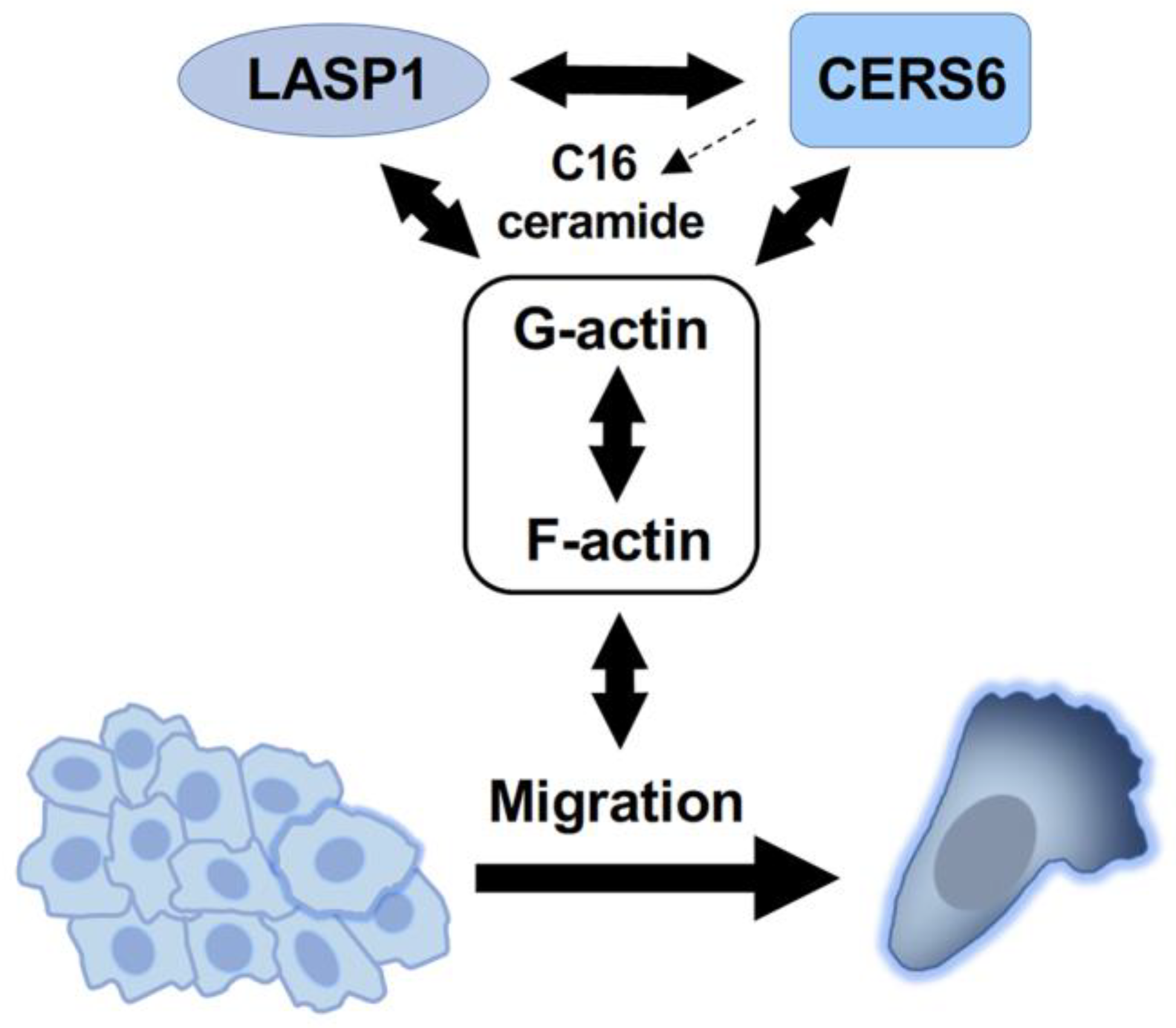
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aoyama, Y.; Sobue, S.; Mizutani, N.; Inoue, C.; Kawamoto, Y.; Nishizawa, Y.; Ichihara, M.; Kyogashima, M.; Suzuki, M.; Nozawa, Y.; et al. Modulation of the sphingolipid rheostat is involved in paclitaxel resistance of the human prostate cancer cell line PC3-PR. Biochem. Biophys. Res. Commun. 2017, 486, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Murakami, M.; Furuhata, A.; Gao, S.; Yoshida, K.; Sobue, S.; Hagiwara, K.; Takagi, A.; Kojima, T.; Suzuki, M.; et al. Transcriptional regulation of neutral sphingomyelinase 2 gene expression of a human breast cancer cell line, MCF-7, induced by the anti-cancer drug, daunorubicin. Biochim. Biophys. Acta 2009, 1789, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, N.; Inoue, M.; Omori, Y.; Ito, H.; Tamiya-Koizumi, K.; Takagi, A.; Kojima, T.; Nakamura, M.; Iwaki, S.; Nakatochi, M.; et al. Increased acid ceramidase expression depends on upregulation of androgen-dependent deubiquitinases, USP2, in a human prostate cancer cell line, LNCaP. J. Biochem. 2015, 158, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, N.; Omori, Y.; Kawamoto, Y.; Sobue, S.; Ichihara, M.; Suzuki, M.; Kyogashima, M.; Nakamura, M.; Tamiya-Koizumi, K.; Nozawa, Y.; et al. Resveratrol-induced transcriptional up-regulation of ASMase (SMPD1) of human leukemia and cancer cells. Biochem. Biophys. Res. Commun. 2016, 470, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Sobue, S.; Nemoto, S.; Murakami, M.; Ito, H.; Kimura, A.; Gao, S.; Furuhata, A.; Takagi, A.; Kojima, T.; Nakamura, M.; et al. Implications of sphingosine kinase 1 expression level for the cellular sphingolipid rheostat: Relevance as a marker for daunorubicin sensitivity of leukemia cells. Int. J. Hematol. 2008, 87, 266–275. [Google Scholar] [CrossRef]
- Karahatay, S.; Thomas, K.; Koybasi, S.; Senkal, C.E.; Elojeimy, S.; Liu, X.; Bielawski, J.; Day, T.A.; Gillespie, M.B.; Sinha, D.; et al. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): Attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007, 256, 101–111. [Google Scholar] [CrossRef]
- Erez-Roman, R.; Pienik, R.; Futerman, A.H. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem. Biophys. Res. Commun. 2010, 391, 219–223. [Google Scholar] [CrossRef]
- Suzuki, M.; Cao, K.; Kato, S.; Mizutani, N.; Tanaka, K.; Arima, C.; Tai, M.C.; Nakatani, N.; Yanagisawa, K.; Takeuchi, T.; et al. CERS6 required for cell migration and metastasis in lung cancer. J. Cell. Mol. Med. 2020, 24, 11949–11959. [Google Scholar] [CrossRef]
- Shi, H.; Niimi, A.; Takeuchi, T.; Shiogama, K.; Mizutani, Y.; Kajino, T.; Inada, K.; Hase, T.; Hatta, T.; Shibata, H.; et al. CEBPgamma facilitates lamellipodia formation and cancer cell migration through CERS6 upregulation. Cancer Sci. 2021, 112, 2770–2780. [Google Scholar] [CrossRef]
- Senkal, C.E.; Ponnusamy, S.; Bielawski, J.; Hannun, Y.A.; Ogretmen, B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010, 24, 296–308. [Google Scholar] [CrossRef]
- Chen, H.; He, B.; Ke, F. Ceramide Synthase 6 Mediates Triple-Negative Breast Cancer Response to Chemotherapy through RhoA- and EGFR-Mediated Signaling Pathways. J. Breast Cancer 2022, 25, 500–512. [Google Scholar] [CrossRef]
- Tomizawa, S.; Tamori, M.; Tanaka, A.; Utsumi, N.; Sato, H.; Hatakeyama, H.; Hisaka, A.; Kohama, T.; Yamagata, K.; Honda, T.; et al. Inhibitory effects of ceramide kinase on Rac1 activation, lamellipodium formation, cell migration, and metastasis of A549 lung cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158675. [Google Scholar] [CrossRef]
- Panaviene, Z.; Moncman, C.L. Linker region of nebulin family members plays an important role in targeting these molecules to cellular structures. Cell Tissue Res. 2007, 327, 353–369. [Google Scholar] [CrossRef]
- Schreiber, V.; Moog-Lutz, C.; Regnier, C.H.; Chenard, M.P.; Boeuf, H.; Vonesch, J.L.; Tomasetto, C.; Rio, M.C. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol. Med. 1998, 4, 675–687. [Google Scholar] [CrossRef]
- Rachlin, A.S.; Otey, C.A. Identification of palladin isoforms and characterization of an isoform-specific interaction between Lasp-1 and palladin. J. Cell Sci. 2006, 119, 995–1004. [Google Scholar] [CrossRef]
- Raman, D.; Sai, J.; Neel, N.F.; Chew, C.S.; Richmond, A. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS ONE 2010, 5, e10050. [Google Scholar] [CrossRef]
- Beckmann, D.; Römer-Hillmann, A.; Krause, A.; Hansen, U.; Wehmeyer, C.; Intemann, J.; de Gorter, D.J.J.; Dankbar, B.; Hillen, J.; Heitzmann, M.; et al. Lasp1 regulates adherens junction dynamics and fibroblast transformation in destructive arthritis. Nat. Commun. 2021, 12, 3624. [Google Scholar] [CrossRef]
- Kozaki, K.; Miyaishi, O.; Tsukamoto, T.; Tatematsu, Y.; Hida, T.; Takahashi, T.; Takahashi, T. Establishment and characterization of a human lung cancer cell line NCI-H460-LNM35 with consistent lymphogenous metastasis via both subcutaneous and orthotopic propagation. Cancer Res. 2000, 60, 2535–2540. [Google Scholar]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Mizutani, Y.; Kihara, A.; Igarashi, Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005, 390, 263–271. [Google Scholar] [CrossRef]
- Kano, K.; Noda, S.; Sato, S.; Kuwata, K.; Mishiro-Sato, E. An efficient in-gel digestion method on small amounts of protein sample from large intact gel pieces. Sep. Sci. Plus 2023, 6, 2200121. [Google Scholar] [CrossRef]
- Mizutani, Y.; Tsuge, S.; Takeda, H.; Hasegawa, Y.; Shiogama, K.; Onouchi, T.; Inada, K.; Sawasaki, T.; Tsutsumi, Y. In situ visualization of plasma cells producing antibodies reactive to Porphyromonas gingivalis in periodontitis: The application of the enzyme-labeled antigen method. Mol. Oral Microbiol. 2014, 29, 156–173. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.M.; Tomida, S.; Masuda, Y.; Arima, C.; Cao, K.; Kasahara, T.A.; Osada, H.; Yatabe, Y.; Akashi, T.; Kamiya, K.; et al. Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer. Cancer Res. 2010, 70, 8407–8416. [Google Scholar] [CrossRef] [PubMed]
- Butt, E.; Raman, D. New Frontiers for the Cytoskeletal Protein LASP1. Front. Oncol. 2018, 8, 391. [Google Scholar] [CrossRef]
- Ruggieri, V.; Agriesti, F.; Tataranni, T.; Perris, R.; Mangieri, D. Paving the path for invasion: The polyedric role of LASP1 in cancer. Tumour Biol. 2017, 39, 1010428317705757. [Google Scholar] [CrossRef]
- Orth, M.F.; Cazes, A.; Butt, E.; Grunewald, T.G. An update on the LIM and SH3 domain protein 1 (LASP1): A versatile structural, signaling, and biomarker protein. Oncotarget 2015, 6, 26–42. [Google Scholar] [CrossRef]
- Tomasetto, C.; Regnier, C.; Moog-Lutz, C.; Mattei, M.G.; Chenard, M.P.; Lidereau, R.; Basset, P.; Rio, M.C. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11–q21.3 region of chromosome 17. Genomics 1995, 28, 367–376. [Google Scholar] [CrossRef]
- Butt, E.; Gambaryan, S.; Gottfert, N.; Galler, A.; Marcus, K.; Meyer, H.E. Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J. Biol. Chem. 2003, 278, 15601–15607. [Google Scholar] [CrossRef]
- Nakagawa, H.; Terasaki, A.G.; Suzuki, H.; Ohashi, K.; Miyamoto, S. Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett. 2006, 580, 3223–3228. [Google Scholar] [CrossRef]
- Zhao, T.; Ren, H.; Li, J.; Chen, J.; Zhang, H.; Xin, W.; Sun, Y.; Sun, L.; Yang, Y.; Sun, J.; et al. LASP1 Is a HIF1α Target Gene Critical for Metastasis of Pancreatic Cancer. Cancer Res. 2015, 75, 111–119. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, H.; Liu, C.; Liu, Y.; Wang, X.; Wang, S.; Sun, X.; Li, J.; Deng, Y.; Jiang, Y.; et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut 2010, 59, 1226–1235. [Google Scholar] [CrossRef]
- Lin, X.; Liu, X.; Fang, Y.; Weng, X. LIM and SH3 protein 1 promotes tumor proliferation and metastasis in lung carcinoma. Oncol. Lett. 2016, 12, 4756–4760. [Google Scholar] [CrossRef]
- Yang, F.; Zhou, X.; Du, S.; Zhao, Y.; Ren, W.; Deng, Q.; Wang, F.; Yuan, J. LIM and SH3 domain protein 1 (LASP-1) overexpression was associated with aggressive phenotype and poor prognosis in clear cell renal cell cancer. PLoS ONE 2014, 9, e100557. [Google Scholar] [CrossRef]
- Frietsch, J.J.; Grunewald, T.G.; Jasper, S.; Kammerer, U.; Herterich, S.; Kapp, M.; Honig, A.; Butt, E. Nuclear localisation of LASP-1 correlates with poor long-term survival in female breast cancer. Br. J. Cancer 2010, 102, 1645–1653. [Google Scholar] [CrossRef]
- Chew, C.S.; Chen, X.; Parente, J.A.; Tarrer, S.; Okamoto, C.; Qin, H.-Y. Lasp-1 binds to non-muscle F-actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J. Cell Sci. 2002, 115, 4787–4799. [Google Scholar] [CrossRef]
- Li, B.; Zhuang, L.; Trueb, B. Zyxin Interacts with the SH3 Domains of the Cytoskeletal Proteins LIM-nebulette and Lasp-1. J. Biol. Chem. 2004, 279, 20401–20410. [Google Scholar] [CrossRef]
- Spence, H.J.; McGarry, L.; Chew, C.S.; Carragher, N.O.; Scott-Carragher, L.A.; Yuan, Z.; Croft, D.R.; Olson, M.F.; Frame, M.; Ozanne, B.W. AP-1 differentially expressed proteins Krp1 and fibronectin cooperatively enhance Rho-ROCK-independent mesenchymal invasion by altering the function, localization, and activity of nondifferentially expressed proteins. Mol. Cell. Biol. 2006, 26, 1480–1495. [Google Scholar] [CrossRef]
- Salvi, A.; Bongarzone, I.; Ferrari, L.; Abeni, E.; Arici, B.; De Bortoli, M.; Scuri, S.; Bonini, D.; Grossi, I.; Benetti, A.; et al. Molecular characterization of LASP-1 expression reveals vimentin as its new partner in human hepatocellular carcinoma cells. Int. J. Oncol. 2015, 46, 1901–1912. [Google Scholar] [CrossRef]
- Zhu, W.; Smith, J.W.; Huang, C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 2010, 840518. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niimi, A.; Limsirichaikul, S.; Kano, K.; Mizutani, Y.; Takeuchi, T.; Sawangsri, P.; Tran, D.Q.; Kawamoto, Y.; Suzuki, M. LASP1, CERS6, and Actin Form a Ternary Complex That Promotes Cancer Cell Migration. Cancers 2023, 15, 2781. https://doi.org/10.3390/cancers15102781
Niimi A, Limsirichaikul S, Kano K, Mizutani Y, Takeuchi T, Sawangsri P, Tran DQ, Kawamoto Y, Suzuki M. LASP1, CERS6, and Actin Form a Ternary Complex That Promotes Cancer Cell Migration. Cancers. 2023; 15(10):2781. https://doi.org/10.3390/cancers15102781
Chicago/Turabian StyleNiimi, Atsuko, Siripan Limsirichaikul, Keiko Kano, Yasuyoshi Mizutani, Toshiyuki Takeuchi, Patinya Sawangsri, Dat Quoc Tran, Yoshiyuki Kawamoto, and Motoshi Suzuki. 2023. "LASP1, CERS6, and Actin Form a Ternary Complex That Promotes Cancer Cell Migration" Cancers 15, no. 10: 2781. https://doi.org/10.3390/cancers15102781
APA StyleNiimi, A., Limsirichaikul, S., Kano, K., Mizutani, Y., Takeuchi, T., Sawangsri, P., Tran, D. Q., Kawamoto, Y., & Suzuki, M. (2023). LASP1, CERS6, and Actin Form a Ternary Complex That Promotes Cancer Cell Migration. Cancers, 15(10), 2781. https://doi.org/10.3390/cancers15102781