Cellular and Molecular Mechanisms of Liver Fibrosis in Patients with NAFLD
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Liver Biopsy Samples
2.2. RNA Isolation and Targeted Gene Expression Profiling
2.3. Multiplex Immunofluorescence (mIF) and Digital Image Analysis
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the NAFLD Patient Cohort
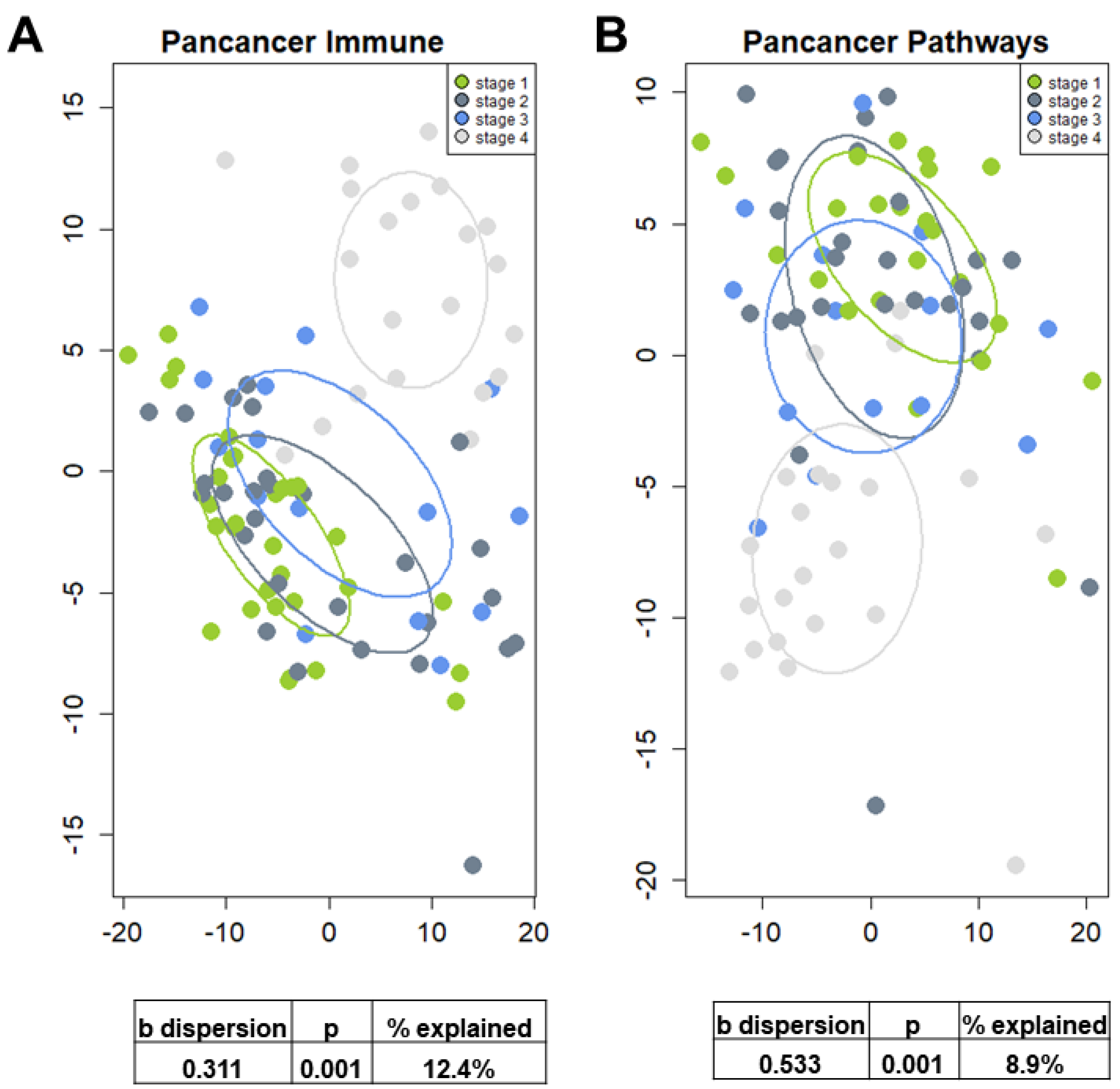
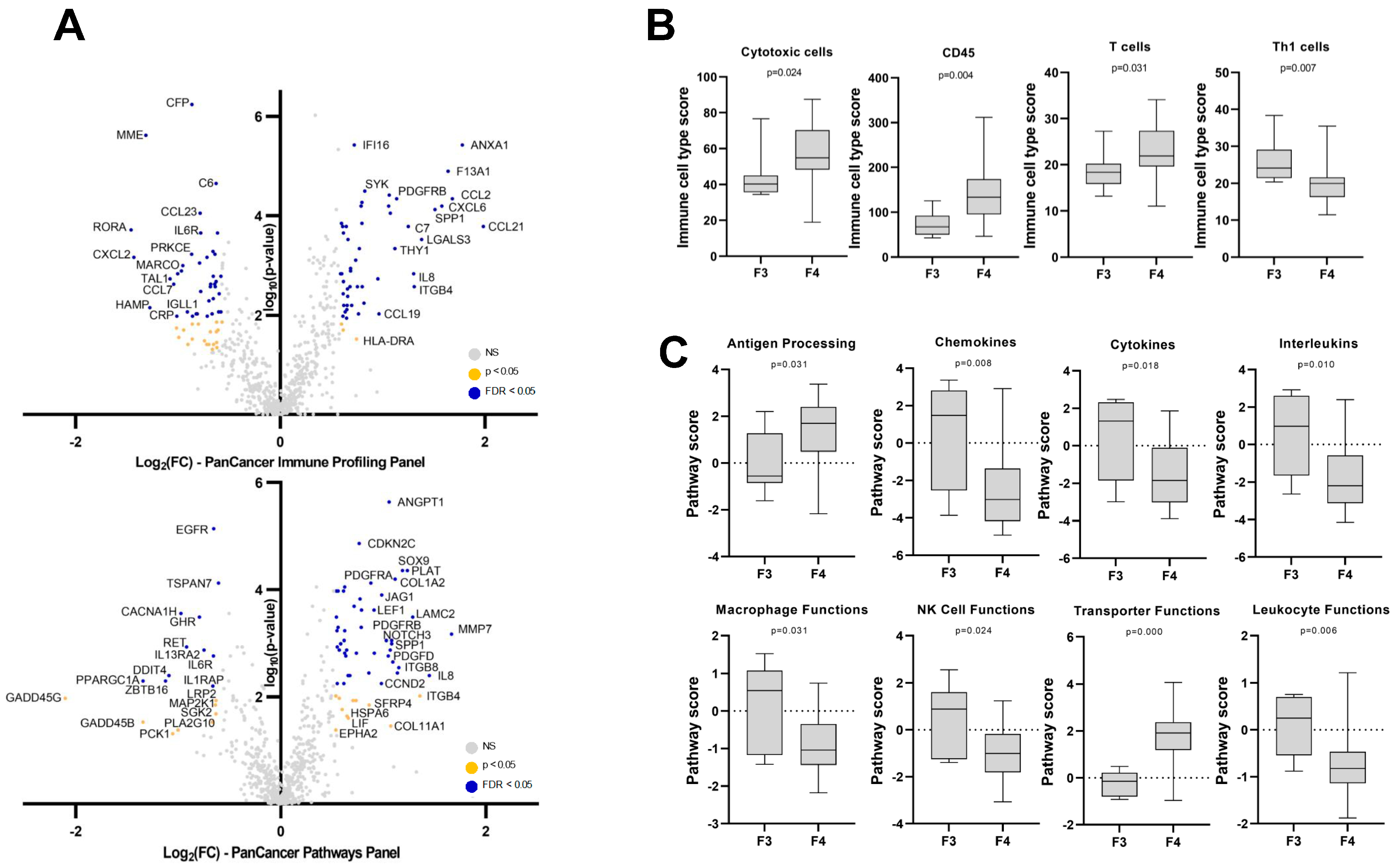
3.2. Hepatic Gene Expression Changes in NAFLD Patients with Cirrhosis
3.3. Hepatic Gene Expression Changes Associated with Fibrosis Progression from F1 to F4 in NAFLD Patients
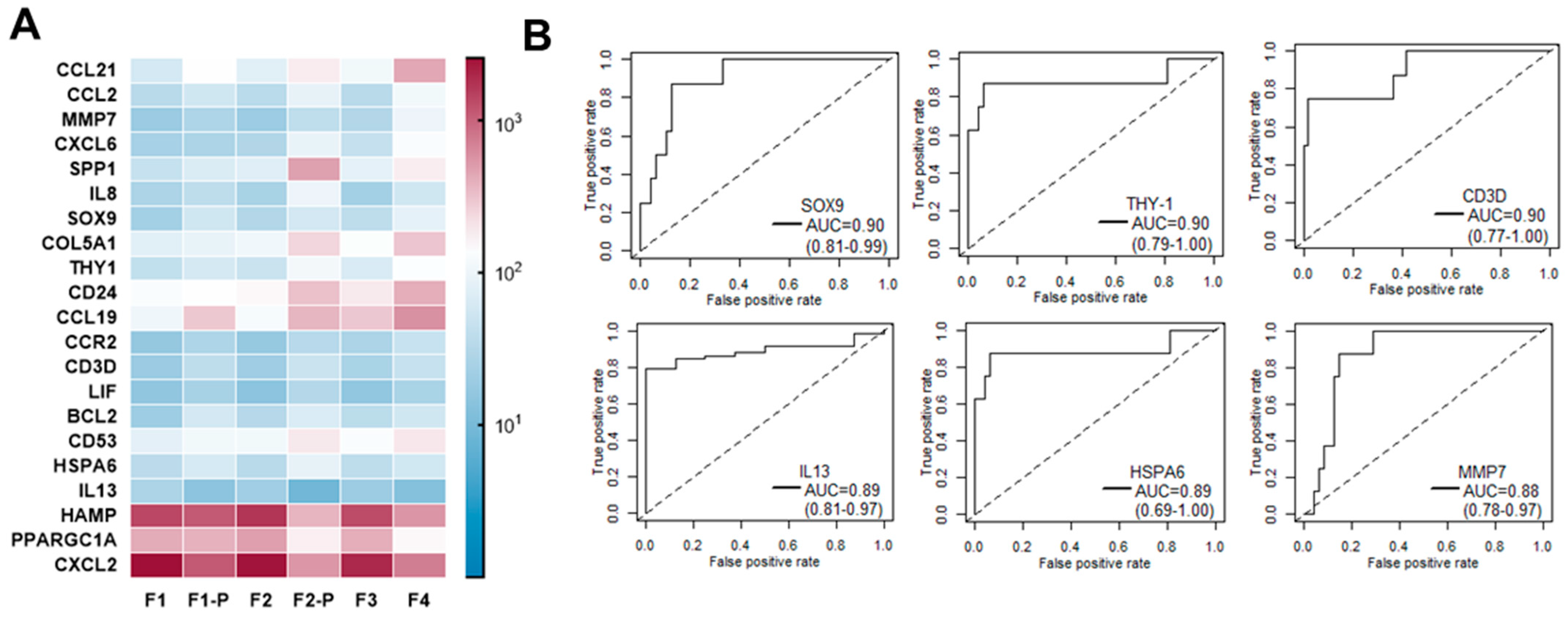
3.4. Early Gene Expression Changes Predicting Fast Liver Fibrosis Progression: A Pilot Study
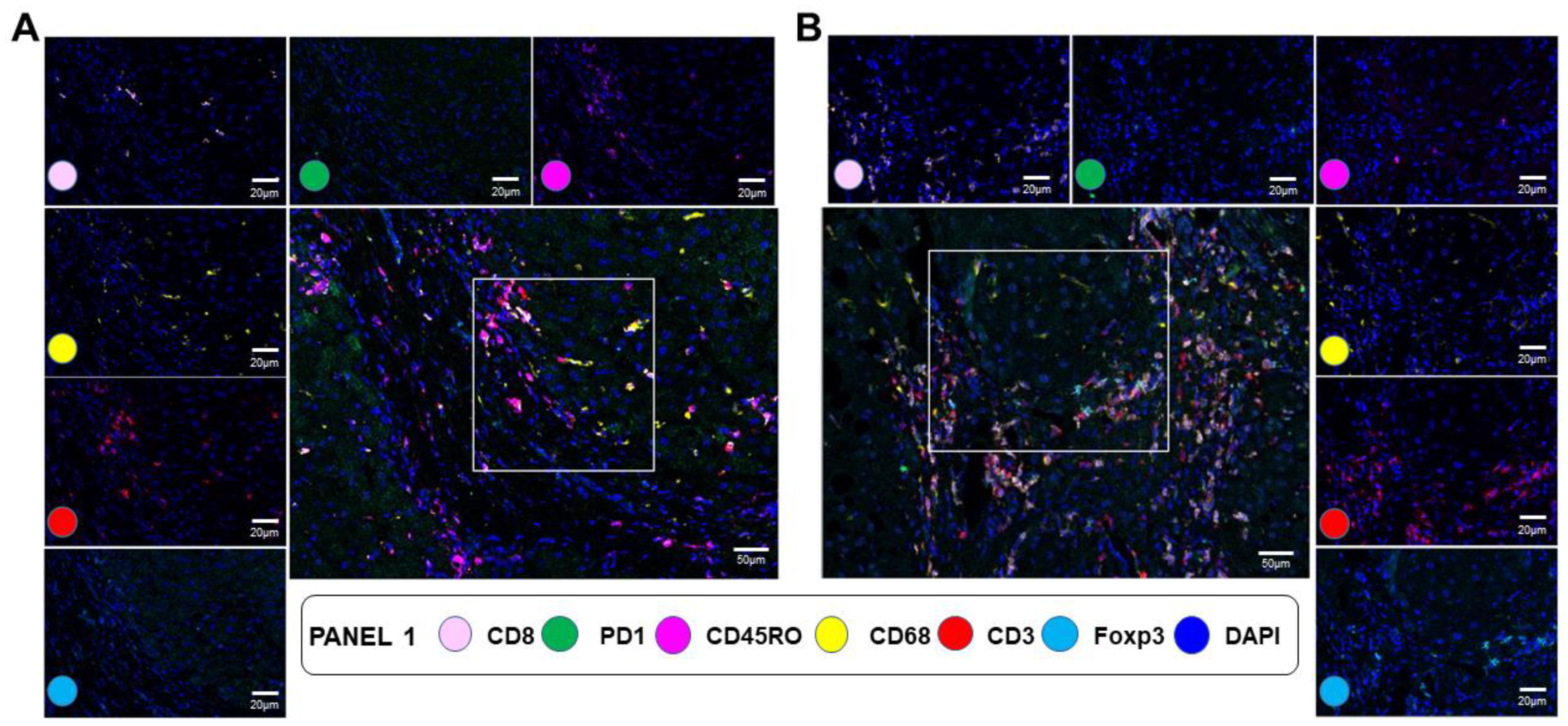
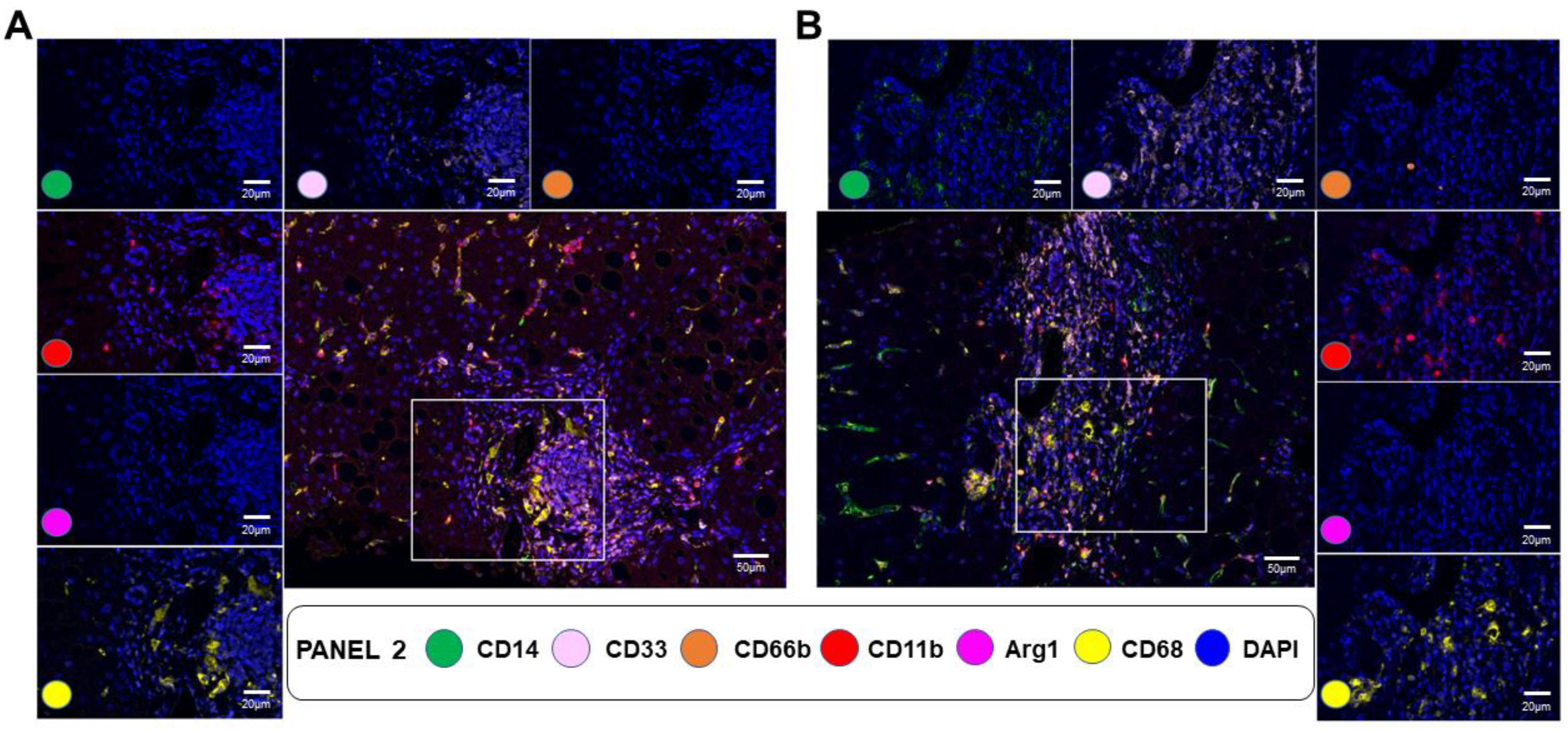
3.5. mIF Staining for Immune Cell Type Distribution in NAFLD Fibrotic Livers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotter, T.G.; Rinella, M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Stepanova, M.; Younossi, Y.; Golabi, P.; Mishra, A.; Rafiq, N.; Henry, L. Epidemiology of chronic liver diseases in the USA in the past three decades. Gut 2020, 69, 564–568. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Stepanova, M.; Ong, J.; Trimble, G.; AlQahtani, S.; Younossi, I.; Ahmed, A.; Racila, A.; Henry, L. Nonalcoholic Steatohepatitis Is the Most Rapidly Increasing Indication for Liver Transplantation in the United States. Clin. Gastroenterol. Hepatol. 2021, 19, 580–589.e5. [Google Scholar] [CrossRef] [PubMed]
- Doycheva, I.; Issa, D.; Watt, K.D.; Lopez, R.; Rifai, G.; Alkhouri, N. Nonalcoholic Steatohepatitis is the Most Rapidly Increasing Indication for Liver Transplantation in Young Adults in the United States. J. Clin. Gastroenterol. 2018, 52, 339–346. [Google Scholar] [CrossRef]
- Paik, J.M.; Golabi, P.; Younossi, Y.; Saleh, N.; Nhyira, A.; Younossi, Z.M. The Growing Burden of Disability Related to Chronic Liver Disease in the United States: Data From the Global Burden of Disease Study 2007–2017. Hepatol. Commun. 2021, 5, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Dulai, P.S.; Singh, S.; Patel, J.; Soni, M.; Prokop, L.J.; Younossi, Z.; Sebastiani, G.; Ekstedt, M.; Hagstrom, H.; Nasr, P.; et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 2017, 65, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.S.; Taylor, R.J.; Bayliss, S.; Hagstrom, H.; Nasr, P.; Schattenberg, J.M.; Ishigami, M.; Toyoda, H.; Wai-Sun Wong, V.; Peleg, N.; et al. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology 2020, 158, 1611–1625.e1612. [Google Scholar] [CrossRef]
- Lee, Y.A.; Friedman, S.L. Inflammatory and fibrotic mechanisms in NAFLD-Implications for new treatment strategies. J. Intern. Med. 2022, 291, 11–31. [Google Scholar] [CrossRef]
- He, W.; Huang, C.; Zhang, X.; Wang, D.; Chen, Y.; Zhao, Y.; Li, X. Identification of transcriptomic signatures and crucial pathways involved in non-alcoholic steatohepatitis. Endocrine 2021, 73, 52–64. [Google Scholar] [CrossRef]
- Zeng, F.; Zhang, Y.; Han, X.; Zeng, M.; Gao, Y.; Weng, J. Predicting Non-Alcoholic Fatty Liver Disease Progression and Immune Deregulations by Specific Gene Expression Patterns. Front. Immunol. 2020, 11, 609900. [Google Scholar] [CrossRef] [PubMed]
- Hasin-Brumshtein, Y.; Sakaram, S.; Khatri, P.; He, Y.D.; Sweeney, T.E. A robust gene expression signature for NASH in liver expression data. Sci. Rep. 2022, 12, 2571. [Google Scholar] [CrossRef]
- Govaere, O.; Cockell, S.; Tiniakos, D.; Queen, R.; Younes, R.; Vacca, M.; Alexander, L.; Ravaioli, F.; Palmer, J.; Petta, S.; et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 2020, 12, eaba4448. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Keogh, A.; Waldt, A.; Cuttat, R.; Neri, M.; Zhu, S.; Schuierer, S.; Ruchti, A.; Crochemore, C.; Knehr, J.; et al. Single-cell and bulk transcriptomics of the liver reveals potential targets of NASH with fibrosis. Sci. Rep. 2021, 11, 19396. [Google Scholar] [CrossRef]
- Pantano, L.; Agyapong, G.; Shen, Y.; Zhuo, Z.; Fernandez-Albert, F.; Rust, W.; Knebel, D.; Hill, J.; Boustany-Kari, C.M.; Doerner, J.F.; et al. Molecular characterization and cell type composition deconvolution of fibrosis in NAFLD. Sci. Rep. 2021, 11, 18045. [Google Scholar] [CrossRef]
- Fred, R.G.; Steen Pedersen, J.; Thompson, J.J.; Lee, J.; Timshel, P.N.; Stender, S.; Opseth Rygg, M.; Gluud, L.L.; Bjerregaard Kristiansen, V.; Bendtsen, F.; et al. Single-cell transcriptome and cell type-specific molecular pathways of human non-alcoholic steatohepatitis. Sci. Rep. 2022, 12, 13484. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Huang, C.; Shi, X.; Wu, M.; Li, H.; Liu, Q.; Zhang, X.; Zhao, Y.; Li, X. Single-cell transcriptomics of hepatic stellate cells uncover crucial pathways and key regulators involved in non-alcoholic steatohepatitis. Endocr. Connect. 2023, 12, e220502. [Google Scholar] [CrossRef]
- Jiao, J.; Sanchez, J.I.; Saldarriaga, O.A.; Solis, L.M.; Tweardy, D.J.; Maru, D.M.; Stevenson, H.L.; Beretta, L. Spatial molecular and cellular determinants of STAT3 activation in liver fibrosis progression in non-alcoholic fatty liver disease. JHEP Rep. 2023, 5, 100628. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Perkins, J.R.; Dawes, J.M.; McMahon, S.B.; Bennett, D.L.; Orengo, C.; Kohl, M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genom. 2012, 13, 296. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Dennis, L.; D'Amico, L.; White, A.; Disis, M.L.; Geller, M.A.; Odunsi, K.; Beechem, J.; Fling, S.P. Gene expression markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer. 2017, 5, 18. [Google Scholar] [CrossRef]
- Tomfohr, J.; Lu, J.; Kepler, T.B. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinform. 2005, 6, 225. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R.; Ferrufino-Schmidt, M.C.; Tamegnon, A.; Zhang, J.; Solis, L.; Jiang, M.; Ibarguen, H.; Haymaker, C.; Lee, J.J.; Bernatchez, C.; et al. Immuno-profiling and cellular spatial analysis using five immune oncology multiplex immunofluorescence panels for paraffin tumor tissue. Sci. Rep. 2021, 11, 8511. [Google Scholar] [CrossRef]
- Haukeland, J.W.; Damas, J.K.; Konopski, Z.; Loberg, E.M.; Haaland, T.; Goverud, I.; Torjesen, P.A.; Birkeland, K.; Bjoro, K.; Aukrust, P. Systemic inflammation in nonalcoholic fatty liver disease is characterized by elevated levels of CCL2. J. Hepatol. 2006, 44, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chiwanda Kaminga, A.; Liu, A.; Wen, S.W.; Chen, J.; Luo, J. Chemokines in Non-alcoholic Fatty Liver Disease: A Systematic Review and Network Meta-Analysis. Front. Immunol. 2020, 11, 1802. [Google Scholar] [CrossRef] [PubMed]
- She, S.; Ren, L.; Chen, P.; Wang, M.; Chen, D.; Wang, Y.; Chen, H. Functional Roles of Chemokine Receptor CCR2 and Its Ligands in Liver Disease. Front. Immunol. 2022, 13, 812431. [Google Scholar] [CrossRef]
- Kado, A.; Tsutsumi, T.; Enooku, K.; Fujinaga, H.; Ikeuchi, K.; Okushin, K.; Moriya, K.; Yotsuyanagi, H.; Koike, K. Noninvasive diagnostic criteria for nonalcoholic steatohepatitis based on gene expression levels in peripheral blood mononuclear cells. J. Gastroenterol. 2019, 54, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, B.; Lu, M.; Wang, Y.; Ding, N.; Liu, C.; Gao, M.; Yao, Z.; Zhang, S.; Zhao, Y.; et al. The methyltransferase METTL3 negatively regulates nonalcoholic steatohepatitis (NASH) progression. Nat. Commun. 2021, 12, 7213. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Zhuge, F.; Ni, L.; Nagata, N.; Yamashita, T.; Mukaida, N.; Kaneko, S.; Ota, T.; Nagashimada, M. CX3CL1/CX3CR1 interaction protects against lipotoxicity-induced nonalcoholic steatohepatitis by regulating macrophage migration and M1/M2 status. Metabolism 2022, 136, 155272. [Google Scholar] [CrossRef]
- Kang, J.; Postigo-Fernandez, J.; Kim, K.; Zhu, C.; Yu, J.; Meroni, M.; Mayfield, B.; Bartolome, A.; Dapito, D.H.; Ferrante, A.W., Jr.; et al. Notch-mediated hepatocyte MCP-1 secretion causes liver fibrosis. JCI Insight 2023, 8, e165369. [Google Scholar] [CrossRef]
- Bonacchi, A.; Petrai, I.; Defranco, R.M.; Lazzeri, E.; Annunziato, F.; Efsen, E.; Cosmi, L.; Romagnani, P.; Milani, S.; Failli, P.; et al. The chemokine CCL21 modulates lymphocyte recruitment and fibrosis in chronic hepatitis C. Gastroenterology 2003, 125, 1060–1076. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, B.A.J.; Finlon, J.M.; Gillen, A.E.; Kriss, M.S.; Riemondy, K.A.; Fu, R.; Schuyler, R.P.; Hesselberth, J.R.; Rosen, H.R.; Burchill, M.A. Chronic Liver Disease in Humans Causes Expansion and Differentiation of Liver Lymphatic Endothelial Cells. Front. Immunol. 2019, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, Y.; Wu, X.; Tong, P.; Yue, Y.; Gao, S.; Huang, D.; Huang, J. Inhibition of CCL19 benefits non-alcoholic fatty liver disease by inhibiting TLR4/NF-kappaB-p65 signaling. Mol. Med. Rep. 2018, 18, 4635–4642. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Iwashita, M.; Nagayasu, S.; Yamashita, A.; Shinjo, T.; Hashikata, A.; Asano, T.; Kushiyama, A.; Ishimaru, N.; Takahama, Y.; et al. Protection from diet-induced obesity and insulin resistance in mice lacking CCL19-CCR7 signaling. Obesity 2015, 23, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Y.; Qu, Y.; Li, Z.; Li, F.; Xiao, C.Y.; Lu, L.G. A 6 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis B. Front. Biosci. Landmark Ed. 2016, 21, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiao, J.; Cermelli, S.; Muir, K.; Jung, K.H.; Zou, R.; Rashid, A.; Gagea, M.; Zabludoff, S.; Kalluri, R.; et al. miR-21 Inhibition Reduces Liver Fibrosis and Prevents Tumor Development by Inducing Apoptosis of CD24+ Progenitor Cells. Cancer Res. 2015, 75, 1859–1867. [Google Scholar] [CrossRef]
- Li, D.; Zheng, L.; Jin, L.; Zhou, Y.; Li, H.; Fu, J.; Shi, M.; Du, P.; Wang, L.; Wu, H.; et al. CD24 polymorphisms affect risk and progression of chronic hepatitis B virus infection. Hepatology 2009, 50, 735–742. [Google Scholar] [CrossRef]
- Athwal, V.S.; Pritchett, J.; Llewellyn, J.; Martin, K.; Camacho, E.; Raza, S.M.; Phythian-Adams, A.; Birchall, L.J.; Mullan, A.F.; Su, K.; et al. SOX9 predicts progression toward cirrhosis in patients while its loss protects against liver fibrosis. EMBO Mol. Med. 2017, 9, 1696–1710. [Google Scholar] [CrossRef]
- Athwal, V.S.; Pritchett, J.; Martin, K.; Llewellyn, J.; Scott, J.; Harvey, E.; Zaitoun, A.M.; Mullan, A.F.; Zeef, L.A.H.; Friedman, S.L.; et al. SOX9 regulated matrix proteins are increased in patients serum and correlate with severity of liver fibrosis. Sci. Rep. 2018, 8, 17905. [Google Scholar] [CrossRef]
- Chatterjee, A.; Basu, A.; Das, K.; Singh, P.; Mondal, D.; Bhattacharya, B.; Roychoudhury, S.; Majumder, P.P.; Chowdhury, A.; Basu, P. Hepatic transcriptome signature correlated with HOMA-IR explains early nonalcoholic fatty liver disease pathogenesis. Ann. Hepatol. 2020, 19, 472–481. [Google Scholar] [CrossRef]
- Suzuki, Y.; Sasaki, T.; Kakisaka, K.; Abe, H.; Takikawa, Y. Evaluation of SOX9-Positive Hepatocytes in Human Liver Specimens and Mature Mouse Hepatocytes. Methods Mol. Biol. 2022, 2544, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Daemen, S.; Gainullina, A.; Kalugotla, G.; He, L.; Chan, M.M.; Beals, J.W.; Liss, K.H.; Klein, S.; Feldstein, A.E.; Finck, B.N.; et al. Dynamic Shifts in the Composition of Resident and Recruited Macrophages Influence Tissue Remodeling in NASH. Cell Rep. 2021, 34, 108626. [Google Scholar] [CrossRef] [PubMed]
- Roeb, E. Diagnostic and Therapy of Nonalcoholic Fatty Liver Disease: A Narrative Review. Visc. Med. 2022, 38, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ratziu, V.; Sanyal, A.; Harrison, S.A.; Wong, V.W.; Francque, S.; Goodman, Z.; Aithal, G.P.; Kowdley, K.V.; Seyedkazemi, S.; Fischer, L.; et al. Cenicriviroc Treatment for Adults With Nonalcoholic Steatohepatitis and Fibrosis: Final Analysis of the Phase 2b CENTAUR Study. Hepatology 2020, 72, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Weiskirchen, R. Non-alcoholic fatty liver disease (NAFLD)/non-alcoholic steatohepatitis (NASH)-related liver fibrosis: Mechanisms, treatment and prevention. Ann. Transl. Med. 2021, 9, 729. [Google Scholar] [CrossRef]
- Irvine, K.M.; Okano, S.; Patel, P.J.; Horsfall, L.U.; Williams, S.; Russell, A.; Powell, E.E. Serum matrix metalloproteinase 7 (MMP7) is a biomarker of fibrosis in patients with non-alcoholic fatty liver disease. Sci. Rep. 2021, 11, 2858. [Google Scholar] [CrossRef]
- Nassir, F.; Ibdah, J.A. Role of mitochondria in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 8713–8742. [Google Scholar] [CrossRef]
- Chimienti, G.; Orlando, A.; Russo, F.; D'Attoma, B.; Aragno, M.; Aimaretti, E.; Lezza, A.M.S.; Pesce, V. The Mitochondrial Trigger in an Animal Model of Nonalcoholic Fatty Liver Disease. Genes 2021, 12, 1439. [Google Scholar] [CrossRef]
- Yoneda, M.; Hotta, K.; Nozaki, Y.; Endo, H.; Uchiyama, T.; Mawatari, H.; Iida, H.; Kato, S.; Hosono, K.; Fujita, K.; et al. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD). BMC Gastroenterol. 2008, 8, 27. [Google Scholar] [CrossRef]
- Lin, Y.C.; Chang, P.F.; Chang, M.H.; Ni, Y.H. A common variant in the peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene is associated with nonalcoholic fatty liver disease in obese children. Am. J. Clin. Nutr. 2013, 97, 326–331. [Google Scholar] [CrossRef]
- Zhang, R.N.; Shen, F.; Pan, Q.; Cao, H.X.; Chen, G.Y.; Fan, J.G. PPARGC1A rs8192678 G>A polymorphism affects the severity of hepatic histological features and nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease. World J. Gastroenterol. 2021, 27, 3863–3876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Chang, X.; Zhang, X. Imbalance in mitochondrial dynamics induced by low PGC-1alpha expression contributes to hepatocyte EMT and liver fibrosis. Cell Death Dis. 2020, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Besse-Patin, A.; Leveille, M.; Oropeza, D.; Nguyen, B.N.; Prat, A.; Estall, J.L. Estrogen Signals Through Peroxisome Proliferator-Activated Receptor-gamma Coactivator 1alpha to Reduce Oxidative Damage Associated With Diet-Induced Fatty Liver Disease. Gastroenterology 2017, 152, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, W.; Yan, X.; Huang, T.; Yang, A. Overexpression of Hepcidin Alleviates Steatohepatitis and Fibrosis in a Diet-induced Nonalcoholic Steatohepatitis. J. Clin. Transl. Hepatol. 2022, 10, 577–588. [Google Scholar] [CrossRef]
- Peiseler, M.; Schwabe, R.; Hampe, J.; Kubes, P.; Heikenwalder, M.; Tacke, F. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease—Novel insights into cellular communication circuits. J. Hepatol. 2022, 77, 1136–1160. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, Y.; Wu, X.; Wang, X.; Niu, J. Identification of Immune Microenvironment Changes and the Expression of Immune-Related Genes in Liver Cirrhosis. Front. Immunol. 2022, 13, 918445. [Google Scholar] [CrossRef]
- Wu, K.J.; Qian, Q.F.; Zhou, J.R.; Sun, D.L.; Duan, Y.F.; Zhu, X.; Sartorius, K.; Lu, Y.J. Regulatory T cells (Tregs) in liver fibrosis. Cell Death Discov. 2023, 9, 53. [Google Scholar] [CrossRef]
- Elchaninov, A.V.; Fatkhudinov, T.K.; Vishnyakova, P.A.; Lokhonina, A.V.; Sukhikh, G.T. Phenotypical and Functional Polymorphism of Liver Resident Macrophages. Cells 2019, 8, 1032. [Google Scholar] [CrossRef]
- Wallace, S.J.; Tacke, F.; Schwabe, R.F.; Henderson, N.C. Understanding the cellular interactome of non-alcoholic fatty liver disease. JHEP Rep. 2022, 4, 100524. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez, J.I.; Parra, E.R.; Jiao, J.; Solis Soto, L.M.; Ledesma, D.A.; Saldarriaga, O.A.; Stevenson, H.L.; Beretta, L. Cellular and Molecular Mechanisms of Liver Fibrosis in Patients with NAFLD. Cancers 2023, 15, 2871. https://doi.org/10.3390/cancers15112871
Sanchez JI, Parra ER, Jiao J, Solis Soto LM, Ledesma DA, Saldarriaga OA, Stevenson HL, Beretta L. Cellular and Molecular Mechanisms of Liver Fibrosis in Patients with NAFLD. Cancers. 2023; 15(11):2871. https://doi.org/10.3390/cancers15112871
Chicago/Turabian StyleSanchez, Jessica I., Edwin R. Parra, Jingjing Jiao, Luisa M. Solis Soto, Debora A. Ledesma, Omar A. Saldarriaga, Heather L. Stevenson, and Laura Beretta. 2023. "Cellular and Molecular Mechanisms of Liver Fibrosis in Patients with NAFLD" Cancers 15, no. 11: 2871. https://doi.org/10.3390/cancers15112871
APA StyleSanchez, J. I., Parra, E. R., Jiao, J., Solis Soto, L. M., Ledesma, D. A., Saldarriaga, O. A., Stevenson, H. L., & Beretta, L. (2023). Cellular and Molecular Mechanisms of Liver Fibrosis in Patients with NAFLD. Cancers, 15(11), 2871. https://doi.org/10.3390/cancers15112871







.png)
