Anoikis-Related Gene Signature for Prognostication of Pancreatic Adenocarcinoma: A Multi-Omics Exploration and Verification Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Construction and Validation of a Risk-Scoring Model with Transcriptomic Data
2.3. Proteomics Analysis for Further Validation
2.4. Clustering and Analysis of Somatic Mutations
2.5. Immune Infiltration in the Low-Risk and High-Risk Groups
2.6. Enrichment Analysis
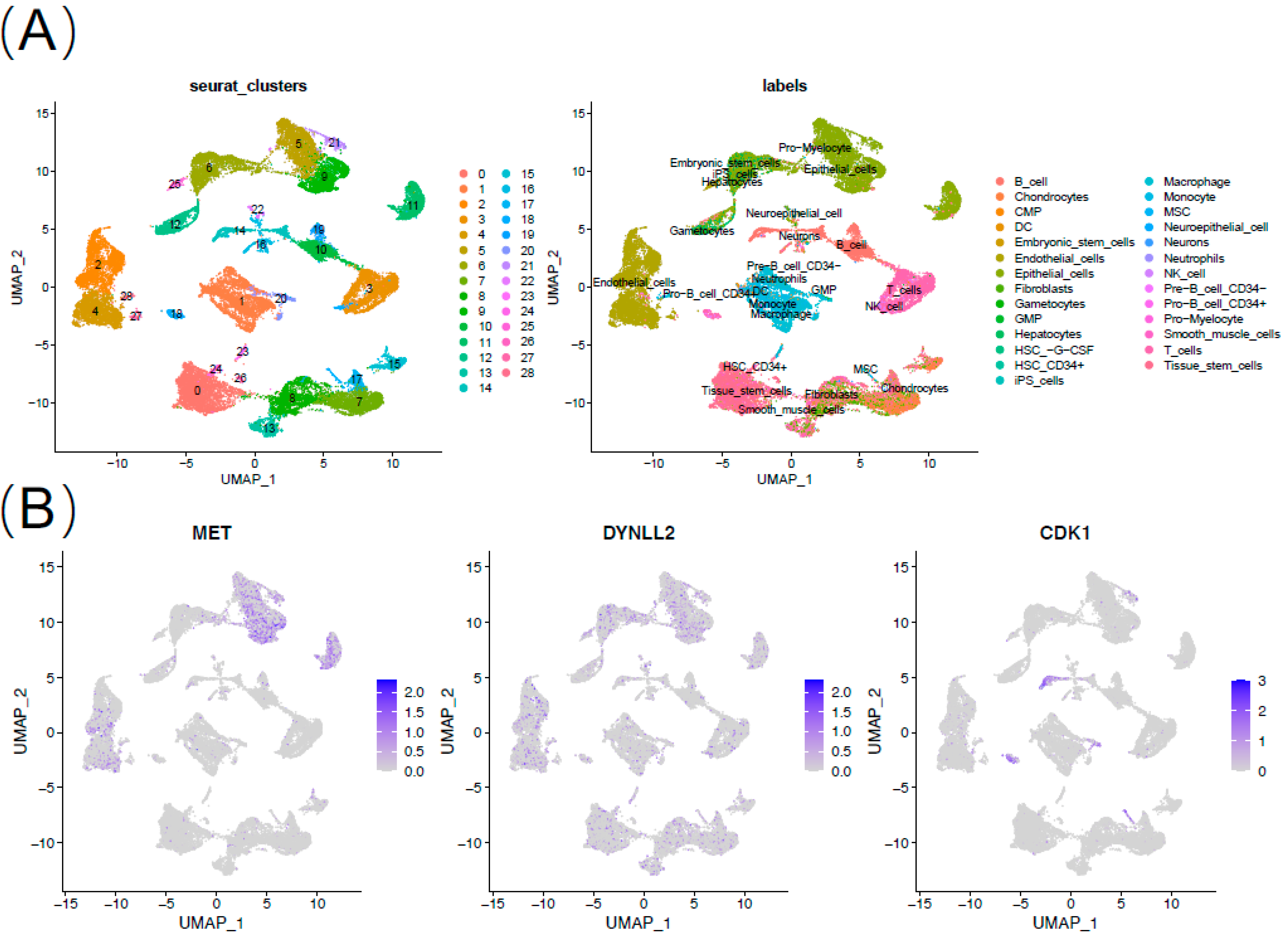
2.7. Validation in Single Cell Sequencing
2.8. Drug Sensitivity Analysis
2.9. Cell Culture and Real-Time Quantitative PCR
2.10. Statistical Analysis
3. Results
3.1. Constructing a Risk-Scoring Model Using Anoikis-Related Genes
3.2. Immune Infiltration in the Low-Risk and High-Risk Groups
3.3. Analysis of Chemo-Sensitivity in High- and Low-Risk Groups
3.4. Analysis of Differential Gene Enrichment between High- and Low-Risk Groups
3.5. Cluster Analysis and Somatic Mutations Analysis between Different Clusters
3.6. 7 Genes Validation in Single Cell Sequencing
3.7. Real-Time Quantitative PCR of Seven Genes in Pancreatic Cancer Cells and Validation in CCLE
3.8. Prognostic Analysis Using Proteomic Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Singhi, A.D.; Koay, E.J.; Chari, S.T.; Maitra, A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology 2019, 156, 2024–2040. [Google Scholar] [CrossRef]
- Chen, S.; Gu, J.; Zhang, Q.; Hu, Y.; Ge, Y. Development of Biomarker Signatures Associated with Anoikis to Predict Prognosis in Endometrial Carcinoma Patients. J. Oncol. 2021, 2021, 3375297. [Google Scholar] [CrossRef]
- Ye, G.; Yang, Q.; Lei, X.; Zhu, X.; Li, F.; He, J.; Chen, H.; Ling, R.; Zhang, H.; Lin, T.; et al. Nuclear MYH9-induced CTNNB1 transcription, targeted by staurosporin, promotes gastric cancer cell anoikis resistance and metastasis. Theranostics 2020, 10, 7545–7560. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chun, J.; Pan, C.; Kumar, A.; Zhang, G.; Ha, Y.; Li, D.; Alesi, G.N.; Kang, Y.; Zhou, L.; et al. The PLAG1-GDH1 Axis Promotes Anoikis Resistance and Tumor Metastasis through CamKK2-AMPK Signaling in LKB1-Deficient Lung Cancer. Mol. Cell 2018, 69, 87–99.e87. [Google Scholar] [CrossRef] [PubMed]
- Buchheit, C.L.; Angarola, B.L.; Steiner, A.; Weigel, K.J.; Schafer, Z.T. Anoikis evasion in inflammatory breast cancer cells is mediated by Bim-EL sequestration. Cell Death Differ. 2015, 22, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, F.; Liu, N. L1CAM promotes epithelial to mesenchymal transition and formation of cancer initiating cells in human endometrial cancer. Exp. Ther. Med. 2018, 15, 2792–2797. [Google Scholar] [CrossRef]
- Kou, F.; Sun, H.; Wu, L.; Li, B.; Zhang, B.; Wang, X.; Yang, L. TOP2A Promotes Lung Adenocarcinoma Cells’ Malignant Progression and Predicts Poor Prognosis in Lung Adenocarcinoma. J. Cancer 2020, 11, 2496–2508. [Google Scholar] [CrossRef]
- Lu, Y.X.; Ju, H.Q.; Liu, Z.X.; Chen, D.L.; Wang, Y.; Zhao, Q.; Wu, Q.N.; Zeng, Z.L.; Qiu, H.B.; Hu, P.S.; et al. ME1 Regulates NADPH Homeostasis to Promote Gastric Cancer Growth and Metastasis. Cancer Res. 2018, 78, 1972–1985. [Google Scholar] [CrossRef]
- Xu, J.; Song, J.; Yang, X.; Guo, J.; Wang, T.; Zhuo, W. ProNGF siRNA inhibits cell proliferation and invasion of pancreatic cancer cells and promotes anoikis. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 111, 1066–1073. [Google Scholar] [CrossRef]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Taddei, M.L.; Giannoni, E.; Fiaschi, T.; Chiarugi, P. Anoikis: An emerging hallmark in health and diseases. J. Pathol. 2012, 226, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, J.; Qian, S.; Wu, X.; Sun, L.; Ling, T.; Jin, Y.; Li, W.; Sun, L.; Lai, M.; et al. S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-β/USF2 axis in colorectal cancer. Cancer Commun. 2021, 41, 154–170. [Google Scholar] [CrossRef]
- Liao, Y.H.; Chiang, K.H.; Shieh, J.M.; Huang, C.R.; Shen, C.J.; Huang, W.C.; Chen, B.K. Epidermal growth factor-induced ANGPTL4 enhances anoikis resistance and tumour metastasis in head and neck squamous cell carcinoma. Oncogene 2017, 36, 2228–2242. [Google Scholar] [CrossRef] [PubMed]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, G.; Tang, T.; Liang, T. Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development. Mol. Cancer 2021, 20, 44. [Google Scholar] [CrossRef]
- Wu, M.; Li, X.; Zhang, T.; Liu, Z.; Zhao, Y. Identification of a Nine-Gene Signature and Establishment of a Prognostic Nomogram Predicting Overall Survival of Pancreatic Cancer. Front. Oncol. 2019, 9, 996. [Google Scholar] [CrossRef]
- Odle, R.I.; Florey, O.; Ktistakis, N.T.; Cook, S.J. CDK1, the Other ‘Master Regulator’ of Autophagy. Trends Cell Biol. 2021, 31, 95–107. [Google Scholar] [CrossRef]
- Xie, B.; Wang, S.; Jiang, N.; Li, J.J. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019, 443, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Wijnen, R.; Pecoraro, C.; Carbone, D.; Fiuji, H.; Avan, A.; Peters, G.J.; Giovannetti, E.; Diana, P. Cyclin Dependent Kinase-1 (CDK-1) Inhibition as a Novel Therapeutic Strategy against Pancreatic Ductal Adenocarcinoma (PDAC). Cancers 2021, 13, 4389. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Shah, K. TRAIL of Hope Meeting Resistance in Cancer. Trends Cancer 2020, 6, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Klampatsa, A.; Dimou, V.; Albelda, S.M. Mesothelin-targeted CAR-T cell therapy for solid tumors. Expert Opin. Biol. Ther. 2021, 21, 473–486. [Google Scholar] [CrossRef]
- Alarcón-Millán, J.; Martínez-Carrillo, D.N.; Peralta-Zaragoza, O.; Fernández-Tilapa, G. Regulation of GKN1 expression in gastric carcinogenesis: A problem to resolve (Review). Int. J. Oncol. 2019, 55, 555–569. [Google Scholar] [CrossRef]
- Rapali, P.; Szenes, Á.; Radnai, L.; Bakos, A.; Pál, G.; Nyitray, L. DYNLL/LC8: A light chain subunit of the dynein motor complex and beyond. FEBS J. 2011, 278, 2980–2996. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.M.; Lin, S.H.; Yu, M.C.; Hsieh, S.Y. CLIC1 recruits PIP5K1A/C to induce cell-matrix adhesions for tumor metastasis. J. Clin. Investig. 2021, 131, e133525. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca di Magliano, M.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733.e739. [Google Scholar] [CrossRef]
- Fujikura, K.; Hosoda, W.; Felsenstein, M.; Song, Q.; Reiter, J.G.; Zheng, L.; Beleva Guthrie, V.; Rincon, N.; Dal Molin, M.; Dudley, J.; et al. Multiregion whole-exome sequencing of intraductal papillary mucinous neoplasms reveals frequent somatic KLF4 mutations predominantly in low-grade regions. Gut 2021, 70, 928–939. [Google Scholar] [CrossRef]
- Collins, M.A.; Bednar, F.; Zhang, Y.; Brisset, J.C.; Galbán, S.; Galbán, C.J.; Rakshit, S.; Flannagan, K.S.; Adsay, N.V.; Pasca di Magliano, M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Investig. 2012, 122, 639–653. [Google Scholar] [CrossRef]
- Collins, M.A.; Brisset, J.C.; Zhang, Y.; Bednar, F.; Pierre, J.; Heist, K.A.; Galbán, C.J.; Galbán, S.; di Magliano, M.P. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS ONE 2012, 7, e49707. [Google Scholar] [CrossRef]
- Ying, H.; Kimmelman, A.C.; Lyssiotis, C.A.; Hua, S.; Chu, G.C.; Fletcher-Sananikone, E.; Locasale, J.W.; Son, J.; Zhang, H.; Coloff, J.L.; et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012, 149, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Yao, W.; Ying, H.; Hua, S.; Liewen, A.; Wang, Q.; Zhong, Y.; Wu, C.J.; Sadanandam, A.; Hu, B.; et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014, 158, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.P.t.; Yashinskie, J.J.; Koche, R.; Chandwani, R.; Tian, S.; Chen, C.C.; Baslan, T.; Marinkovic, Z.S.; Sánchez-Rivera, F.J.; Leach, S.D.; et al. α-Ketoglutarate links p53 to cell fate during tumour suppression. Nature 2019, 573, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Weissmueller, S.; Manchado, E.; Saborowski, M.; Morris, J.P.t.; Wagenblast, E.; Davis, C.A.; Moon, S.H.; Pfister, N.T.; Tschaharganeh, D.F.; Kitzing, T.; et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 2014, 157, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Barriga, F.M.; Tsanov, K.M.; Ho, Y.J.; Sohail, N.; Zhang, A.; Baslan, T.; Wuest, A.N.; Del Priore, I.; Meškauskaitė, B.; Livshits, G.; et al. MACHETE identifies interferon-encompassing chromosome 9p21.3 deletions as mediators of immune evasion and metastasis. Nat. Cancer 2022, 3, 1367–1385. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, X.; Lu, Y.; Wang, G.; Ma, Y. Anoikis-Related Gene Signature for Prognostication of Pancreatic Adenocarcinoma: A Multi-Omics Exploration and Verification Study. Cancers 2023, 15, 3146. https://doi.org/10.3390/cancers15123146
Zhang J, Li X, Lu Y, Wang G, Ma Y. Anoikis-Related Gene Signature for Prognostication of Pancreatic Adenocarcinoma: A Multi-Omics Exploration and Verification Study. Cancers. 2023; 15(12):3146. https://doi.org/10.3390/cancers15123146
Chicago/Turabian StyleZhang, Jin, Xuesong Li, Yi Lu, Guowen Wang, and Ying Ma. 2023. "Anoikis-Related Gene Signature for Prognostication of Pancreatic Adenocarcinoma: A Multi-Omics Exploration and Verification Study" Cancers 15, no. 12: 3146. https://doi.org/10.3390/cancers15123146
APA StyleZhang, J., Li, X., Lu, Y., Wang, G., & Ma, Y. (2023). Anoikis-Related Gene Signature for Prognostication of Pancreatic Adenocarcinoma: A Multi-Omics Exploration and Verification Study. Cancers, 15(12), 3146. https://doi.org/10.3390/cancers15123146





