Unsupervised Hierarchical Clustering of Head and Neck Cancer Patients by Pre-Treatment Plasma Metabolomics Creates Prognostic Metabolic Subtypes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Overall and Progression-Free Survival
2.3. High-Resolution Untargeted Metabolomics (HRM) of Blood Plasma
2.4. Statistical Analysis
2.5. Sensitivity Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winquist, E.; Head and Neck Disease Site Group; Agbassi, C.; Meyers, B.M.; Yoo, J.; Chan, K.K.W. Systemic therapy in the curative treatment of head and neck squamous cell cancer: A systematic review. J. Otolaryngol.-Head Neck Surg. 2017, 46, 29. [Google Scholar] [CrossRef] [PubMed]
- Trotti, A.; Bellm, L.A.; Epstein, J.B.; Frame, D.; Fuchs, H.J.; Gwede, C.K.; Komaroff, E.; Nalysnyk, L.; Zilberberg, M.D. Mucositis incidence, severity and associated outcomes in patients with head and neck cancer receiving radiotherapy with or without chemotherapy: A systematic literature review. Radiother. Oncol. 2003, 66, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Manikantan, K.; Khode, S.; Sayed, S.I.; Roe, J.; Nutting, C.M.; Rhys-Evans, P.; Harrington, K.J.; Kazi, R. Dysphagia in head and neck cancer. Cancer Treat. Rev. 2009, 35, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Ridner, S.H.; Dietrich, M.S.; Wells, N.; Wallston, K.A.; Sinard, R.J.; Cmelak, A.J.; Murphy, B.A. Prevalence of Secondary Lymphedema in Patients with Head and Neck Cancer. J. Pain Symptom Manag. 2012, 43, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Van Deudekom, F.J.; Schimberg, A.S.; Kallenberg, M.H.; Slingerland, M.; van der Velden, L.-A.; Mooijaart, S.P. Functional and cognitive impairment, social environment, frailty and adverse health outcomes in older patients with head and neck cancer, a systematic review. Oral Oncol. 2017, 64, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Capuano, G.; Gentile, P.C.; Bianciardi, F.; Tosti, M.; Palladino, A.; Di Palma, M. Prevalence and influence of malnutrition on quality of life and performance status in patients with locally advanced head and neck cancer before treatment. Support. Care Cancer 2010, 18, 433–437. [Google Scholar] [CrossRef]
- Jacobi, I.; van der Molen, L.; Huiskens, H.; van Rossum, M.A.; Hilgers, F.J.M. Voice and speech outcomes of chemoradiation for advanced head and neck cancer: A systematic review. Eur. Arch. Oto-Rhino-Laryngol. 2010, 267, 1495–1505. [Google Scholar] [CrossRef]
- Denis, F.; Garaud, P.; Bardet, E.; Alfonsi, M.; Sire, C.; Germain, T.; Bergerot, P.; Rhein, B.; Tortochaux, J.; Calais, G. Final Results of the 94–01 French Head and Neck Oncology and Radiotherapy Group Randomized Trial Comparing Radiotherapy Alone with Concomitant Radiochemotherapy in Advanced-Stage Oropharynx Carcinoma. J. Clin. Oncol. 2004, 22, 69–76. [Google Scholar] [CrossRef]
- Rosenberg, A.J.; Vokes, E.E. Optimizing Treatment De-Escalation in Head and Neck Cancer: Current and Future Perspectives. Oncologist 2021, 26, 40–48. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Gillison, M.L.; Trotti, A.M.; Harris, J.; Eisbruch, A.; Harari, P.M.; Adelstein, D.J.; Jordan, R.C.K.; Zhao, W.; Sturgis, E.M.; Burtness, B.; et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): A randomized, multicentre, non-inferiority trial. Lancet 2019, 393, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Mehanna, H.; Robinson, M.; Hartley, A.; Kong, A.; Foran, B.; Fulton-Lieuw, T.; Dalby, M.; Mistry, P.; Sen, M.; O’Toole, L.; et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): An open-label randomized controlled phase 3 trial. Lancet 2019, 393, 51–60. [Google Scholar] [CrossRef]
- Bigelow, E.O.; Seiwert, T.Y.; Fakhry, C. Deintensification of treatment for human papillomavirus-related oropharyngeal cancer: Current state and future directions. Oral Oncol. 2020, 105, 104652. [Google Scholar] [CrossRef]
- Yom, S.S.; Torres-Saavedra, P.; Caudell, J.J.; Waldron, J.N.; Gillison, M.L.; Xia, P.; Truong, M.T.; Kong, C.; Jordan, R.; Le, Q.T.; et al. Faculty Opinions recommendation of Reduced-Dose Radiation Therapy for HPV-Associated Oropharyngeal Carcinoma (NRG Oncology HN002). J. Clin. Oncol. 2021, 39, 956–965. [Google Scholar] [CrossRef]
- Siravegna, G.; O’Boyle, C.J.; Varmeh, S.; Queenan, N.; Michel, A.; Stein, J.; Thierauf, J.; Sadow, P.M.; Faquin, W.C.; Perry, S.K.; et al. Cell-Free HPV DNA Provides an Accurate and Rapid Diagnosis of HPV-Associated Head and Neck Cancer. Clin. Cancer Res. 2021, 28, 719–727. [Google Scholar] [CrossRef]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; de Sanjosé, S.; Trottier, H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: A systematic review and meta-analysis. Lancet Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- Neskey, D.M.; Osman, A.A.; Ow, T.J.; Katsonis, P.; McDonald, T.; Hicks, S.C.; Hsu, T.-K.; Pickering, C.R.; Ward, A.; Patel, A.; et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res. 2015, 75, 1527–1536. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Dassonville, O.; Formento, J.L.; Francoual, M.; Ramaioli, A.; Santini, J.; Schneider, M.; Demard, F.; Milano, G. Expression of epidermal growth factor receptor and survival in upper aerodigestive tract cancer. J. Clin. Oncol. 1993, 11, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Licitra, L.; Störkel, S.; Kerr, K.M.; Van Cutsem, E.; Pirker, R.; Hirsch, F.R.; Vermorken, J.B.; von Heydebreck, A.; Esser, R.; Celik, I.; et al. Predictive value of epidermal growth factor receptor expression for first-line chemotherapy plus cetuximab in patients with head and neck and colorectal cancer: Analysis of data from the EXTREME and CRYSTAL studies. Eur. J. Cancer 2013, 49, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.D.; Burtness, B.; Ferris, R.L. Immunotherapy for head and neck cancer: Recent advances and future directions. Oral Oncol. 2019, 99, 104460. [Google Scholar] [CrossRef]
- Cottrell, T.R.; Taube, J.M. PD-L1 and Emerging Biomarkers in Immune Checkpoint Blockade Therapy. Cancer J. 2018, 24, 41–46. [Google Scholar] [CrossRef]
- Lenouvel, D.; González-Moles, M.Á.; Talbaoui, A.; Ramos-García, P.; González-Ruiz, L.; Ruiz-Ávila, I.; Gil-Montoya, J.A. An update of knowledge on PD-L1 in head and neck cancers: Physiologic, prognostic and therapeutic perspectives. Oral Dis. 2020, 26, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, N.; Mizuno-Kamiya, M. The Role of Immune Modulatory Cytokines in the Tumor Microenvironments of Head and Neck Squamous Cell Carcinomas. Cancers 2022, 14, 2884. [Google Scholar] [CrossRef]
- Kim, S.Y.; Beer, M.; Vogel, D.W.T. Imaging in head and neck cancers: Update for non-radiologist. Oral Oncol. 2021, 120, 105434. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.L.; Harris, J.; Yao, M.; Rosenthal, D.I.; Opanowski, A.; Levering, A.; Ang, K.K.; Trotti, A.M.; Garden, A.S.; Jones, C.U.; et al. Metabolic Tumor Volume as a Prognostic Imaging-Based Biomarker for Head-and-Neck Cancer: Pilot Results from Radiation Therapy Oncology Group Protocol 0522. Int. J. Radiat. Oncol. 2015, 91, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Krupar, R.; Hautmann, M.G.; Pathak, R.R.; Varier, I.; McLaren, C.; Gaag, D.; Hellerbrand, C.; Evert, M.; Laban, S.; Idel, C.; et al. Immunometabolic Determinants of Chemoradiotherapy Response and Survival in Head and Neck Squamous Cell Carcinoma. Am. J. Pathol. 2018, 188, 72–83. [Google Scholar] [CrossRef]
- Sandulache, V.C.; Myers, J.N. Altered metabolism in head and neck squamous cell carcinoma: An opportunity for identification of novel biomarkers and drug targets. Head Neck 2012, 34, 282–290. [Google Scholar] [CrossRef]
- Wu, Z.-H.; Tang, Y.; Zhou, Y. A Metabolic Gene Signature to Predict Overall Survival in Head and Neck Squamous Cell Carcinoma. Mediat. Inflamm. 2020, 2020, 6716908. [Google Scholar] [CrossRef]
- Wu, X.; Yao, Y.; Li, Z.; Ge, H.; Wang, D.; Wang, Y. Identification of a Transcriptional Prognostic Signature from Five Metabolic Pathways in Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 572919. [Google Scholar] [CrossRef]
- Xing, L.; Guo, M.; Zhang, X.; Liu, F. A transcriptional metabolic gene-set based prognostic signature is associated with clinical and mutational features in head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2020, 146, 621–630. [Google Scholar] [CrossRef]
- Carenzo, A.; Serafini, M.S.; Roca, E.; Paderno, A.; Mattavelli, D.; Romani, C.; Saintigny, P.; Koljenović, S.; Licitra, L.; De Cecco, L.; et al. Gene Expression Clustering and Selected Head and Neck Cancer Gene Signatures Highlight Risk Probability Differences in Oral Premalignant Lesions. Cells 2020, 9, 1828. [Google Scholar] [CrossRef]
- Luo, X.-J.; Zheng, M.; Cao, M.-X.; Zhang, W.-L.; Huang, M.-C.; Dai, L.; Tang, Y.-L.; Liang, X.-H. Distinguishable Prognostic miRNA Signatures of Head and Neck Squamous Cell Cancer With or Without HPV Infection. Front. Oncol. 2021, 10, 614487. [Google Scholar] [CrossRef]
- Bossche, V.V.D.; Zaryouh, H.; Vara-Messler, M.; Vignau, J.; Machiels, J.-P.; Wouters, A.; Schmitz, S.; Corbet, C. Microenvironment-driven intratumoral heterogeneity in head and neck cancers: Clinical challenges and opportunities for precision medicine. Drug Resist. Updates 2022, 60, 100806. [Google Scholar] [CrossRef] [PubMed]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Kulasinghe, A.; Kenny, L.; Punyadeera, C. The development of a liquid biopsy for head and neck cancers. Oral Oncol. 2016, 61, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Xiao, C.; Eldridge, R.C.; Beitler, J.J.; Higgins, K.A.; Chico, C.E.; Felger, J.C.; Wommack, E.C.; Knobf, T.; Saba, N.F.; Shin, D.M.; et al. Association Among Glucocorticoid Receptor Sensitivity, Fatigue, and Inflammation in Patients with Head and Neck Cancer. Psychosom. Med. 2020, 82, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Accardi, C.J.P.; Walker, D.I.B.; Uppal, K.; Quyyumi, A.A.; Rohrbeck, P.D.; Pennell, K.D.; Mallon, C.T.M.; Jones, D.P. High-Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. J. Occup. Environ. Med. 2016, 58 (Suppl. S8), S80–S88. [Google Scholar] [CrossRef]
- Chandler, J.D.; Hu, X.; Ko, E.-J.; Park, S.; Lee, Y.-T.; Orr, M.; Fernandes, J.; Uppal, K.; Kang, S.-M.; Jones, D.P.; et al. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef]
- Yu, T.; Park, Y.; Li, S.; Jones, D.P. Hybrid Feature Detection and Information Accumulation Using High-Resolution LC–MS Metabolomics Data. J. Proteome Res. 2013, 12, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Nellis, M.; Uppal, K.; Ma, C.; Tran, V.; Liang, Y.; Walker, D.I.; Jones, D.P. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem. 2020, 92, 8836–8844. [Google Scholar] [CrossRef] [PubMed]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 1–3. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38 (Suppl. S2), W71–W77. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 28 February 2023).
- R Studio Team. R Studio: Integrated Development Environment for R. R Studio, PBC; R Studio Team: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 28 February 2023).
- SEER Cancer Stat Facts: Oral Cavity and Pharynx Cancer; National Cancer Institute: Bethesda, MD, USA, 2022. Available online: https://seer.cancer.gov/statfacts/html/oralcav.html (accessed on 28 February 2023).
- Zhou, J.; Xu, B.; Huang, J.; Jia, X.; Xue, J.; Shi, X.; Xiao, L.; Li, W. 1H NMR-based metabonomic and pattern recognition analysis for detection of oral squamous cell carcinoma. Clin. Chim. Acta 2009, 401, 8–13. [Google Scholar] [CrossRef]
- Tiziani, S.; Lopes, V.; Günther, U.L. Early Stage Diagnosis of Oral Cancer Using 1H NMR–Based Metabolomics. Neoplasia 2009, 11, 269–276, IN7–IN10. [Google Scholar] [CrossRef]
- Yonezawa, K.; Nishiumi, S.; Kitamoto-Matsuda, J.; Fujita, T.; Morimoto, K.; Yamashita, D.; Saito, M.; Otsuki, N.; Irino, Y.; Yoshida, M.; et al. Serum and tissue metabolomics of head and neck cancer. Cancer Genom.-Proteom. 2013, 10, 233–238. [Google Scholar]
- Gupta, A.; Gupta, S.; Mahdi, A.A. 1H NMR-derived serum metabolomics of leukoplakia and squamous cell carcinoma. Clin. Chim. Acta 2015, 441, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Li, N.; Jia, Q.; Wang, X.; Zuo, L.; Long, J.; Xue, P.; Sun, Z.; Zhao, H. Metabolomics based plasma biomarkers for diagnosis of oral squamous cell carcinoma and oral erosive lichen planus. J. Cancer 2022, 13, 76–87. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Chen, Z.; Lam AK, Y.; Li, Z.; Liu, X.; Zhu, B.; Qiao, B. Analysis of metabolomics and transcriptomics data in head and neck squamous cell carcinoma. Oral Dis. 2022, 29, 1464–1479. [Google Scholar] [CrossRef] [PubMed]
- Jelonek, K.; Krzywon, A.; Jablonska, P.; Slominska, E.M.; Smolenski, R.T.; Polanska, J.; Rutkowski, T.; Mrochem-Kwarciak, J.; Skladowski, K.; Widlak, P. Systemic Effects of Radiotherapy and Concurrent Chemo-Radiotherapy in Head and Neck Cancer Patients—Comparison of Serum Metabolome Profiles. Metabolites 2020, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Tomàs, E.; Arguis, M.; Arenas, M.; Royo, L.T.; Murcia, M.; Sabater, S.; Torres, L.; Baiges-Gayà, G.; Hernández-Aguilera, A.; Camps, J.; et al. Alterations in plasma concentrations of energy-balance-related metabolites in patients with lung, or head & neck, cancers: Effects of radiotherapy. J. Proteom. 2020, 213, 103605. [Google Scholar] [CrossRef]
- Wojakowska, A.; Zebrowska, A.; Skowronek, A.; Rutkowski, T.; Polanski, K.; Widlak, P.; Marczak, L.; Pietrowska, M. Metabolic Profiles of Whole Serum and Serum-Derived Exosomes Are Different in Head and Neck Cancer Patients Treated by Radiotherapy. J. Pers. Med. 2020, 10, 229. [Google Scholar] [CrossRef]
- Boguszewicz, Ł.; Bieleń, A.; Ciszek, M.; Wendykier, J.; Szczepanik, K.; Skorupa, A.; Mrochem-Kwarciak, J.; Składowski, K.; Sokół, M. NMR-Based Metabolomics in Investigation of the Radiation Induced Changes in Blood Serum of Head and Neck Cancer Patients and Its Correlation with the Tissue Volumes Exposed to the Particulate Doses. Int. J. Mol. Sci. 2021, 22, 6310. [Google Scholar] [CrossRef]
- Tsai, C.-K.; Lin, C.-Y.; Kang, C.-J.; Liao, C.-T.; Wang, W.-L.; Chiang, M.-H.; Yen, T.-C.; Lin, G. Nuclear Magnetic Resonance Metabolomics Biomarkers for Identifying High Risk Patients with Extranodal Extension in Oral Squamous Cell Carcinoma. J. Clin. Med. 2020, 9, 951. [Google Scholar] [CrossRef]
- Silva, S.; Cunha, I.; Younes, R.; Soares, F.A.; Kowalski, L.P.; Graner, E. ErbB receptors and fatty acid synthase expression in aggressive head and neck squamous cell carcinomas. Oral Dis. 2010, 16, 774–780. [Google Scholar] [CrossRef]
- Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef]
- Burtness, B.; Bauman, J.E.; Galloway, T. Novel targets in HPV-negative head and neck cancer: Overcoming resistance to EGFR inhibition. Lancet Oncol. 2013, 14, e302–e309. [Google Scholar] [CrossRef]
- Kumar, D. Regulation of glycolysis in head and neck squamous cell carcinoma. Postdoc J. A J. Postdr. Res. Postdr. Aff. 2017, 5, 14–28. [Google Scholar] [CrossRef]
- Eldridge, R.C.; Uppal, K.; Hayes, D.N.; Smith, M.R.; Hu, X.; Qin, Z.S.; Beitler, J.J.; Miller, A.H.; Wommack, E.C.; Higgins, K.A.; et al. Plasma Metabolic Phenotypes of HPV-Associated versus Smoking-Associated Head and Neck Cancer and Patient Survival. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1858–1866. [Google Scholar] [CrossRef]
- Meijer, T.W.; Kaanders, J.H.; Span, P.N.; Bussink, J. Targeting Hypoxia, HIF-1, and Tumor Glucose Metabolism to Improve Radiotherapy Efficacy. Clin. Cancer Res. 2012, 18, 5585–5594. [Google Scholar] [CrossRef]
- Tantiwongkosi, B.; Yu, F.; Kanard, A.; Miller, F.R. Role of18F-FDG PET/CT in pre and post treatment evaluation in head and neck carcinoma. World J. Radiol. 2014, 6, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Gutkind, J.S.; Molinolo, A.A.; Wu, X.; Wang, Z.; Nachmanson, D.; Harismendy, O.; Alexandrov, L.B.; Wuertz, B.R.; Ondrey, F.G.; Laronde, D.; et al. Inhibition of mTOR signaling and clinical activity of metformin in oral premalignant lesions. J. Clin. Investig. 2021, 6, e147096. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Richard, D.; Laplante, M. The Roles of mTOR Complexes in Lipid Metabolism. Annu. Rev. Nutr. 2015, 35, 321–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qian, J.; He, Q.; Zhao, H.; Toral-Barza, L.; Shi, C.; Zhang, X.; Wu, J.; Yu, K. mTOR complex-2 stimulates acetyl-CoA and de novo lipogenesis through ATP citrate lyase in HER2/PIK3CA-hyperactive breast cancer. Oncotarget 2016, 7, 25224–25240. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef]
- Sandulache, V. A Phase I/II Study of Metformin in Combination with Cisplatin and Radiation in Head and Neck Squamous Cell Carcinoma. Identifier NCT02949700. 26 January 2017–7 July 2022. Available online: https://beta.clinicaltrials.gov/study/NCT02949700 (accessed on 28 February 2023).
- Wise-Draper, T. A Phase 2 Feasibility Study Combining Pembrolizumab and Metformin to Harness the Natural Killer Cytotoxic Response in Metastatic Head and Neck Cancer Patients. Identifier NCT04414540. 31 August 2020. Available online: https://beta.clinicaltrials.gov/study/NCT04414540 (accessed on 28 February 2023).
- Marur, S.; Li, S.; Cmelak, A.J.; Gillison, M.L.; Zhao, W.J.; Ferris, R.L.; Westra, W.H.; Gilbert, J.; Bauman, J.E.; Wagner, L.I.; et al. E1308: Phase II Trial of Induction Chemotherapy Followed by Reduced-Dose Radiation and Weekly Cetuximab in Patients with HPV-Associated Resectable Squamous Cell Carcinoma of the Oropharynx—ECOG-ACRIN Cancer Research Group. J. Clin. Oncol. 2017, 35, 490–497. [Google Scholar] [CrossRef]
- Couch, M.E.; Dittus, K.; Toth, M.J.; Willis, M.S.; Guttridge, D.C.; George, J.R.; Barnes, C.A.; Gourin, C.G.; Der-Torossian, H. Cancer cachexia update in head and neck cancer: Definitions and diagnostic features. Head Neck 2015, 37, 594–604. [Google Scholar] [CrossRef]
- Boguszewicz, Ł.; Bieleń, A.; Mrochem-Kwarciak, J.; Skorupa, A.; Ciszek, M.; Heyda, A.; Wygoda, A.; Kotylak, A.; Składowski, K.; Sokół, M. NMR-based metabolomics in real-time monitoring of treatment induced toxicity and cachexia in head and neck cancer: A method for early detection of high risk patients. Metabolomics 2019, 15, 110. [Google Scholar] [CrossRef]
- De Cecco, L.; Nicolau, M.; Giannoccaro, M.; Daidone, M.G.; Bossi, P.; Locati, L.; Licitra, L.; Canevari, S. Head and neck cancer subtypes with biological and clinical relevance: Meta-analysis of gene-expression data. Oncotarget 2015, 6, 9627–9642. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.M.; Roback, J.D.; Uppal, K.; Yu, T.; Jones, D.P.; Josephson, C.D. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion 2015, 55, 544–552. [Google Scholar] [CrossRef]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
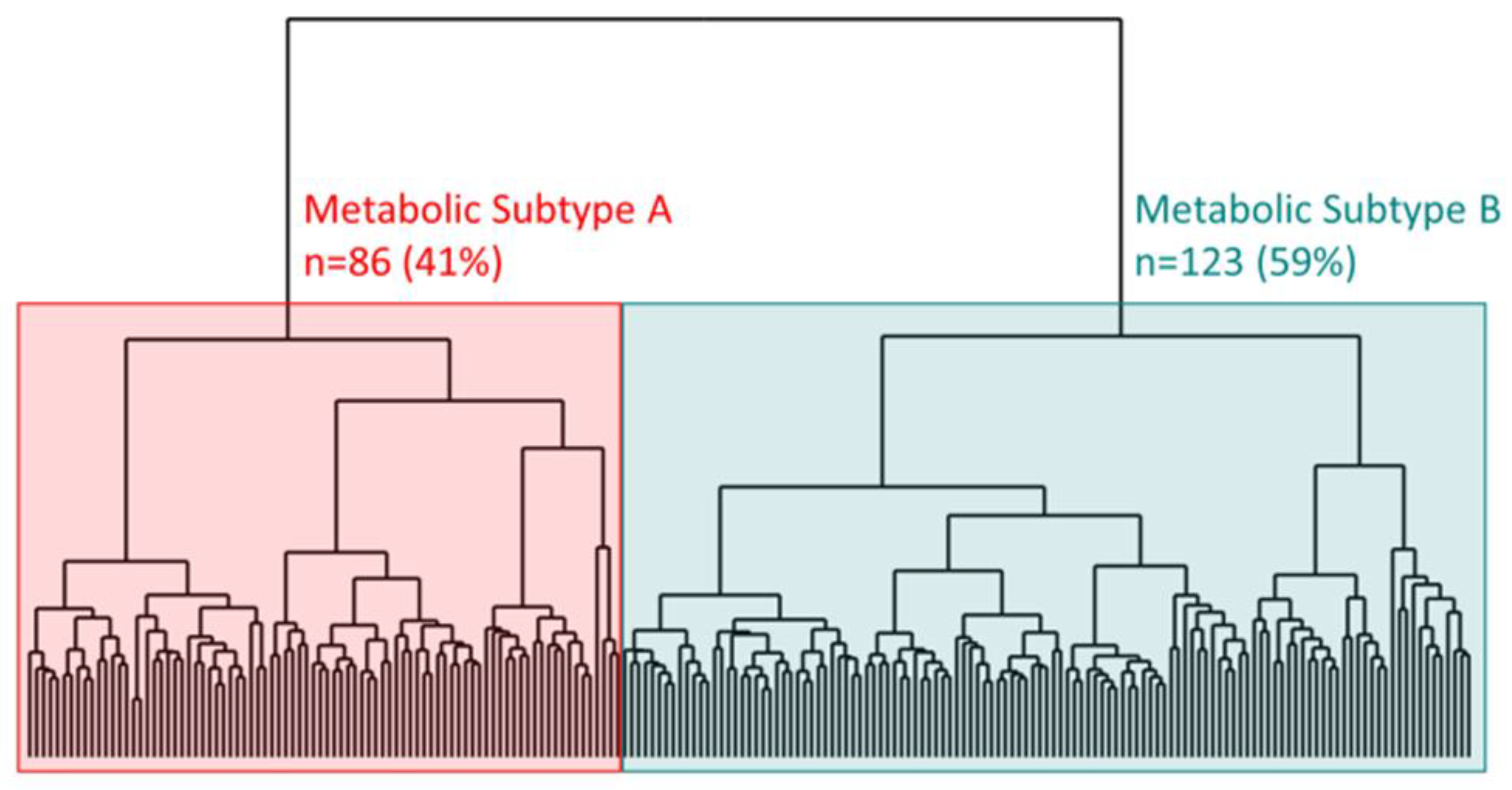

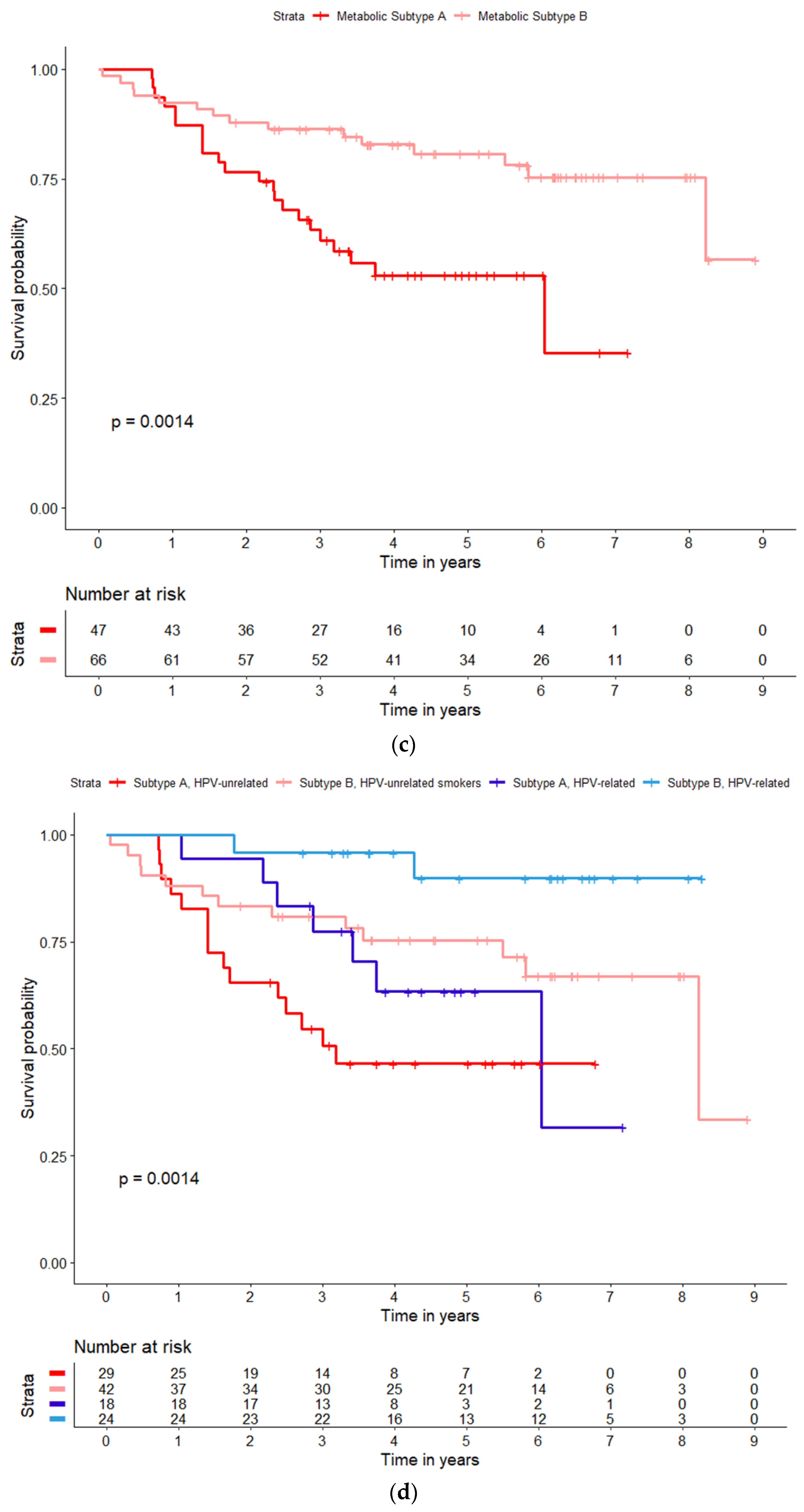
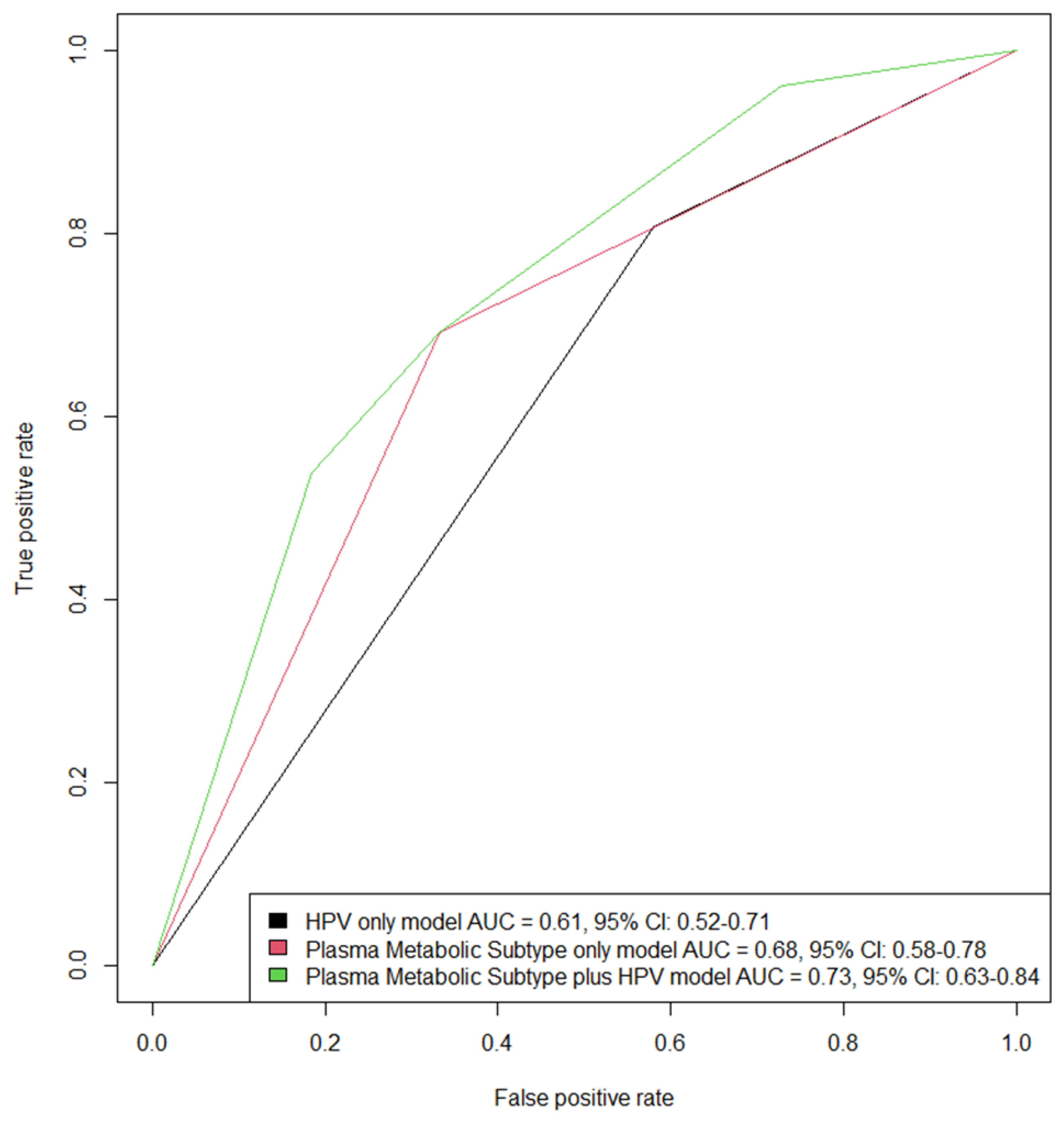
| All (N = 209) | Subtype A (N = 86) | Subtype B (N = 123) | p Value | ||
|---|---|---|---|---|---|
| Age | mean (SD) | 59.3 (10.1) | 61.7 (10.3) | 57.6 (9.7) | 0.004 |
| BMI | mean (SD) | 27.5 (5.2) | 28.1 (5.8) | 27.1 (4.7) | 0.18 |
| Sex | Male | 156 (75%) | 65 (76%) | 91 (74%) | 0.92 |
| Female | 53 (25%) | 21 (24%) | 32 (26%) | ||
| Race | White | 170 (81%) | 73 (85%) | 97 (79%) | 0.36 |
| Black | 39 (19%) | 13 (15%) | 26 (21%) | ||
| HPV status | Negative | 108 (52%) | 43 (50%) | 65 (53%) | 0.79 |
| Positive | 101 (48%) | 43 (50%) | 58 (47%) | ||
| Smoking history | Never | 81 (39%) | 33 (39%) | 48 (39%) | 0.83 |
| Former | 67 (33%) | 29 (35%) | 38 (31%) | ||
| Current | 58 (28%) | 22 (26%) | 36 (30%) | ||
| Alcohol | <1 drink/week | 114 (55%) | 47 (55%) | 67 (55%) | 0.99 |
| 1+ drink/week | 92 (45%) | 38 (45%) | 54 (45%) | ||
| Marital status | Married or partnered | 148 (71%) | 62 (72%) | 86 (70%) | 0.85 |
| Single | 61 (29%) | 24 (28%) | 37 (30%) | ||
| Tumor site | Oropharynx | 106 (51%) | 45 (53%) | 61 (50%) | 0.56 |
| Oral cavity | 31 (15%) | 12 (14%) | 19 (15%) | ||
| Larynx | 35 (17%) | 11 (13%) | 24 (20%) | ||
| Other | 36 (17%) | 17 (20%) | 19 (15%) | ||
| Tumor stage * | I | 10 (5%) | 5 (6%) | 5 (4%) | 0.77 |
| II | 16 (8%) | 5 (6%) | 11 (9%) | ||
| III | 83 (40%) | 36 (42%) | 47 (39%) | ||
| IV | 98 (47%) | 40 (47%) | 58 (48%) | ||
| T | 1 | 37 (18%) | 19 (22%) | 18 (15%) | 0.43 |
| 2 | 51 (25%) | 20 (24%) | 31 (26%) | ||
| 3 | 47 (23%) | 21 (25%) | 26 (22%) | ||
| 4 | 70 (34%) | 25 (29%) | 45 (37%) | ||
| N | 0 | 46 (22%) | 18 (21%) | 28 (23%) | 0.09 |
| 1 | 22 (11%) | 4 (5%) | 18 (15%) | ||
| 2 | 130 (63%) | 60 (70%) | 70 (58%) | ||
| 3 | 8 (4%) | 4 (5%) | 4 (3%) | ||
| Treatment | Radiotherapy | 45 (22%) | 20 (23%) | 25 (20%) | 0.87 |
| Chemoradiotherapy with Cisplatin | 118 (56%) | 48 (56%) | 70 (57%) | ||
| Chemoradiotherapy with Carboplatin and Paclitaxel | 46 (22%) | 18 (21%) | 28 (23%) | ||
| Feeding tube | No | 80 (40%) | 35 (43%) | 45 (39%) | 0.60 |
| Yes | 118 (60%) | 46 (57%) | 72 (62%) | ||
| ECOG performance | Active | 100 (50%) | 39 (48%) | 61 (52%) | 0.89 |
| Restricted | 73 (37%) | 31 (38%) | 42 (36%) | ||
| Non-working | 26 (13%) | 11 (14%) | 15 (13%) | ||
| Prior comorbidity | Yes | 154 (75%) | 66 (77%) | 88 (73%) | 0.69 |
| No | 52 (25%) | 20 (23%) | 32 (27%) | ||
| Albumin | mean (SD) | 3.95 (0.42) | 3.97 (0.39) | 3.94 (0.43) | 0.60 |
| Hemoglobin | mean (SD) | 13.18 (1.80) | 13.22 (1.71) | 13.15 (1.87) | 0.80 |
| Neutrophil-to-lymphocyte ratio | mean (SD) | 3.18 (2.07) | 3.32 (1.98) | 3.08 (2.13) | 0.45 |
| Platelet-to-lymphocyte ratio | mean (SD) | 170,347 (98,859) | 161,891 (64,755) | 176,207 (116,774) | 0.35 |
| Name | mz | rt | ESI | Adduct | HMDB# | Z-Score * A | Z-Score * B | p-Value ** |
|---|---|---|---|---|---|---|---|---|
| Fatty acid biosynthesis (p = 0.004) | ||||||||
| Acetoacetate | 101.0244 | 25 | C18- | (M − H) | HMDB0000060 | 0.53 | −0.37 | 2.0 × 10−9 |
| β-hydroxybutyrate | 103.0401 | 22 | C18- | (M − H) | HMDB0000357 | 0.43 | −0.30 | 4.3 × 10−6 |
| FA 16:0 (Palmitate) | 255.2329 | 231 | C18- | (M − H) | HMDB00220 | 0.38 | −0.27 | 4.6 × 10−6 |
| FA 14:0 (Myristate) | 227.2016 | 212 | C18- | (M − H) | HMDB00806 | 0.38 | −0.27 | 7.1 × 10−6 |
| Transfer of acetyl groups into the mitochondria (p = 0.03) | ||||||||
| Glucose | 215.0328 | 21 | C18- | (M + Cl) | HMDB0000122 | 0.40 | −0.28 | 3.1 × 10−6 |
| Citric acid | 191.0197 | 19 | C18- | (M − H) | HMDB0000094 | 0.41 | −0.28 | 7.5 × 10−6 |
| Malic acid | 133.0143 | 20 | C18- | (M − H) | HMDB0000156 | 0.37 | −0.26 | 7.6 × 10−6 |
| Arginine and Proline metabolism (p = 0.06) | ||||||||
| S-adenosylmethionine | 399.1445 | 162 | HILIC+ | (M + H) | HMDB0001185 | 0.47 | −0.33 | 1.6 × 10−9 |
| Proline | 116.0706 | 87 | HILIC+ | (M + H) | HMDB0000162 | 0.49 | −0.35 | 8.3 × 10−8 |
| Ornithine | 133.0972 | 125 | HILIC+ | (M + H) | HMDB0000214 | 0.45 | −0.32 | 5.1 × 10−7 |
| Citrulline | 176.103 | 109 | HILIC+ | (M + H) | HMDB0000904 | 0.40 | −0.28 | 8.4 × 10−7 |
| Guanidinoacetate | 118.0617 | 89 | HILIC+ | (M + H) | HMDB0000128 | 0.41 | −0.29 | 8.8 × 10−7 |
| Galactose metabolism (p = 0.07) | ||||||||
| Fructose | 219.0265 | 73 | HILIC+ | (M + K) | HMDB0000660 | 0.60 | −0.42 | 3.6 × 10−11 |
| Galactose | 203.0526 | 71 | HILIC+ | (M + Na) | HMDB0000143 | 0.59 | −0.41 | 3.6 × 10−11 |
| Glucose | 215.0328 | 21 | C18- | (M + Cl) | HMDB0000122 | 0.40 | −0.28 | 3.1 × 10−6 |
| HR | 95% CI | p Value | |
|---|---|---|---|
| Full Population (N = 189, Ndeaths = 47) | |||
| Unadjusted model | 2.33 | (1.30, 4.16) | 0.004 |
| Age, sex, HPV, smoking adjusted model | 2.38 | (1.30, 4.33) | 0.005 |
| Fully adjusted model a | 2.76 | (1.32, 5.77) | 0.007 |
| Among Ever Smokers (N = 113, Ndeaths = 37) | |||
| Unadjusted model | 2.91 | (1.47, 5.78) | 0.002 |
| Age, sex, HPV adjusted model | 2.98 | (1.48, 5.99) | 0.002 |
| Fully adjusted model a | 3.58 | (1.46, 8.78) | 0.005 |
| Among Never Smokers (N = 76, Ndeaths = 10) | |||
| Unadjusted model | 1.60 | (0.46, 5.51) | 0.46 |
| Age, sex, HPV adjusted model | 1.13 | (0.30, 4.24) | 0.85 |
| Fully adjusted model a | 0.92 | (0.06, 15.3) | 0.95 |
| HR | 95% CI | p Value | |
|---|---|---|---|
| Full Population (Nevents = 62) | |||
| Unadjusted model | 1.62 | (0.98, 2.67) | 0.06 |
| Age, sex, HPV, smoking adjusted model | 1.65 | (0.99, 2.75) | 0.06 |
| Fully adjusted model a | 1.70 | (0.93, 3.11) | 0.08 |
| Among Ever Smokers (Nevents = 47) | |||
| Unadjusted model | 2.06 | (1.14, 3.71) | 0.02 |
| Age, sex, HPV adjusted model | 2.13 | (1.17, 3.89) | 0.01 |
| Fully adjusted model a | 2.11 | (1.03, 4.32) | 0.04 |
| Among Never Smokers (Nevents = 15) | |||
| Unadjusted model | 1.00 | (0.35, 2.80) | 0.99 |
| Age, sex, HPV adjusted model | 0.80 | (0.27, 2.37) | 0.68 |
| Fully adjusted model a | 0.48 | (0.08, 3.03) | 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eldridge, R.C.; Qin, Z.S.; Saba, N.F.; Houser, M.C.; Hayes, D.N.; Miller, A.H.; Bruner, D.W.; Jones, D.P.; Xiao, C. Unsupervised Hierarchical Clustering of Head and Neck Cancer Patients by Pre-Treatment Plasma Metabolomics Creates Prognostic Metabolic Subtypes. Cancers 2023, 15, 3184. https://doi.org/10.3390/cancers15123184
Eldridge RC, Qin ZS, Saba NF, Houser MC, Hayes DN, Miller AH, Bruner DW, Jones DP, Xiao C. Unsupervised Hierarchical Clustering of Head and Neck Cancer Patients by Pre-Treatment Plasma Metabolomics Creates Prognostic Metabolic Subtypes. Cancers. 2023; 15(12):3184. https://doi.org/10.3390/cancers15123184
Chicago/Turabian StyleEldridge, Ronald C., Zhaohui S. Qin, Nabil F. Saba, Madelyn C. Houser, D. Neil Hayes, Andrew H. Miller, Deborah W. Bruner, Dean P. Jones, and Canhua Xiao. 2023. "Unsupervised Hierarchical Clustering of Head and Neck Cancer Patients by Pre-Treatment Plasma Metabolomics Creates Prognostic Metabolic Subtypes" Cancers 15, no. 12: 3184. https://doi.org/10.3390/cancers15123184






