Taxifolin Inhibits Breast Cancer Growth by Facilitating CD8+ T Cell Infiltration and Inducing a Novel Set of Genes including Potential Tumor Suppressor Genes in 1q21.3
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Programs and Websites
2.3. Profiling BC-Associated Immune Cells
2.4. Model Size Optimization
2.5. Signature Score Assignment for Individual Tumors
2.6. Immunohistochemistry (IHC)
2.7. Culture of 4T-1 Cells, Generation of 4T-1 Tumors, and Taxifolin Treatment
2.8. RNA Sequencing Analysis
2.9. Quantitative Real-Time PCR
2.10. Statistical Analysis
3. Results
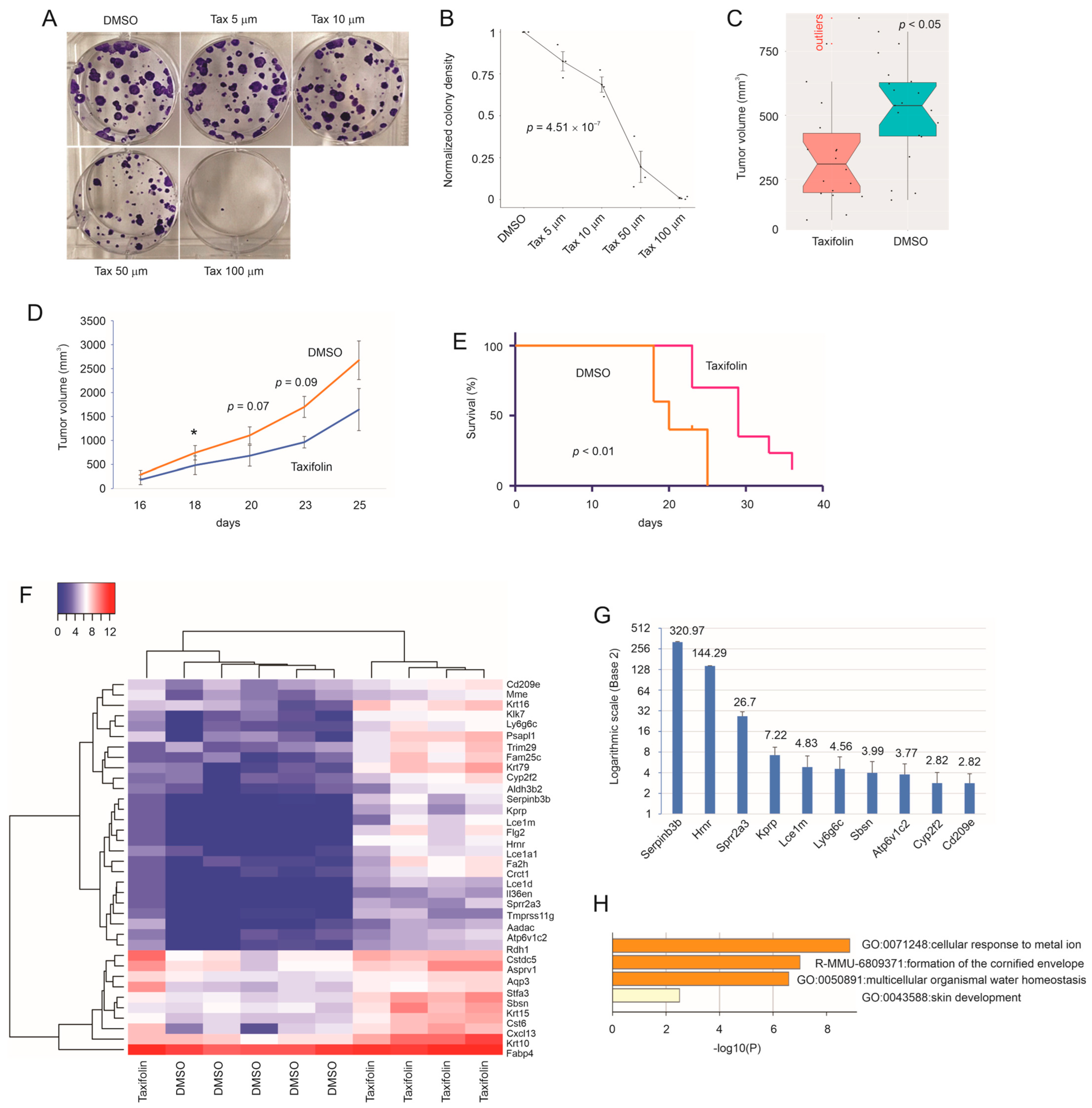
3.1. Taxifolin Inhibits Breast Cancer Progression
3.2. The Presence of Multiple Tumor-Suppressive Components in Taxifolin-Induced DEGs
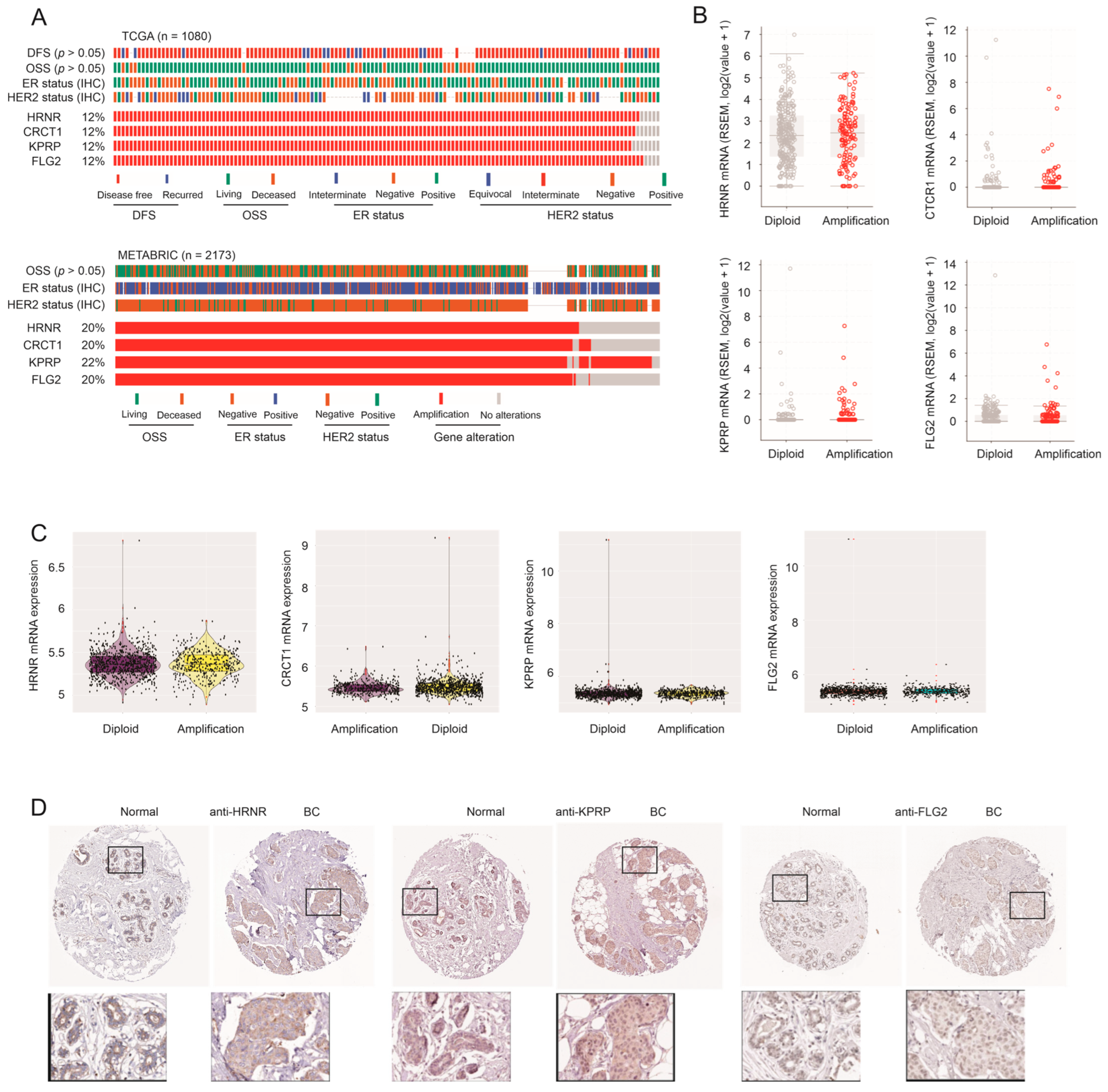
3.3. Amplification of HRNR, CRCT1, KPRP, and FLG2 Genes Does Not Lead to Their Upregulation
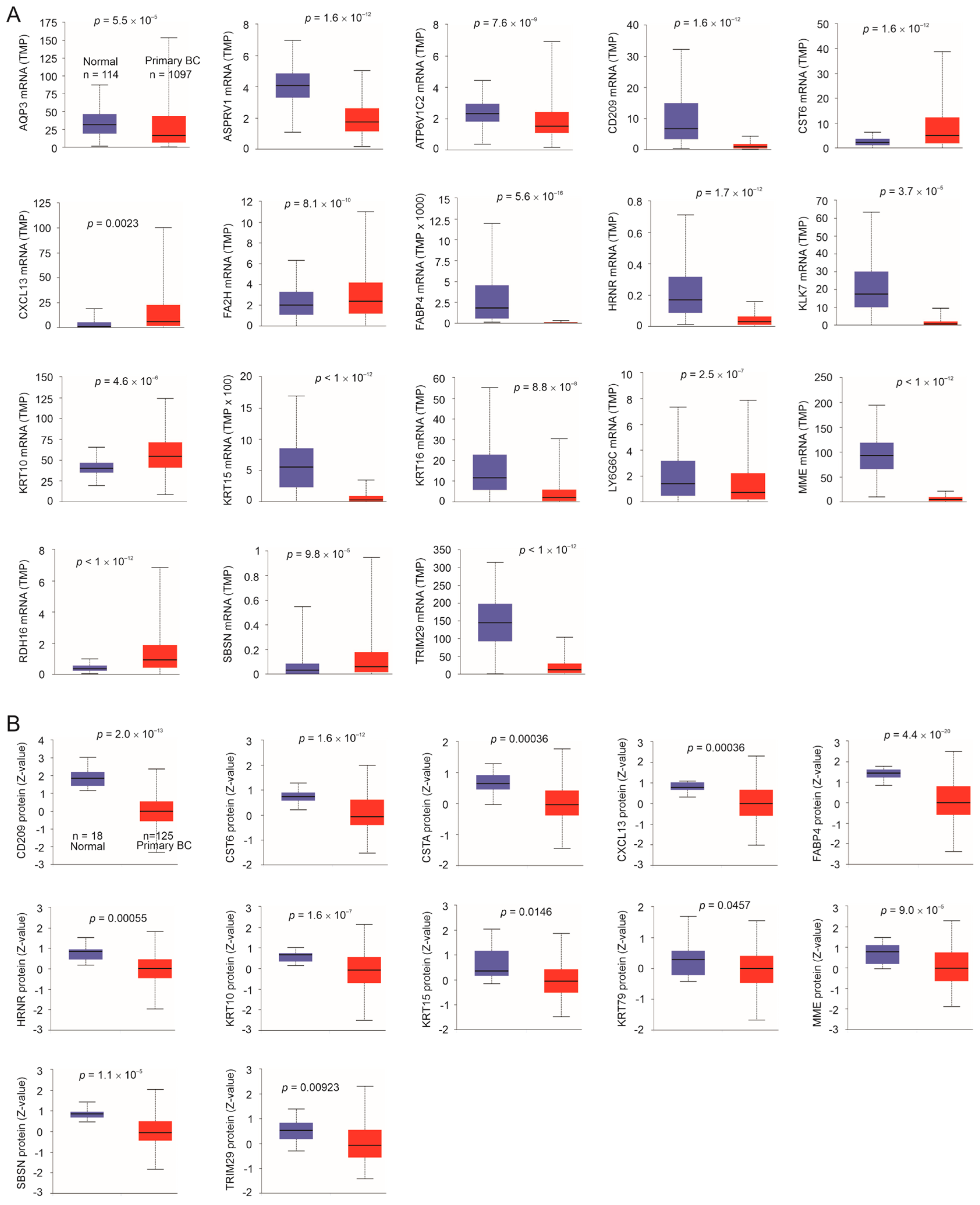
3.4. Downregulation of Taxifolin-Induced DEGs Following BC Pathogenesis
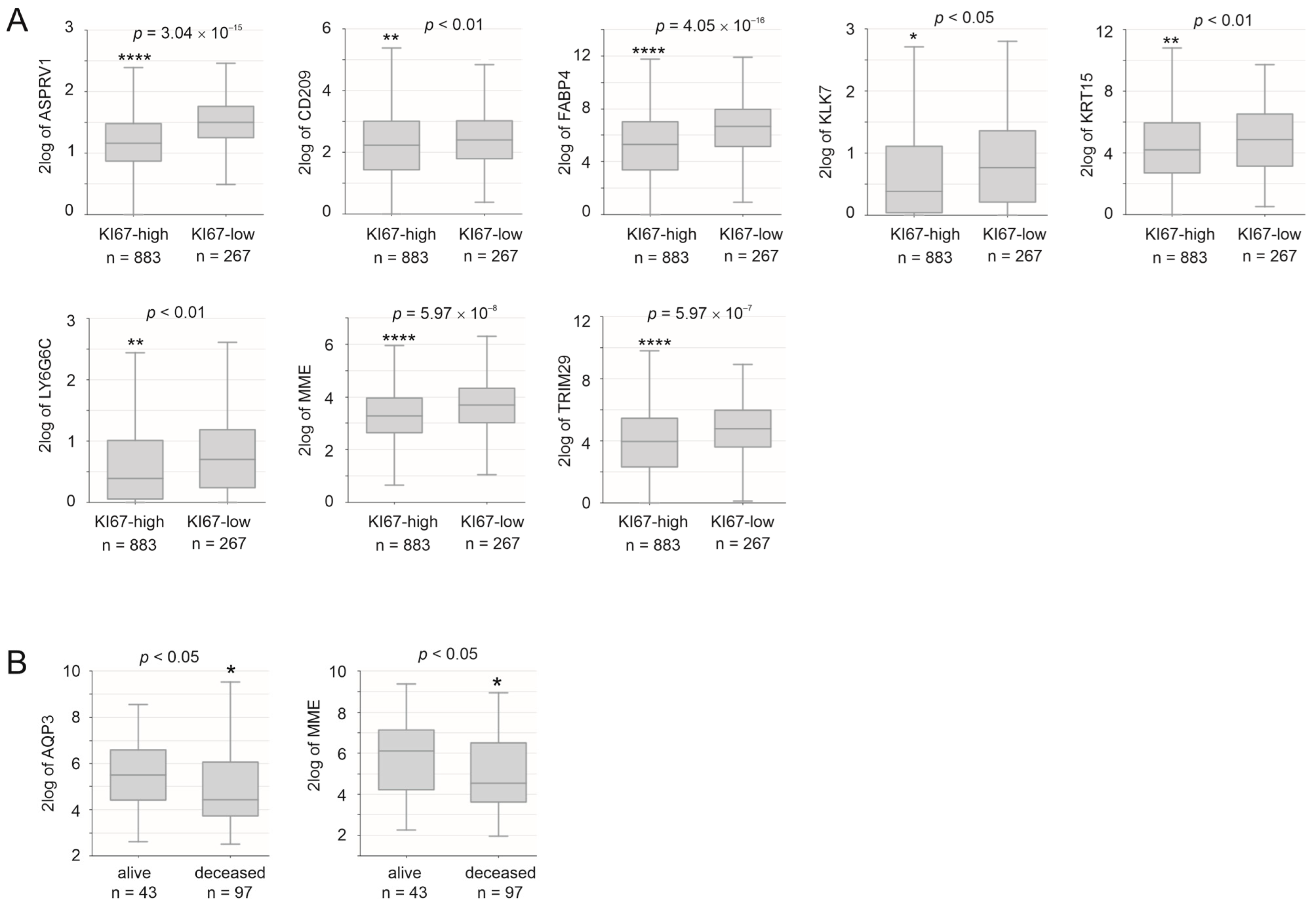
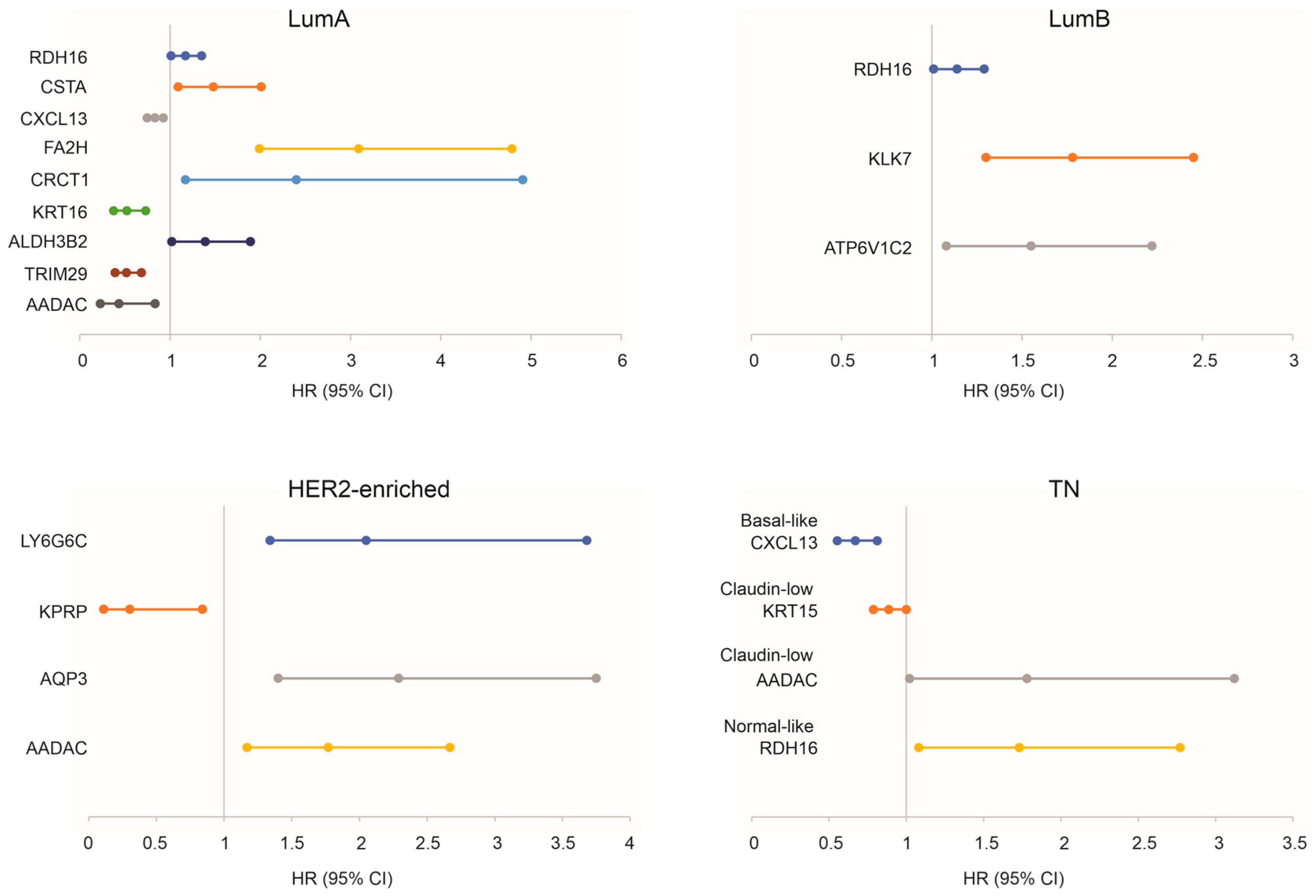
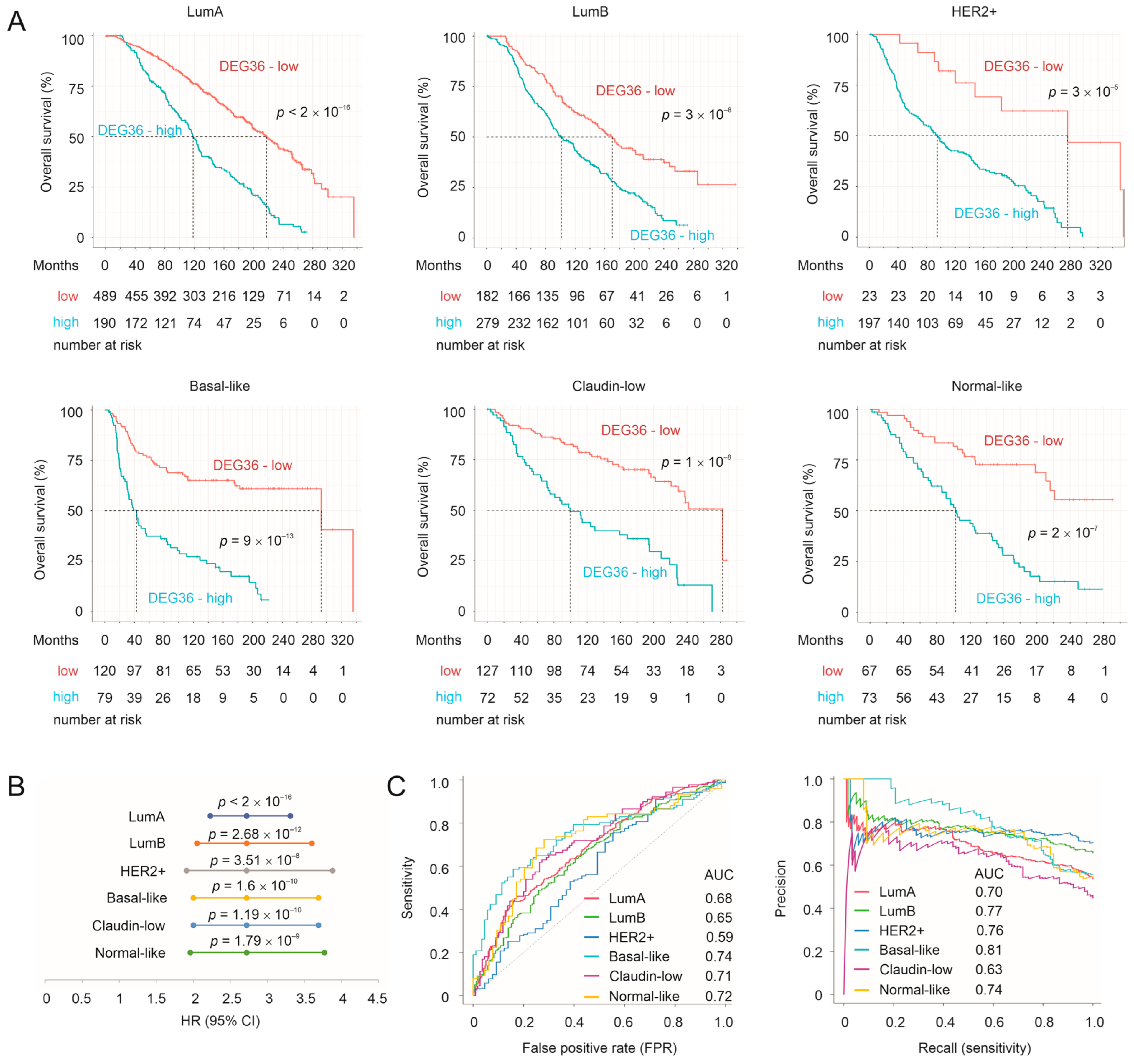
3.5. Prediction of Survival Probability by DEGs Relevant to Taxifolin Treatment
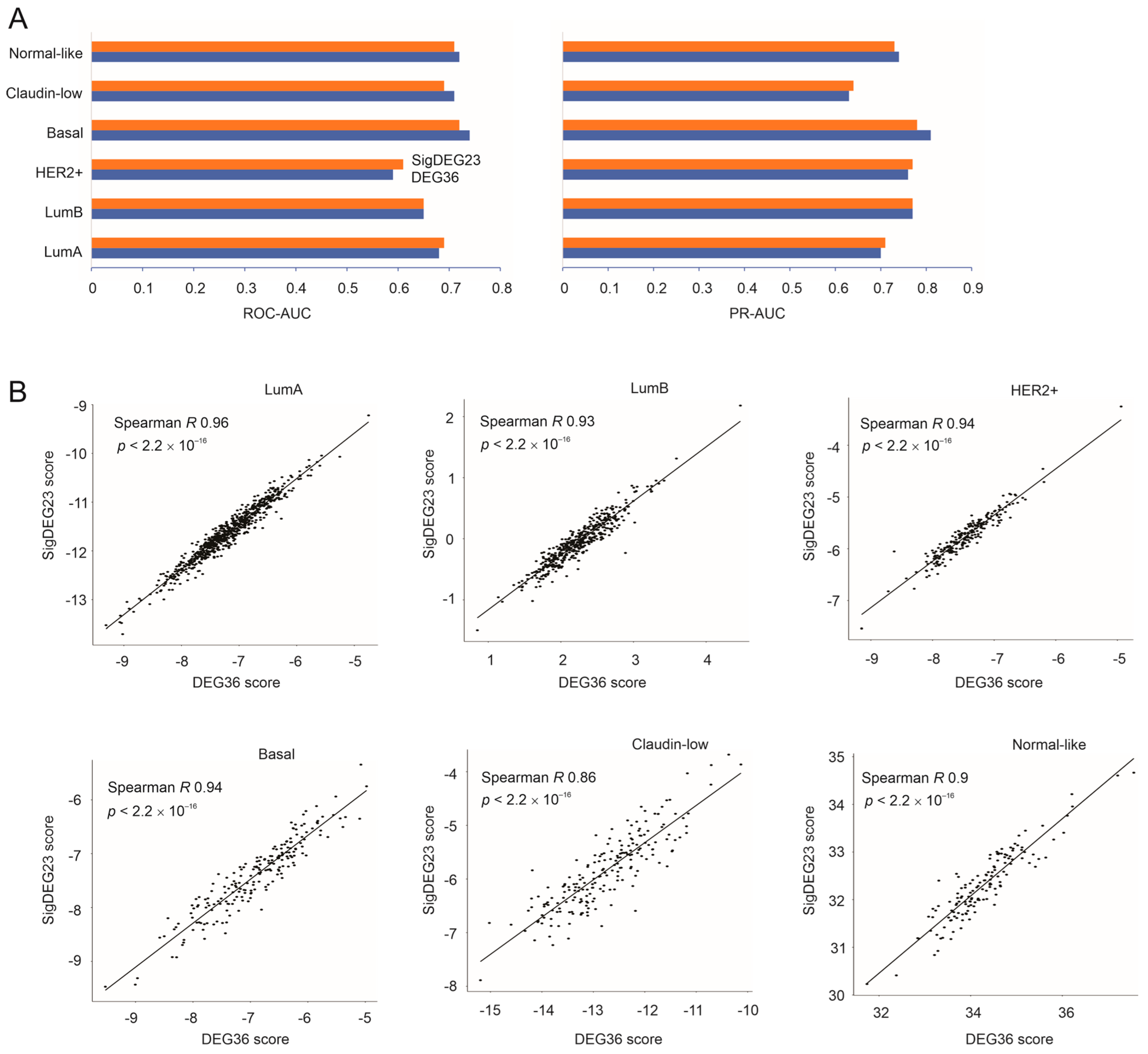
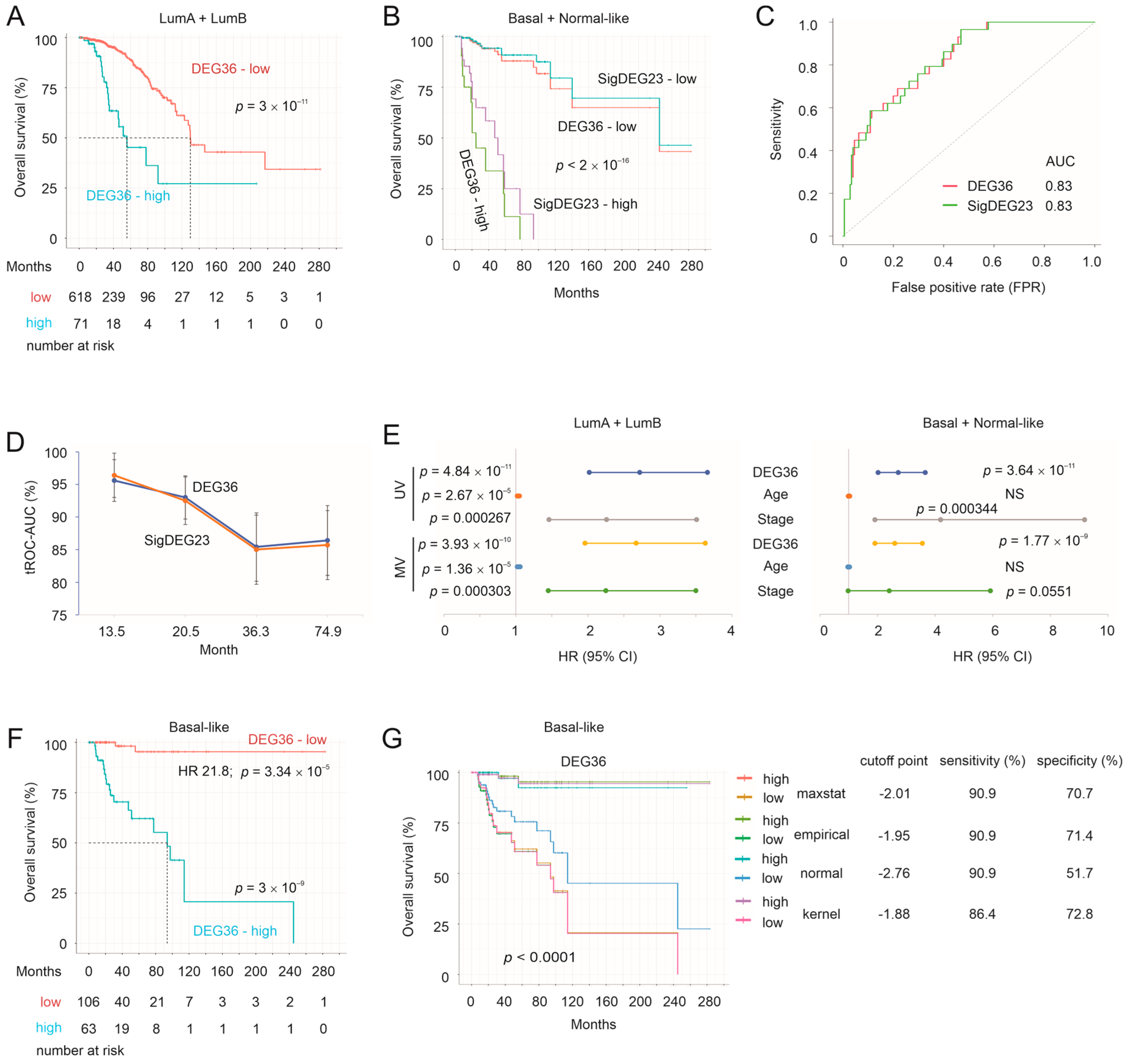
3.6. Validation of the DEG36 and SigDEG23
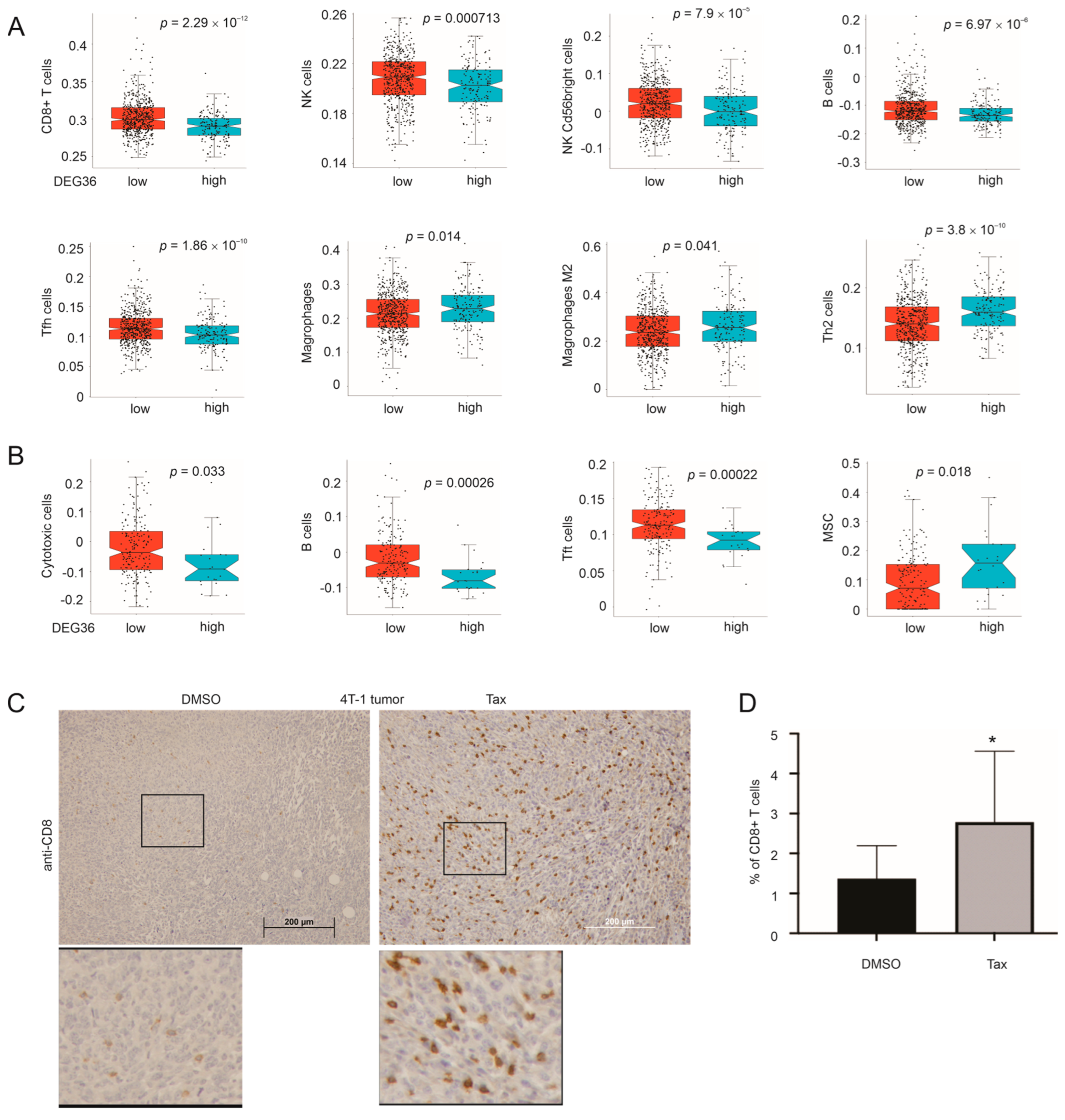
3.7. DEG36 Predicts the Exclusion of Cytotoxic Lymphocytes from BC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast cancer treatment: A review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Harbeck, N.; Salem, M.; Nitz, U.; Gluz, O.; Liedtke, C. Personalized treatment of early-stage breast cancer: Present concepts and future directions. Cancer Treat. Rev. 2010, 36, 584–594. [Google Scholar] [CrossRef]
- Pilie, P.G.; Gay, C.M.; Byers, L.A.; O’Connor, M.J.; Yap, T.A. Parp inhibitors: Extending benefit beyond brca-mutant cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3759–3771. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Haakensen, V.D.; Lingjaerde, O.C.; Luders, T.; Riis, M.; Prat, A.; Troester, M.A.; Holmen, M.M.; Frantzen, J.O.; Romundstad, L.; Navjord, D.; et al. Gene expression profiles of breast biopsies from healthy women identify a group with claudin-low features. BMC Med. Genom. 2011, 4, 77. [Google Scholar] [CrossRef]
- Li, X.; Oprea-Ilies, G.M.; Krishnamurti, U. New developments in breast cancer and their impact on daily practice in pathology. Arch. Pathol. Lab. Med. 2017, 141, 490–498. [Google Scholar] [CrossRef]
- Ghareghomi, S.; Habibi-Rezaei, M.; Arese, M.; Saso, L.; Moosavi-Movahedi, A.A. Nrf2 modulation in breast cancer. Biomedicines 2022, 10, 2668. [Google Scholar] [CrossRef]
- Bernatova, I.; Liskova, S. Mechanisms modified by (-)-epicatechin and taxifolin relevant for the treatment of hypertension and viral infection: Knowledge from preclinical studies. Antioxidants 2021, 10, 467. [Google Scholar] [CrossRef]
- Weidmann, A.E. Dihydroquercetin: More than just an impurity? Eur. J. Pharmacol. 2012, 684, 19–26. [Google Scholar] [CrossRef]
- Sunil, C.; Xu, B. An insight into the health-promoting effects of taxifolin (dihydroquercetin). Phytochemistry 2019, 166, 112066. [Google Scholar] [CrossRef]
- Das, A.; Baidya, R.; Chakraborty, T.; Samanta, A.K.; Roy, S. Pharmacological basis and new insights of taxifolin: A comprehensive review. Biomed. Pharmacother. 2021, 142, 112004. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, X.; Wang, Q.; Li, X.; Wang, E.; Zhao, Q.; Wang, Q.; Cao, H. The anti-tumor effect of taxifolin on lung cancer via suppressing stemness and epithelial-mesenchymal transition in vitro and oncogenesis in nude mice. Ann. Transl. Med. 2020, 8, 590. [Google Scholar] [CrossRef]
- Razak, S.; Afsar, T.; Ullah, A.; Almajwal, A.; Alkholief, M.; Alshamsan, A.; Jahan, S. Taxifolin, a natural flavonoid interacts with cell cycle regulators causes cell cycle arrest and causes tumor regression by activating wnt/beta -catenin signaling pathway. BMC Cancer 2018, 18, 1043. [Google Scholar] [CrossRef]
- Woo, Y.; Shin, S.Y.; Hyun, J.; Lee, S.D.; Lee, Y.H.; Lim, Y. Flavanones inhibit the clonogenicity of hct116 cololectal cancer cells. Int. J. Mol. Med. 2012, 29, 403–408. [Google Scholar]
- Lee, S.B.; Cha, K.H.; Selenge, D.; Solongo, A.; Nho, C.W. The chemopreventive effect of taxifolin is exerted through are-dependent gene regulation. Biol. Pharm. Bull. 2007, 30, 1074–1079. [Google Scholar] [CrossRef]
- Dostal, Z.; Sebera, M.; Srovnal, J.; Staffova, K.; Modriansky, M. Dual effect of taxifolin on zeb2 cancer signaling in hepg2 cells. Molecules 2021, 26, 1476. [Google Scholar] [CrossRef]
- Oi, N.; Chen, H.; Ok Kim, M.; Lubet, R.A.; Bode, A.M.; Dong, Z. Taxifolin suppresses uv-induced skin carcinogenesis by targeting egfr and pi3k. Cancer Prev. Res. 2012, 5, 1103–1114. [Google Scholar] [CrossRef]
- Chen, X.; Gu, N.; Xue, C.; Li, B.R. Plant flavonoid taxifolin inhibits the growth, migration and invasion of human osteosarcoma cells. Mol. Med. Rep. 2018, 17, 3239–3245. [Google Scholar] [CrossRef]
- Xie, J.; Pang, Y.; Wu, X. Taxifolin suppresses the malignant progression of gastric cancer by regulating the ahr/cyp1a1 signaling pathway. Int. J. Mol. Med. 2021, 48, 197. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Gong, H.; Mei, H.; Shi, L.; Yu, J.; Hu, Y. Taxifolin targets pi3k and mtor and inhibits glioblastoma multiforme. J. Oncol. 2021, 2021, 5560915. [Google Scholar] [CrossRef]
- Zhang, Z.R.; Al Zaharna, M.; Wong, M.M.; Chiu, S.K.; Cheung, H.Y. Taxifolin enhances andrographolide-induced mitotic arrest and apoptosis in human prostate cancer cells via spindle assembly checkpoint activation. PLoS ONE 2013, 8, e54577. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.; Yaduvanshi, S.; Silvestre, S.; Duarte, A.P.; Santos, A.O.; Soares, C.P.; Kumar, V.; Passarinha, L.; Sousa, A. Taxifolin and lucidin as potential e6 protein inhibitors: P53 function re-establishment and apoptosis induction in cervical cancer cells. Cancers 2022, 14, 2834. [Google Scholar] [CrossRef] [PubMed]
- Alzaharna, M.; Alqouqa, I.; Cheung, H.Y. Taxifolin synergizes andrographolide-induced cell death by attenuation of autophagy and augmentation of caspase dependent and independent cell death in hela cells. PLoS ONE 2017, 12, e0171325. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.W.; Pattanayak, S.P. Taxifolin inhibits 7,12-dimethylbenz(a)anthracene-induced breast carcinogenesis by regulating ahr/cyp1a1 signaling pathway. Pharmacogn. Mag. 2018, 13, S749–S755. [Google Scholar] [PubMed]
- Hossain, M.M.; Ray, S.K. Ews knockdown and taxifolin treatment induced differentiation and removed DNA methylation from p53 promoter to promote expression of puma and noxa for apoptosis in ewing’s sarcoma. J. Cancer Ther. 2014, 5, 1092–1113. [Google Scholar] [CrossRef]
- Li, J.; Hu, L.; Zhou, T.; Gong, X.; Jiang, R.; Li, H.; Kuang, G.; Wan, J.; Li, H. Taxifolin inhibits breast cancer cells proliferation, migration and invasion by promoting mesenchymal to epithelial transition via beta-catenin signaling. Life Sci. 2019, 232, 116617. [Google Scholar] [CrossRef]
- Goh, J.Y.; Feng, M.; Wang, W.; Oguz, G.; Yatim, S.; Lee, P.L.; Bao, Y.; Lim, T.H.; Wang, P.; Tam, W.L.; et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017, 23, 1319–1330. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of rna-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. Xcell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Yan, J.; Ojo, D.; Kapoor, A.; Lin, X.; Pinthus, J.H.; Aziz, T.; Bismar, T.A.; Wei, F.; Wong, N.; De Melo, J.; et al. Neural cell adhesion protein cntn1 promotes the metastatic progression of prostate cancer. Cancer Res. 2016, 76, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.N.; Dussaq, A.M.; Kennell, T., Jr.; Willey, C.D.; Hjelmeland, A.B. Hpaanalyze: An r package that facilitates the retrieval and analysis of the human protein atlas data. BMC Bioinform. 2019, 20, 463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lin, X.; Kapoor, A.; He, L.; Wei, F.; Gu, Y.; Mei, W.; Zhao, K.; Yang, H.; Tang, D. Fam84b promotes prostate tumorigenesis through a network alteration. Ther. Adv. Med. Oncol. 2019, 11, 1758835919846372. [Google Scholar] [CrossRef] [PubMed]
- Eckhart, L.; Lippens, S.; Tschachler, E.; Declercq, W. Cell death by cornification. Biochim. Biophys. Acta 2013, 1833, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhang, Y.; Li, H.; Yao, L.; Fu, D.; Yao, X.; Xu, L.X.; Hu, X.; Hu, G. Differential secretome analysis reveals cst6 as a suppressor of breast cancer bone metastasis. Cell Res. 2012, 22, 1356–1373. [Google Scholar] [CrossRef]
- Ai, L.; Kim, W.J.; Alpay, M.; Tang, M.; Pardo, C.E.; Hatakeyama, S.; May, W.S.; Kladde, M.P.; Heldermon, C.D.; Siegel, E.M.; et al. Trim29 suppresses twist1 and invasive breast cancer behavior. Cancer Res. 2014, 74, 4875–4887. [Google Scholar] [CrossRef]
- John Mary, D.J.S.; Sikarwar, G.; Kumar, A.; Limaye, A.M. Interplay of eralpha binding and DNA methylation in the intron-2 determines the expression and estrogen regulation of cystatin a in breast cancer cells. Mol. Cell. Endocrinol. 2020, 504, 110701. [Google Scholar] [CrossRef]
- Zhong, P.; Shu, R.; Wu, H.; Liu, Z.; Shen, X.; Hu, Y. Low krt15 expression is associated with poor prognosis in patients with breast invasive carcinoma. Exp. Ther. Med. 2021, 21, 305. [Google Scholar] [CrossRef]
- Geng, X.; Babayeva, L.; Walch, A.; Aubele, M.; Gross, E.; Kiechle, M.; Bronger, H.; Dreyer, T.; Magdolen, V.; Dorn, J. High levels of klk7 protein expression are related to a favorable prognosis in triple-negative breast cancer patients. Am. J. Cancer Res. 2020, 10, 1785–1792. [Google Scholar]
- Ren, Q.; Khoo, W.H.; Corr, A.P.; Phan, T.G.; Croucher, P.I.; Stewart, S.A. Gene expression predicts dormant metastatic breast cancer cell phenotype. Breast Cancer Res. BCR 2022, 24, 10. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, H.; Qi, Y.; Li, S.; Geng, C. Epigenetic study of early breast cancer (ebc) based on DNA methylation and gene integration analysis. Sci. Rep. 2022, 12, 1989. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Chen, Y.; Qin, Q.; Guo, F.; Wang, Y.S.; Li, D. Cxcl13 expression in mouse 4t1 breast cancer microenvironment elicits antitumor immune response by regulating immune cell infiltration. Precis. Clin. Med. 2021, 4, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, U.; Alshehri, B.; Khan, A.A.; Bhat, A.A.; Bagga, P.; Wani, N.A.; Mir, M.A. Expression pattern and prognostic significance of chemokines in breast cancer: An integrated bioinformatics analysis. Clin. Breast Cancer 2022, 22, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, Z.; Li, B.; Li, J.; Ou, Y.; Yu, X.; Zhang, Z.; Liu, S.; Fu, X.; Jin, H.; et al. Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunology 2022, 11, 2085432. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Jiao, L.; Li, T.; Wang, H.; Wei, W.; Qian, H. Expression of aqp3 and aqp5 as a prognostic marker in triple-negative breast cancer. Oncol. Lett. 2018, 16, 2661–2667. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, Y.; Gao, S.; Wang, Y.; Qu, C.; Wu, Y.; Ding, N.; Dai, Y.; Jiang, L.; Liu, S. Prognostic signature and therapeutic value based on membrane lipid biosynthesis-related genes in breast cancer. J. Oncol. 2022, 2022, 7204415. [Google Scholar] [CrossRef]
- Gao, C.; Zhuang, J.; Li, H.; Liu, C.; Zhou, C.; Liu, L.; Feng, F.; Sun, C.; Wu, J. Development of a risk scoring system for evaluating the prognosis of patients with her2-positive breast cancer. Cancer Cell Int. 2020, 20, 121. [Google Scholar] [CrossRef]
- Dai, X.; Zhang, S.; Cheng, H.; Cai, D.; Chen, X.; Huang, Z. Fa2h exhibits tumor suppressive roles on breast cancers via cancer stemness control. Front. Oncol. 2019, 9, 1089. [Google Scholar] [CrossRef]
- Kim, J.; Villadsen, R. The expression pattern of epidermal differentiation marker keratin 10 in the normal human breast and breast cancer cells. J. Histochem. Cytochem. 2020, 68, 561–570. [Google Scholar] [CrossRef]
- Elazezy, M.; Schwentesius, S.; Stegat, L.; Wikman, H.; Werner, S.; Mansour, W.Y.; Failla, A.V.; Peine, S.; Muller, V.; Thiery, J.P.; et al. Emerging insights into keratin 16 expression during metastatic progression of breast cancer. Cancers 2021, 13, 3869. [Google Scholar] [CrossRef]
- Sheshadri, N.; Catanzaro, J.M.; Bott, A.J.; Sun, Y.; Ullman, E.; Chen, E.I.; Pan, J.A.; Wu, S.; Crawford, H.C.; Zhang, J.; et al. Scca1/serpinb3 promotes oncogenesis and epithelial-mesenchymal transition via the unfolded protein response and il6 signaling. Cancer Res. 2014, 74, 6318–6329. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, Z.; Shi, W.; Liu, H.; Pang, W. Breast cancer survival prediction using seven prognostic biomarker genes. Oncol. Lett. 2019, 18, 2907–2916. [Google Scholar] [CrossRef] [PubMed]
- Calaf, G.M.; Roy, D. Human drug metabolism genes in parathion-and estrogen-treated breast cells. Int. J. Mol. Med. 2007, 20, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.M.; Ginsburg, E.; Oliver, S.D.; Goldsmith, P.; Vonderhaar, B.K. Hornerin, an s100 family protein, is functional in breast cells and aberrantly expressed in breast cancer. BMC Cancer 2012, 12, 266. [Google Scholar] [CrossRef]
- Ademuyiwa, F.O.; Chen, I.; Luo, J.; Rimawi, M.F.; Hagemann, I.S.; Fisk, B.; Jeffers, G.; Skidmore, Z.L.; Basu, A.; Richters, M.; et al. Immunogenomic profiling and pathological response results from a clinical trial of docetaxel and carboplatin in triple-negative breast cancer. Breast Cancer Res. Treat. 2021, 189, 187–202. [Google Scholar] [CrossRef]
- Luo, R.; Chong, W.; Wei, Q.; Zhang, Z.; Wang, C.; Ye, Z.; Abu-Khalaf, M.M.; Silver, D.P.; Stapp, R.T.; Jiang, W.; et al. Whole-exome sequencing identifies somatic mutations and intratumor heterogeneity in inflammatory breast cancer. NPJ Breast Cancer 2021, 7, 72. [Google Scholar] [CrossRef]
- McConnell, M.; Feng, S.; Chen, W.; Zhu, G.; Shen, D.; Ponnazhagan, S.; Deng, L.; Li, Y.P. Osteoclast proton pump regulator atp6v1c1 enhances breast cancer growth by activating the mtorc1 pathway and bone metastasis by increasing v-atpase activity. Oncotarget 2017, 8, 47675–47690. [Google Scholar] [CrossRef]
- Rakha, E.A.; Alsaleem, M.; ElSharawy, K.A.; Toss, M.S.; Raafat, S.; Mihai, R.; Minhas, F.A.; Green, A.R.; Rajpoot, N.M.; Dalton, L.W.; et al. Visual histological assessment of morphological features reflects the underlying molecular profile in invasive breast cancer: A morphomolecular study. Histopathology 2020, 77, 631–645. [Google Scholar] [CrossRef]
- Garreis, F.; Jahn, J.; Wild, K.; Abrar, D.B.; Schicht, M.; Schroder, J.M.; Paulsen, F. Expression and regulation of s100 fused-type protein hornerin at the ocular surface and lacrimal apparatus. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5968–5977. [Google Scholar] [CrossRef]
- Holthaus, K.B.; Lachner, J.; Ebner, B.; Tschachler, E.; Eckhart, L. Gene duplications and gene loss in the epidermal differentiation complex during the evolutionary land-to-water transition of cetaceans. Sci. Rep. 2021, 11, 12334. [Google Scholar] [CrossRef]
- Wu, N.; Song, Y.; Pang, L.; Chen, Z. Crct1 regulated by microrna-520 g inhibits proliferation and induces apoptosis in esophageal squamous cell cancer. Tumour Biol. 2016, 37, 8271–8279. [Google Scholar] [CrossRef] [PubMed]
- Dalal, H.; Dahlgren, M.; Gladchuk, S.; Brueffer, C.; Gruvberger-Saal, S.K.; Saal, L.H. Clinical associations of esr2 (estrogen receptor beta) expression across thousands of primary breast tumors. Sci. Rep. 2022, 12, 4696. [Google Scholar] [CrossRef] [PubMed]
- Sinn, B.V.; Fu, C.; Lau, R.; Litton, J.; Tsai, T.H.; Murthy, R.; Tam, A.; Andreopoulou, E.; Gong, Y.; Murthy, R.; et al. Set(er/pr): A robust 18-gene predictor for sensitivity to endocrine therapy for metastatic breast cancer. NPJ Breast Cancer 2019, 5, 16. [Google Scholar] [CrossRef]
- Parvani, J.G.; Davuluri, G.; Wendt, M.K.; Espinosa, C.; Tian, M.; Danielpour, D.; Sossey-Alaoui, K.; Schiemann, W.P. Deptor enhances triple-negative breast cancer metastasis and chemoresistance through coupling to survivin expression. Neoplasia 2015, 17, 317–328. [Google Scholar] [CrossRef]
- Weigelt, B.; Baehner, F.L.; Reis-Filho, J.S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: A retrospective of the last decade. J. Pathol. 2010, 220, 263–280. [Google Scholar] [CrossRef]
- Hammad, H.; Debeuf, N.; Aegerter, H.; Brown, A.S.; Lambrecht, B.N. Emerging paradigms in type 2 immunity. Annu. Rev. Immunol. 2022, 40, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef]
- Sanchez, N.; Miranda, A.; Funes, J.M.; Hevia, G.; Perez, R.; de Leon, J. Oncogenic transformation tunes the cross-talk between mesenchymal stem cells and t lymphocytes. Cell Immunol. 2014, 289, 174–184. [Google Scholar] [CrossRef]
- Caubet, C.; Jonca, N.; Brattsand, M.; Guerrin, M.; Bernard, D.; Schmidt, R.; Egelrud, T.; Simon, M.; Serre, G. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, scte/klk5/hk5 and scce/klk7/hk7. J. Investig. Dermatol. 2004, 122, 1235–1244. [Google Scholar] [CrossRef]
- Schmidt, C.A.; Fisher-Wellman, K.H.; Neufer, P.D. From ocr and ecar to energy: Perspectives on the design and interpretation of bioenergetics studies. J. Biol. Chem. 2021, 297, 101140. [Google Scholar] [CrossRef]
- Gu, X.; Ma, Y.; Liu, Y.; Wan, Q. Measurement of mitochondrial respiration in adherent cells by seahorse xf96 cell mito stress test. STAR Protoc. 2021, 2, 100245. [Google Scholar] [CrossRef] [PubMed]
- Underwood, E.; Redell, J.B.; Zhao, J.; Moore, A.N.; Dash, P.K. A method for assessing tissue respiration in anatomically defined brain regions. Sci. Rep. 2020, 10, 13179. [Google Scholar] [CrossRef] [PubMed]
- Schniertshauer, D.; Gebhard, D.; Bergemann, J. A new efficient method for measuring oxygen consumption rate directly ex vivo in human epidermal biopsies. Bio Protoc. 2019, 9, e3185. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Ojo, D.; Yan, J.; Tang, D. Pkm2 contributes to cancer metabolism. Cancer Lett. 2015, 356, 184–191. [Google Scholar] [CrossRef]
- Terry, A.R.; Hay, N. Fuelling cancer cells. Nat. Rev. Endocrinol. 2019, 15, 71–72. [Google Scholar] [CrossRef]
| Symbol i | Description | Locus | Pub ii | Function in BC iii | Ref |
|---|---|---|---|---|---|
| FAM25A vi | Family with sequence similarity 25 member A | 10q23.2 | 0 | Unknown | |
| CST6 | Cystatin E/M | 11q13.1 | 37 | Tumor suppressor activities | [45] |
| ALDH3B2 vi | Aldehyde dehydrogenase 3 family member B2 | 11q13.2 | 3 | A component gene in a multigene biomarker of breast cancer | [56] |
| TRIM29 vi | Tripartite motif containing 29 | 11q23.3 | 13 | A tumor suppressor of BC | [46] |
| KRT79 vi | Keratin 79 | 12q13.13 | 0 | Unknown | |
| RDH16 vi | Retinol dehydrogenase 16 | 12q13.3 | 1 | A component gene in a multigene biomarker of Her2+ breast cancer | [57] |
| FA2H vi | Fatty acid 2-hydroxylase | 16q23.1 | 9 | Downregulation in TN BC; tumor-suppressive actions in BC | [58] |
| KRT10 | Keratin 10 | 17q21.2 | 6 | expressed in breast cancer cells | [59] |
| KRT15 | Keratin 15 | 17q21.2 | 12 | Association with good prognosis in BC | [48] |
| KRT16 vi | Keratin 16 | 17q21.2 | 7 | Association with poor prognosis in BC | [60] |
| SERPINB3 | Serpin family B member 3 | 18q21.33 | 3 | Facilitating BC | [61] |
| CD209 iv | CD209 molecule | 19p13.2 | 8 | In a multigene biomarker of BC | [62] |
| SBSN vi | Suprabasin | 19q13.12 | 0 | Unknown | |
| CYP2F1 vi | Cytochrome P450 family 2 subfamily F member 1 | 19q13.2 | 2 | Its expression is potentially affected by estrogen in MCF-10F cells | [63] |
| KLK7 vi | Kallikrein-related peptidase 7 | 19q13.41 | 19 | Favorable prognosis in TNBC | [49] |
| HRNR vi | Hornerin | 1q21.3 | 1 | Reductions in high T stage, LN metastasis, and BC cells with metastatic capacity | [64] |
| CRCT1 | Cysteine rich C-terminal 1 | 1q21.3 | 0 | Unknown | |
| KPRP vi | Keratinocyte proline-rich protein | 1q21.3 | 1 | Mutations of KPRP in TNBCs with pathological response to neoadjuvant trials | [65] |
| FLG2 vi | Filaggrin 2 | 1q21.3 | 2 | Mutations in 4/12 inflammatory BC | [66] |
| ASPRV1 vi | Aspartic peptidase retroviral like 1 | 2p13.3 | 0 | Unknown | |
| ATP6V1C2 vi | ATPase H+ transporting V1 subunit C2 | 2p25.1 | 1 | Facilitating BC | [67] |
| IL36RN vi | Interleukin 36 receptor antagonist | 2q14.1 | 0 | Unknown | |
| CSTA vi | Cystatin A | 3q21.1 | 14 | An ER target gene and a suppressor of BC | [47] |
| AADAC vi | Arylacetamide deacetylase | 3q25.1 | 0 | Unknown | |
| MME vi | Membrane metalloendopeptidase | 3q25.2 | 27 | High expression in dormant BC cell residing in the bone and lung; BCs with high MME expression display better OS. | [50,51] |
| PSAPL1 iv | Prosaposin-like 1 | 4p16.1 | 1 | In a multigene biomarker of BC | [68] |
| TMPRSS11E v,vi | Transmembrane serine protease 11E | 4q13.2 | 0 | Unknown | |
| CXCL13 vi | C-X-C motif chemokine ligand 13 | 4q21.1 | 75 | Facilitating immune response; association with favorable OS | [52,53] |
| LY6G6C vi | Lymphocyte antigen 6 family member G6C | 6p21.33 | 0 | Unknown | |
| FABP4 | Fatty acid binding protein 4 | 8q21.13 | 55 | Facilitating BC progression | [54] |
| AQP3 vi | Aquaporin 3 (Gill blood group) | 9p13.3 | 30 | Association with poor OS in TNBC | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Dong, Y.; Gu, Y.; Kapoor, A.; Peng, J.; Su, Y.; Wei, F.; Wang, Y.; Yang, C.; Gill, A.; et al. Taxifolin Inhibits Breast Cancer Growth by Facilitating CD8+ T Cell Infiltration and Inducing a Novel Set of Genes including Potential Tumor Suppressor Genes in 1q21.3. Cancers 2023, 15, 3203. https://doi.org/10.3390/cancers15123203
Lin X, Dong Y, Gu Y, Kapoor A, Peng J, Su Y, Wei F, Wang Y, Yang C, Gill A, et al. Taxifolin Inhibits Breast Cancer Growth by Facilitating CD8+ T Cell Infiltration and Inducing a Novel Set of Genes including Potential Tumor Suppressor Genes in 1q21.3. Cancers. 2023; 15(12):3203. https://doi.org/10.3390/cancers15123203
Chicago/Turabian StyleLin, Xiaozeng, Ying Dong, Yan Gu, Anil Kapoor, Jingyi Peng, Yingying Su, Fengxiang Wei, Yanjun Wang, Chengzhi Yang, Armaan Gill, and et al. 2023. "Taxifolin Inhibits Breast Cancer Growth by Facilitating CD8+ T Cell Infiltration and Inducing a Novel Set of Genes including Potential Tumor Suppressor Genes in 1q21.3" Cancers 15, no. 12: 3203. https://doi.org/10.3390/cancers15123203
APA StyleLin, X., Dong, Y., Gu, Y., Kapoor, A., Peng, J., Su, Y., Wei, F., Wang, Y., Yang, C., Gill, A., Neira, S. V., & Tang, D. (2023). Taxifolin Inhibits Breast Cancer Growth by Facilitating CD8+ T Cell Infiltration and Inducing a Novel Set of Genes including Potential Tumor Suppressor Genes in 1q21.3. Cancers, 15(12), 3203. https://doi.org/10.3390/cancers15123203







