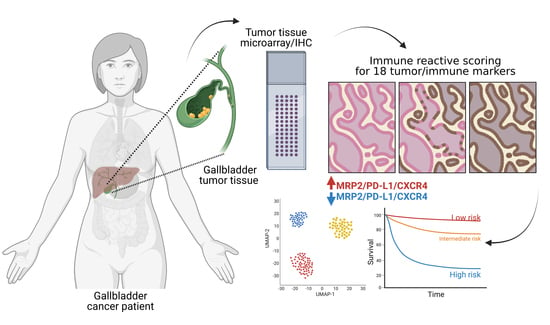
Co-Expression of Immunohistochemical Markers MRP2, CXCR4, and PD-L1 in Gallbladder Tumors Is Associated with Prolonged Patient Survival
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Biopsies
2.2. Tissue Microarrays
2.3. Immunohistochemistry
2.4. Antibodies
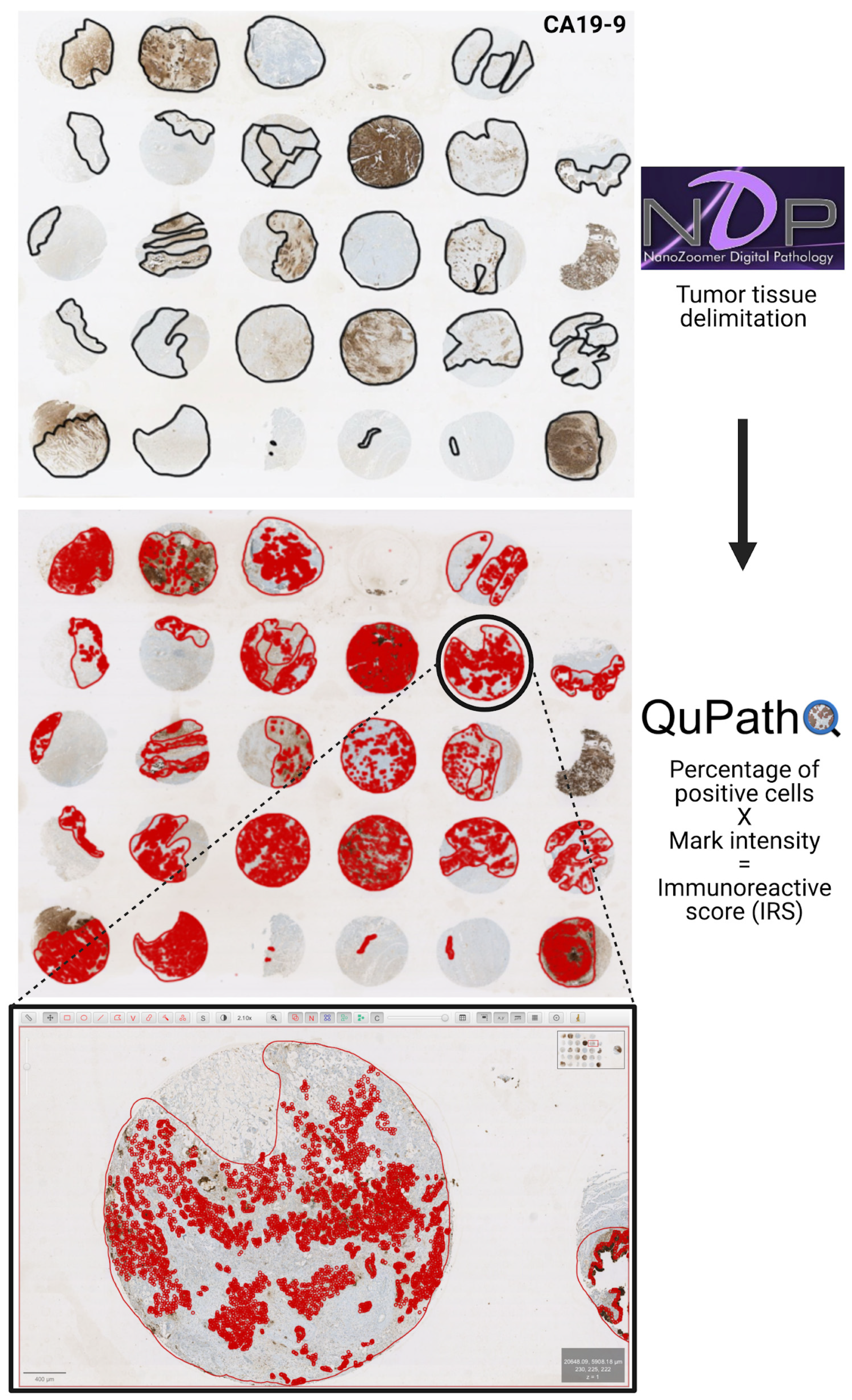
2.5. Determination of Immunoreactive Scores
2.6. Statistical Analysis
3. Results
3.1. Clinicopathological Characteristics of the Gallbladder Cancer Patient Cohort
3.2. Immunohistochemical Evaluation and Immunoreactive Score Analysis of Multiple Markers in Gallbladder Cancer Tissues
3.3. Association of Tumor Markers with Gallbladder Cancer Patient Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roa, I.; Ibacache, M.G.; Muñoz, S.; de Aretxabala, X. Gallbladder cancer in Chile: Pathologic characteristics of survival and prognostic factors: Analysis of 1366 cases. Am. J. Clin. Pathol. 2014, 141, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Randi, G.; Franceschi, S.; La Vecchia, C. Gallbladder cancer worldwide: Geographical distribution and risk factors. Int. J. Cancer 2006, 118, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Patel, H.K.; Boakye, D.; Chandrasekar, V.T.; Koulaouzidis, A.; Iii, D.E.L.-P.; Ngai, C.H.; Pun, C.N.; Bai, Y.; Lok, V.; et al. Worldwide distribution, associated factors, and trends of gallbladder cancer: A global country-level analysis. Cancer Lett. 2021, 521, 238–251. [Google Scholar] [CrossRef]
- Iyer, R.V.; Pokuri, V.K.; Groman, A.; Ma, W.W.; Malhotra, U.; Iancu, D.M.; Grande, C.; Saab, T.B. A Multicenter Phase II Study of Gemcitabine, Capecitabine, and Bevacizumab for Locally Advanced or Metastatic Biliary Tract Cancer. Am. J. Clin. Oncol. 2018, 41, 649–655. [Google Scholar] [CrossRef]
- Lai, J.; Yang, S.; Lin, Z.; Huang, W.; Li, X.; Li, R.; Tan, J.; Wang, W. Update on Chemoresistance Mechanisms to First-Line Chemotherapy for Gallbladder Cancer and Potential Reversal Strategies. Am. J. Clin. Oncol. 2023, 46, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Jansen, H.; Pape, U.-F.; Utku, N. A review of systemic therapy in biliary tract carcinoma. J. Gastrointest. Oncol. 2020, 11, 770–789. [Google Scholar] [CrossRef] [PubMed]
- Nimisha; Saluja, S.S.; Sharma, A.K.; Nekarakanti, P.K.; Apurva; Kumar, A.; Sattar, R.S.A.; Anjum, H.; Batra, V.V.; Husain, S.A. Molecular aspects of ABCB1 and ABCG2 in Gallbladder cancer and its clinical relevance. Mol. Cell. Biochem. 2023, 1–16. [Google Scholar] [CrossRef]
- De Lorenzo, S.; Garajova, I.; Stefanini, B.; Tovoli, F. Targeted therapies for gallbladder cancer: An overview of agents in preclinical and clinical development. Expert Opin. Investig. Drugs 2021, 30, 759–772. [Google Scholar] [CrossRef]
- Javle, M.; Rashid, A.; Churi, C.; Kar, S.P.; Zuo, M.; Eterovic, A.K.; Nogueras-Gonzalez, G.M.; Janku, F.; Shroff, R.T.; Aloia, T.A.; et al. Molecular characterization of gallbladder cancer using somatic mutation profiling. Hum. Pathol. 2014, 45, 701–708. [Google Scholar] [CrossRef]
- Valle, J.W.; Lamarca, A.; Goyal, L.; Barriuso, J.; Zhu, A.X. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017, 7, 943–962. [Google Scholar] [CrossRef]
- D’afonseca, V.; Arencibia, A.D.; Echeverría-Vega, A.; Cerpa, L.; Cayún, J.P.; Varela, N.M.; Salazar, M.; Quiñones, L.A. Identification of Altered Genes in Gallbladder Cancer as Potential Driver Mutations for Diagnostic and Prognostic Purposes: A Computational Approach. Cancer Inform. 2020, 19. [Google Scholar] [CrossRef]
- Arkenau, H.-T.; Martin-Liberal, J.; Calvo, E.; Penel, N.; Krebs, M.G.; Herbst, R.S.; Walgren, R.A.; Widau, R.C.; Mi, G.; Jin, J.; et al. Ramucirumab Plus Pembrolizumab in Patients with Previously Treated Advanced or Metastatic Biliary Tract Cancer: Nonrandomized, Open-Label, Phase I Trial (JVDF). Oncologist 2018, 23, 1407-e136. [Google Scholar] [CrossRef] [PubMed]
- Ueno, M.; Ikeda, M.; Morizane, C.; Kobayashi, S.; Ohno, I.; Kondo, S.; Okano, N.; Kimura, K.; Asada, S.; Namba, Y.; et al. Nivolumab alone or in combination with cisplatin plus gemcitabine in Japanese patients with unresectable or recurrent biliary tract cancer: A non-randomised, multicentre, open-label, phase 1 study. Lancet Gastroenterol. Hepatol. 2019, 4, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.D.; Chung, V.; Alese, O.B.; El-Rayes, B.F.; Li, D.; Al-Toubah, T.E.; Schell, M.J.; Zhou, J.-M.; Mahipal, A.; Kim, B.H.; et al. A Phase 2 Multi-institutional Study of Nivolumab for Patients With Advanced Refractory Biliary Tract Cancer. JAMA Oncol. 2020, 6, 888–894. [Google Scholar] [CrossRef]
- Boilève, A.; Hilmi, M.; Smolenschi, C.; Ducreux, M.; Hollebecque, A.; Malka, D. Immunotherapy in Advanced Biliary Tract Cancers. Cancers 2021, 13, 1569. [Google Scholar] [CrossRef] [PubMed]
- Lepisto, A.J.; Moser, A.J.; Zeh, H.; Lee, K.; Bartlett, D.; McKolanis, J.R.; Geller, B.A.; Schmotzer, A.; Potter, D.P.; Whiteside, T.; et al. A phase I/II study of a MUC1 peptide pulsed autologous dendritic cell vaccine as adjuvant therapy in patients with resected pancreatic and biliary tumors. Cancer Ther. 2008, 6, 955–964. [Google Scholar]
- Kobayashi, M.; Sakabe, T.; Abe, H.; Tanii, M.; Takahashi, H.; Chiba, A.; Yanagida, E.; Shibamoto, Y.; Ogasawara, M.; Tsujitani, S.; et al. Dendritic Cell-Based Immunotherapy Targeting Synthesized Peptides for Advanced Biliary Tract Cancer. J. Gastrointest. Surg. 2013, 17, 1609–1617. [Google Scholar] [CrossRef]
- Rojas-Sepúlveda, D.; Tittarelli, A.; Gleisner, M.A.; Ávalos, I.; Pereda, C.; Gallegos, I.; González, F.E.; López, M.N.; Butte, J.M.; Roa, J.C.; et al. Tumor lysate-based vaccines: On the road to immunotherapy for gallbladder cancer. Cancer Immunol. Immunother. 2018, 67, 1897–1910. [Google Scholar] [CrossRef]
- Schmidt, M.A.; Marcano-Bonilla, L.; Roberts, L.R. Gallbladder cancer: Epidemiology and genetic risk associations. Chin. Clin. Oncol. 2019, 8, 31. [Google Scholar] [CrossRef]
- Rustagi, T.; Dasanu, C.A. Risk Factors for Gallbladder Cancer and Cholangiocarcinoma: Similarities, Differences and Updates. J. Gastrointest. Cancer 2012, 43, 137–147. [Google Scholar] [CrossRef]
- Roa, J.C.; García, P.; Kapoor, V.K.; Maithel, S.K.; Javle, M.; Koshiol, J. Gallbladder cancer. Nat. Rev. Dis. Prim. 2022, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Srivastava, A.; Mittal, B. Potential biomarkers in gallbladder cancer: Present status and future directions. Biomarkers 2013, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Neyaz, A.; Husain, N.; Gupta, S.; Kumari, S.; Arora, A.; Awasthi, N.P.; Malhotra, K.P.; Misra, S. Investigation of targetable predictive and prognostic markers in gallbladder carcinoma. J. Gastrointest. Oncol. 2018, 9, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.; Tittarelli, A.; Paillaleve, N.; Del Pozo, M.; Rojas-Sepúlveda, D.; Barría, O.; Fluxá, P.; Hott, M.; Martin, C.; Quezada, C.; et al. The Evaluation of 17 Gastrointestinal Tumor Markers Reveals Prognosis Value for MUC6, CK17, and CD10 in Gallbladder-Cancer Patients. Diagnostics 2021, 11, 153. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: New York, NY, USA; American Joint Commission on Cancer: Chicago, IL, USA, 2017. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Peng, P.-J.; Li, Y.; Sun, S. On the significance of Tim-3 expression in pancreatic cancer. Saudi J. Biol. Sci. 2017, 24, 1754–1757. [Google Scholar] [CrossRef]
- Neyaz, A.; Husain, N.; Kumari, S.; Gupta, S.; Shukla, S.; Arshad, S.; Anand, N.; Chaturvedi, A. Clinical relevance of PD-L1 expression in gallbladder cancer: A potential target for therapy. Histopathology 2018, 73, 622–633. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, L.; Shi, L.; Song, H.; Shi, P.; Yang, T.; Fan, R.; Jiang, T.; Song, J. Prognostic Impact of Adenosine Receptor 2 (A2aR) and Programmed Cell Death Ligand 1 (PD-L1) Expression in Colorectal Cancer. BioMed Res. Int. 2019, 2019, 8014627. [Google Scholar] [CrossRef]
- Dablain, D.; Krawczyk, B.; Chawla, N.V. DeepSMOTE: Fusing Deep Learning and SMOTE for Imbalanced Data. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–15. [Google Scholar] [CrossRef]
- Abraham, A.; Pedregosa, F.; Eickenberg, M.; Gervais, P.; Mueller, A.; Kossaifi, J.; Gramfort, A.; Thirion, B.; Varoquaux, G. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 2014, 8, 14. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 4765–4774. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Fan, J.; Upadhye, S.; Worster, A. Understanding receiver operating characteristic (ROC) curves. Can. J. Emerg. Med. 2006, 8, 19–20. [Google Scholar] [CrossRef]
- Nohara, Y.; Matsumoto, K.; Soejima, H.; Nakashima, N. Explanation of machine learning models using shapley additive explanation and application for real data in hospital. Comput. Methods Programs Biomed. 2022, 214, 106584. [Google Scholar] [CrossRef]
- Li, M.; Liu, F.; Zhang, F.; Zhou, W.; Jiang, X.; Yang, Y.; Qu, K.; Wang, Y.; Ma, Q.; Wang, T.; et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: A whole-exome sequencing analysis. Gut 2019, 68, 1024–1033. [Google Scholar] [CrossRef]
- Yang, P.; Javle, M.; Pang, F.; Zhao, W.; Abdel-Wahab, R.; Chen, X.; Meric-Bernstam, F.; Chen, H.; Borad, M.J.; Liu, Y.; et al. Somatic genetic aberrations in gallbladder cancer: Comparison between Chinese and US patients. HepatoBiliary Surg. Nutr. 2019, 8, 604–614. [Google Scholar] [CrossRef]
- Roa, I.; de Toro, G.; Schalper, K.; de Aretxabala, X.; Churi, C.; Javle, M. Overexpression of the HER2/neu Gene: A New Therapeutic Possibility for Patients with Advanced Gallbladder Cancer. Gastrointest. Cancer Res. 2014, 7, 42–48. [Google Scholar]
- Fluxá, P.; Rojas-Sepúlveda, D.; Gleisner, M.A.; Tittarelli, A.; Villegas, P.; Tapia, L.; Rivera, M.T.; López, M.N.; Catán, F.; Uribe, M.; et al. High CD8+ and absence of Foxp3+ T lymphocytes infiltration in gallbladder tumors correlate with prolonged patients survival. BMC Cancer 2018, 18, 243. [Google Scholar] [CrossRef]
- Koshiol, J.; Gao, Y.-T.; Corbel, A.; Kemp, T.J.; Shen, M.-C.; Hildesheim, A.; Hsing, A.W.; Rashid, A.; Wang, B.; Pfeiffer, R.M.; et al. Circulating inflammatory proteins and gallbladder cancer: Potential for risk stratification to improve prioritization for cholecystectomy in high-risk regions. Cancer Epidemiol. 2018, 54, 25–30. [Google Scholar] [CrossRef]
- Liu, Z.; Kemp, T.J.; Gao, Y.-T.; Corbel, A.; McGee, E.E.; Roa, J.C.; Wang, B.; Araya, J.C.; Shen, M.-C.; Rashid, A.; et al. Circulating Levels of Inflammatory Proteins and Survival in Patients with Gallbladder Cancer. Sci. Rep. 2018, 8, 5671. [Google Scholar] [CrossRef]
- Liu, Z.; Kemp, T.J.; Gao, Y.-T.; Corbel, A.; McGee, E.E.; Wang, B.; Shen, M.-C.; Rashid, A.; Hsing, A.W.; Hildesheim, A.; et al. Association of circulating inflammation proteins and gallstone disease. J. Gastroenterol. Hepatol. 2018, 33, 1920–1924. [Google Scholar] [CrossRef]
- Gold, P.; Freedman, S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965, 121, 439–462. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef] [PubMed]
- Michl, M.; Stintzing, S.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Vehling-Kaiser, U.; Al-Batran, S.-E.; Heintges, T.; Lerchenmueller, C.; Kahl, C.; et al. CEA response is associated with tumor response and survival in patients with KRAS exon 2 wild-type and extended RAS wild-type metastatic colorectal cancer receiving first-line FOLFIRI plus cetuximab or bevacizumab (FIRE-3 trial). Ann. Oncol. 2016, 27, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Reissfelder, C.; Stamova, S.; Gossmann, C.; Braun, M.; Bonertz, A.; Walliczek, U.; Grimm, M.; Rahbari, N.N.; Koch, M.; Saadati, M.; et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J. Clin. Investig. 2015, 125, 739–751. [Google Scholar] [CrossRef]
- Kim, H.-S.; Kim, N.C.; Chae, K.H.; Kim, G.; Park, W.S.; Park, Y.-K.; Kim, Y.W. Expression of Multidrug Resistance-Associated Protein 2 in Human Gallbladder Carcinoma. BioMed Res. Int. 2013, 2013, 527534. [Google Scholar] [CrossRef] [PubMed]
- Trauner, M.; Boyer, J.L.; Cai, J.; Wang, Z.; Chen, G.; Li, D.; Liu, J.; Hu, H.; Qin, J.; Fiamoncini, J.; et al. Bile Salt Transporters: Molecular Characterization, Function, and Regulation. Physiol. Rev. 2003, 83, 633–671. [Google Scholar] [CrossRef]
- Cirqueira, C.S.; Felipe-Silva, A.S.; Wakamatsu, A.; Marins, L.V.; Rocha, E.C.; de Mello, E.S.; Alves, V.A.F. Immunohistochemical Assessment of the Expression of Biliary Transportation Proteins MRP2 and MRP3 in Hepatocellular Carcinoma and in Cholangiocarcinoma. Pathol. Oncol. Res. 2019, 25, 1363–1371. [Google Scholar] [CrossRef]
- Nengroo, M.A.; Khan, M.A.; Verma, A.; Datta, D. Demystifying the CXCR4 conundrum in cancer biology: Beyond the surface signaling paradigm. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2022, 1877, 188790. [Google Scholar] [CrossRef]
- Yao, X.; Zhou, L.; Han, S.; Chen, Y. High Expression of CXCR4 and CXCR7 Predicts Poor Survival in Gallbladder Cancer. J. Int. Med. Res. 2011, 39, 1253–1264. [Google Scholar] [CrossRef]
- Bao, L.; Lai, Y.; Liu, Y.; Qin, Y.; Zhao, X.; Lu, X.; Jiang, Q.; Lu, J.; Huang, X. CXCR4 is a good survival prognostic indicator in multiple myeloma patients. Leuk. Res. 2013, 37, 1083–1088. [Google Scholar] [CrossRef]
- Liu, F.; Lang, R.; Wei, J.; Fan, Y.; Cui, L.; Gu, F.; Guo, X.; Pringle, G.A.; Zhang, X.; Fu, L. Increased expression of SDF-1/CXCR4 is associated with lymph node metastasis of invasive micropapillary carcinoma of the breast. Histopathology 2009, 54, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-C.; Wu, M.-T.; Huang, C.-J.; Hou, M.-F.; Yang, S.-F.; Chai, C.-Y. Expression of CXCR4 is associated with axillary lymph node status in patients with early breast cancer. Breast 2006, 15, 533–539. [Google Scholar] [CrossRef]
- Wagner, P.L.; Hyjek, E.; Vazquez, M.F.; Meherally, D.; Liu, Y.F.; Chadwick, P.A.; Rengifo, T.; Sica, G.L.; Port, J.L.; Lee, P.C.; et al. CXCL12 and CXCR4 in adenocarcinoma of the lung: Association with metastasis and survival. J. Thorac. Cardiovasc. Surg. 2009, 137, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.A.; Tran, T.; Vilain, R.E.; Madore, J.; Selinger, C.I.; Kohonen-Corish, M.; Yip, P.; Yu, B.; O’toole, S.A.; McCaughan, B.C.; et al. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer 2015, 89, 181–188. [Google Scholar] [CrossRef]
- Schmidt, L.H.; Kümmel, A.; Görlich, D.; Mohr, M.; Bröckling, S.; Mikesch, J.-H.; Grünewald, I.; Marra, A.; Schultheis, A.; Wardelmann, E.; et al. PD-1 and PD-L1 Expression in NSCLC Indicate a Favorable Prognosis in Defined Subgroups. PLoS ONE 2015, 10, e0136023. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.R.; Milne, K.; Kroeger, D.R.; Nelson, B.H. PD-L1 expression is associated with tumor-infiltrating T cells and favorable prognosis in high-grade serous ovarian cancer. Gynecol. Oncol. 2016, 141, 293–302. [Google Scholar] [CrossRef]
- Jung, H.I.; Jeong, D.; Ji, S.; Ahn, T.S.; Bae, S.H.; Chin, S.; Chung, J.C.; Kim, H.C.; Lee, M.S.; Baek, M.-J. Overexpression of PD-L1 and PD-L2 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res. Treat. 2017, 49, 246–254. [Google Scholar] [CrossRef]
- Qin, T.; Zeng, Y.-D.; Qin, G.; Xu, F.; Lu, J.-B.; Fang, W.-F.; Xue, C.; Zhan, J.-H.; Zhang, X.-K.; Zheng, Q.-F.; et al. High PD-L1 expression was associated with poor prognosis in 870 Chinese patients with breast cancer. Oncotarget 2015, 6, 33972–33981. [Google Scholar] [CrossRef]
- Lin, Y.-M.; Sung, W.-W.; Hsieh, M.-J.; Tsai, S.-C.; Lai, H.-W.; Yang, S.-M.; Shen, K.-H.; Chen, M.-K.; Lee, H.; Yeh, K.-T.; et al. High PD-L1 Expression Correlates with Metastasis and Poor Prognosis in Oral Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0142656. [Google Scholar] [CrossRef]



| Gender | Female | 79.2% (n = 191) |
| Male | 20.8% (n = 50) | |
| Mean age (SD) | All | 65.1 (±11.2) |
| Female | 64.4 (±11.4) | |
| Male | 67.6 (±10.4) | |
| Tumor differentiation | Poor | 30.8% (n = 65) |
| Median OS time: 6 months | ||
| Five-year survival rate: 3.1% | ||
| Moderate | 56.9% (n = 120) | |
| Median OS time: 16.5 months | ||
| Five-year survival rate: 20% | ||
| Well | 12.3% (n = 26) | |
| Median OS time: 178.5 months | ||
| Five-year survival rate: 46.1% | ||
| Tumor staging | Early stages (TIS + T1) | 46 patients (22 TIS + 24 T1) |
| 41 female, 5 male | ||
| Mean age: 61.6 (±11.4) | ||
| Median OS time: 220 months | ||
| Five-year survival rate: 52.2% | ||
| Late stages (T2 + T3 + T4) | 195 patients (52 T2 + 137 T3 + 6 T4) | |
| 150 female, 45 male | ||
| Mean age: 65.9 (±11) | ||
| Median OS time: 10 months | ||
| Five-year survival rate: 11.8% |
| Marker | Number (%) of Positive Cases, Distribution Using IRS | |
|---|---|---|
| Early Stage Tumors (TIS + T1) | Late-Stage Tumors (T2–T4) | |
| Epithelial tumor markers | ||
| MUC1 | 38 (100%): 20 Low + 18 High | 171 (98.8%): 60 Low + 111 High |
| CEA | 26 (83.9%): 18 Low + 8 High | 128 (85.3%): 54 Low + 74 High |
| CA19-9 | 31 (86.1%): 11 Low + 20 High | 145 (83.8%): 49 Low + 96 High |
| Multidrug or apoptosis resistance | ||
| MRP2 | 29 (93.5%): 17 Low + 12 High | 159 (98.1%): 124 Low + 35 High |
| MRP3 | 30 (100%): 16 Low + 14 High | 161 (99.4%): 124 Low + 37 High |
| Pg-P | 28 (100%): 17 Low + 11 High | 159 (99.4%): 123 Low + 36 High |
| Survivin | 36 (97.3%) | 145 (92.9%) |
| Immunological markers | ||
| TIM3 | 24 (76%): 11 Low + 13 High | 130 (78.8%): 101 Low + 29 High |
| PD-L1 | 37 (88.1%): 12 Low + 25 High | 148 (82.7%): 82 Low + 66 High |
| A2aR | 39 (88.6%): 27 Low + 12 High | 145 (79.7%): 114 Low + 31 High |
| HLA-ABC | 42 (97.7%): 15 Low + 27 High | 180 (98.4%): 71 Low + 109 High |
| IFN-γ | 39 (97.5%): 34 Low + 5 High | 166 (94.8%): 161 Low + 5 High |
| IL-8 | 34 (80.9%): 26 Low + 8 High | 152 (84.9%): 119 Low + 33 High |
| CXCR4 | 39 (95.1%): 10 Low + 29 High | 153 (85.9%): 63 Low + 90 High |
| TNF-α | 32 (76.2%): 18 Low + 14 High | 143 (81.2%): 115 Low + 28 High |
| CCR7 | 17 (65.4%): 9 Low + 8 High | 105 (66.4%): 59 Low + 46 High |
| TGF-β | 39 (97.5%): 6 Low + 33 High | 169 (97.1%): 56 Low + 113 High |
| IL-6 | 35 (100%): 14 Low + 21 High | 165 (97.6%): 120 Low + 45 High |
| Complete Cohort | MRP2/CXCR4/PD-L1 Co-Expression Pattern | p-Value | |
|---|---|---|---|
| High (n = 15) | Neg/Low (n = 42) | ||
| Mean age (SD) | 66.3 (±7.9) | 64.7 (±11.3) | 0.62 a |
| Gender (F/M) | 12/3 | 54/8 | 0.48 b |
| Tumor differentiation | |||
| Poor | 15.4% (n = 2) | 39% (n = 16) | 0.11 b |
| Moderate | 61.5% (n = 8) | 48.8% (n = 20) | |
| Well | 23.1% (n = 3) | 12.2% (n = 5) | |
| Primary tumor staging (TNM) | |||
| Early stages (TIS + T1) | 33.3% (n = 5) | 4.8% (n = 2) | 0.0038 b |
| Late stages (T2–T4) | 66.7% (n = 10) | 95.2% (n = 40) | |
| Cancer disease stage | |||
| I | 33.3% (n = 5) | 4.8% (n = 2) | 0.0627 b |
| II | 26.7% (n = 4) | 19% (n = 8) | |
| III | 26. 7% (n = 4) | 64.3% (n = 27) | |
| IV | 13.3% (n = 2) | 11.9% (n = 5) | |
| R status | |||
| R0 | 66.7% (n = 10) | 45.2% (n = 19) | 0.38 b |
| R1 | 26.7% (n = 4) | 40.5% (n = 17) | |
| R2 | 6.6% (n = 1) | 14.3% (n = 6) | |
| Late-stage (T2–T4) tumors | High (n = 10) | Neg/Low (n = 40) | |
| Mean age (SD) | 64.3 (±8) | 65.1 (±11.2) | 0.83 a |
| Gender (F/M) | 8/2 | 54/8 | 0.55 b |
| Tumor differentiation | |||
| Poor | 20% (n = 2) | 39% (n = 16) | 0.26 b |
| Moderate | 60% (n = 6) | 48.8% (n = 20) | |
| Well | 20% (n = 2) | 12.2% (n = 5) | |
| Cancer disease stage | |||
| I | 0% (n = 0) | 0% (n = 0) | 0.36 b |
| II | 40% (n = 4) | 20% (n = 8) | |
| III | 40% (n = 4) | 67.5% (n = 27) | |
| IV | 20% (n = 2) | 12.5% (n = 5) | |
| R status | |||
| R0 | 50% (n = 5) | 42.5% (n = 17) | 0.77 b |
| R1 | 40% (n = 4) | 42.5% (n = 17) | |
| R2 | 10% (n = 1) | 15% (n = 6) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tittarelli, A.; Barría, O.; Sanders, E.; Bergqvist, A.; Brange, D.U.; Vidal, M.; Gleisner, M.A.; Vergara, J.R.; Niechi, I.; Flores, I.; et al. Co-Expression of Immunohistochemical Markers MRP2, CXCR4, and PD-L1 in Gallbladder Tumors Is Associated with Prolonged Patient Survival. Cancers 2023, 15, 3440. https://doi.org/10.3390/cancers15133440
Tittarelli A, Barría O, Sanders E, Bergqvist A, Brange DU, Vidal M, Gleisner MA, Vergara JR, Niechi I, Flores I, et al. Co-Expression of Immunohistochemical Markers MRP2, CXCR4, and PD-L1 in Gallbladder Tumors Is Associated with Prolonged Patient Survival. Cancers. 2023; 15(13):3440. https://doi.org/10.3390/cancers15133440
Chicago/Turabian StyleTittarelli, Andrés, Omar Barría, Evy Sanders, Anna Bergqvist, Daniel Uribe Brange, Mabel Vidal, María Alejandra Gleisner, Jorge Ramón Vergara, Ignacio Niechi, Iván Flores, and et al. 2023. "Co-Expression of Immunohistochemical Markers MRP2, CXCR4, and PD-L1 in Gallbladder Tumors Is Associated with Prolonged Patient Survival" Cancers 15, no. 13: 3440. https://doi.org/10.3390/cancers15133440
APA StyleTittarelli, A., Barría, O., Sanders, E., Bergqvist, A., Brange, D. U., Vidal, M., Gleisner, M. A., Vergara, J. R., Niechi, I., Flores, I., Pereda, C., Carrasco, C., Quezada-Monrás, C., & Salazar-Onfray, F. (2023). Co-Expression of Immunohistochemical Markers MRP2, CXCR4, and PD-L1 in Gallbladder Tumors Is Associated with Prolonged Patient Survival. Cancers, 15(13), 3440. https://doi.org/10.3390/cancers15133440