SHARPIN Enhances Ferroptosis in Synovial Sarcoma Cells via NF-κB- and PRMT5-Mediated PGC1α Reduction
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Studies

2.2. Cell Culture
2.3. Antibodies and Reagents
2.4. Immunoblotting
2.5. Gene Silencing
2.6. RNA Extraction and qPCR
2.7. Cell Viability/Cell Death Assays
2.8. Immunohistochemistry
2.9. Detection of Lipid Peroxidation
2.10. Complex I Activity Assay
2.11. ROS Assay
2.12. GSH/GSSG Ratio Assay
2.13. In-Silico Analysis
2.14. Statistics
3. Results
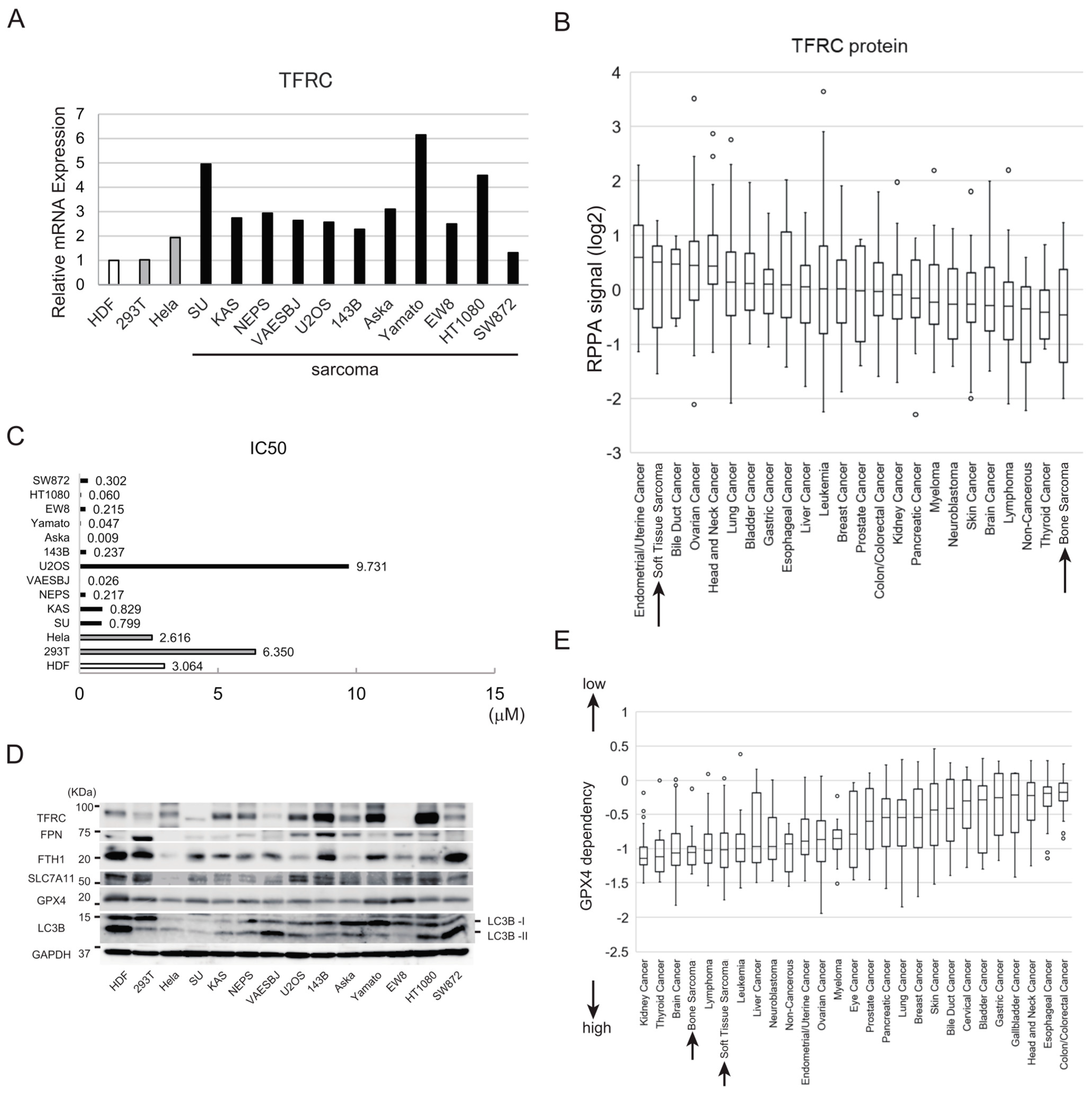
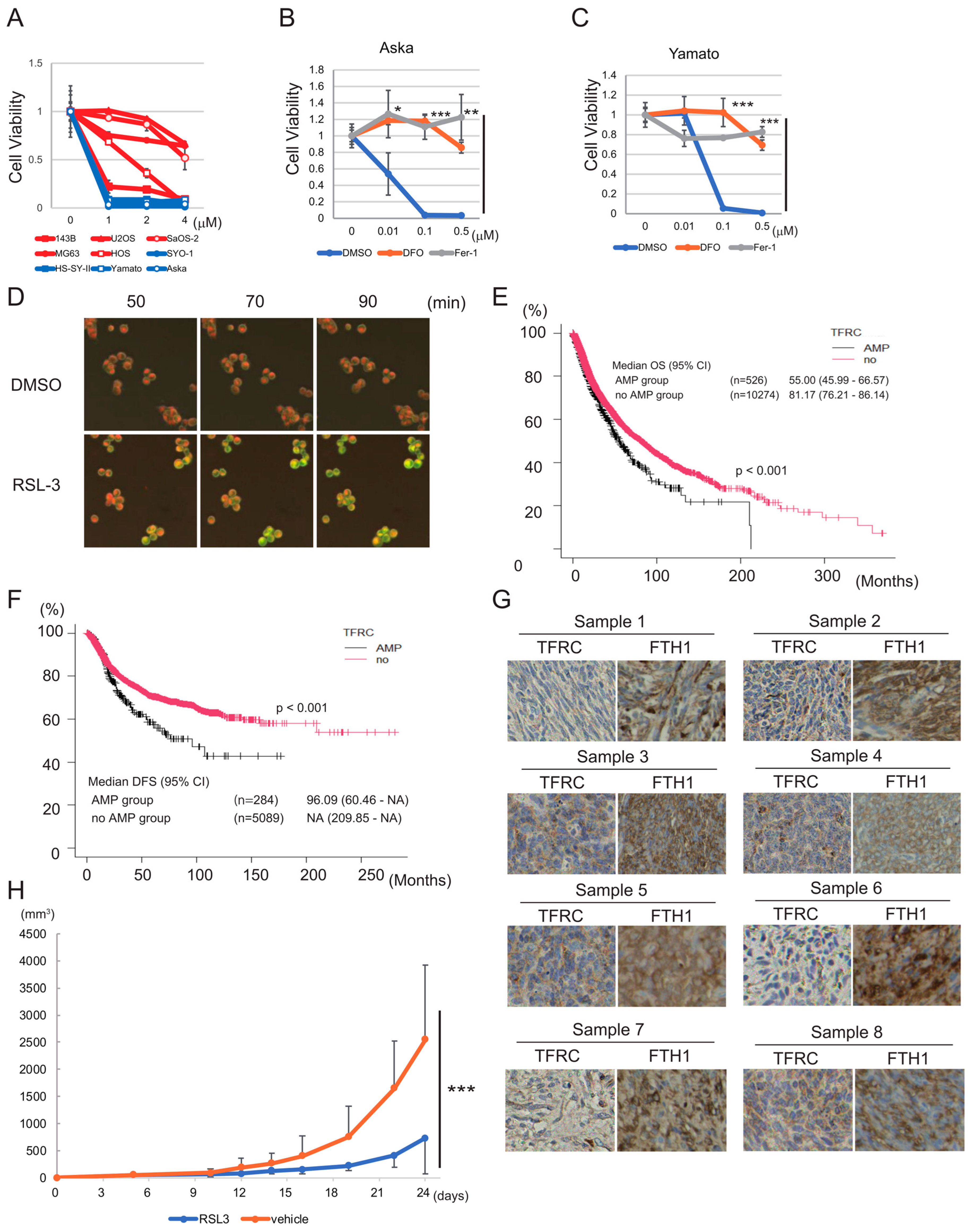
3.1. Sarcoma Cell Lines Are More Sensitive to Ferroptosis than Noncancer or Carcinoma Cell Lines
3.2. Synovial Sarcoma Cells Are Sensitive to Ferroptosis Inducers and Express High Levels of Iron Metabolism-Related Proteins
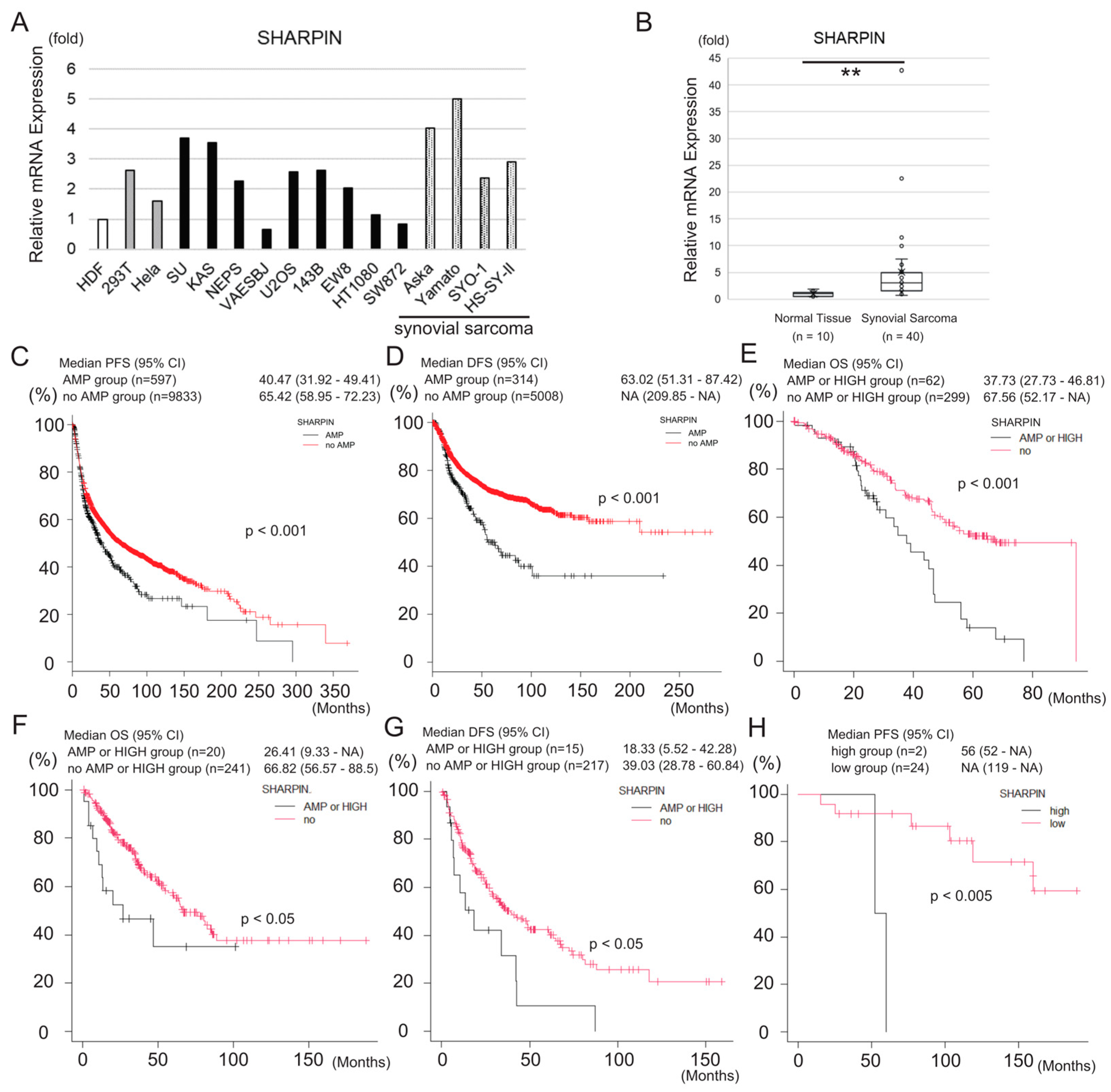
3.3. SHARPIN Is Expressed at a High Level in Synovial Sarcoma and Is Associated with Poor Prognosis of the Disease
3.4. SHARPIN Promotes the Sensitivity of Synovial Sarcoma Cell Lines to Ferroptosis by Inhibiting the PGC1α/SLC7A11 Axis
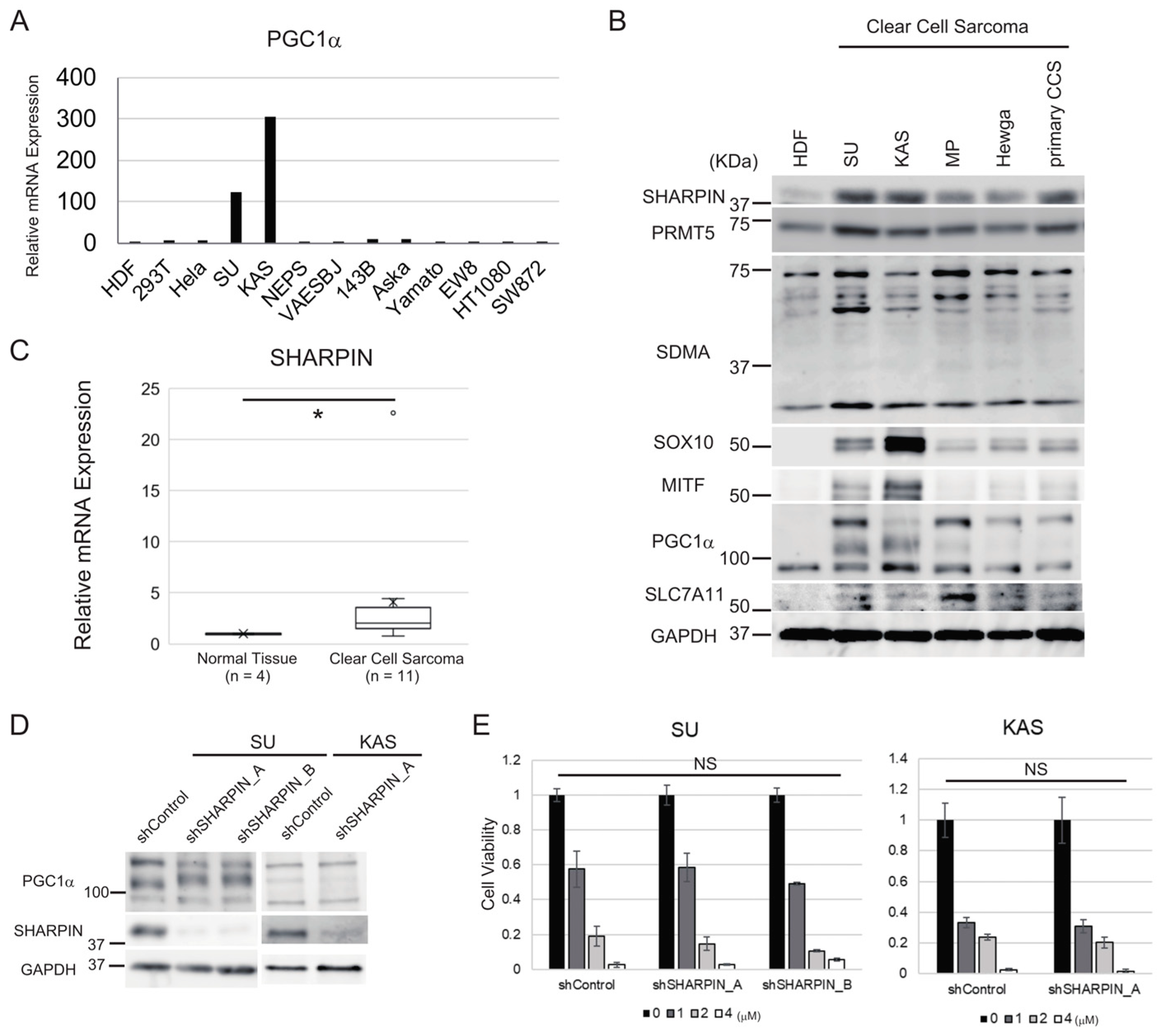
3.5. Clear Cell Sarcoma Cells Expressing High Levels of PGC1α Do Not Respond to SHARPIN-Mediated Regulation of Ferroptosis Sensitivity
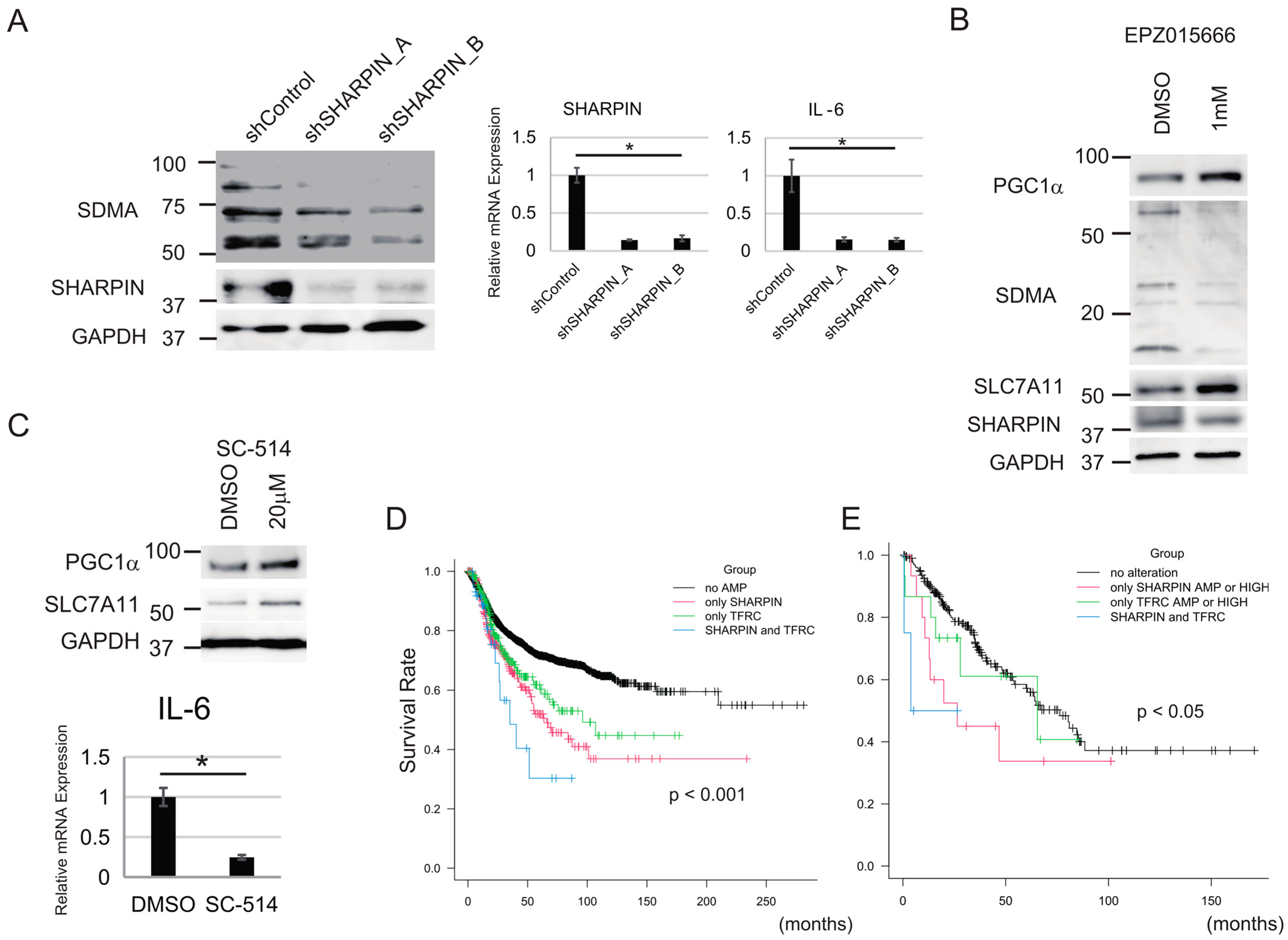
3.6. NF-κΒ and PRMT5 Are Involved in the Regulation of Sensitivity to Ferroptosis by SHARPIN
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jo, V.Y.; Fletcher, C.D. WHO Classification of Soft Tissue Tumours: An Update Based on the 2013 (4th) Edition. Pathology 2014, 46, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Cates, J. The AJCC 8th Edition Staging System for Soft Tissue Sarcoma of the Extremities or Trunk: A Cohort Study of the SEER Database. J. Natl. Compr. Cancer Netw. 2018, 16, 144–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blay, J.-Y.; Schöffski, P.; Bauer, S.; Krarup-Hansen, A.; Benson, C.; D’adamo, D.R.; Jia, Y.; Maki, R.G. Eribulin versus Dacarbazine in Patients with Leiomyosarcoma: Subgroup Analysis from a Phase 3, Open-Label, Randomised Study. Br. J. Cancer 2019, 120, 1026–1032. [Google Scholar] [CrossRef] [Green Version]
- van der Graaf, W.T.; Blay, J.-Y.; Chawla, S.P.; Kim, D.-W.; Bui-Nguyen, B.; Casali, P.G.; Schöffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for Metastatic Soft-Tissue Sarcoma (PALETTE): A Randomised, Double-Blind, Placebo-Controlled Phase 3 Trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Le Cesne, A.; Blay, J.-Y.; Cupissol, D.; Italiano, A.; Delcambre, C.; Penel, N.; Isambert, N.; Chevreau, C.; Bompas, E.; Bertucci, F.; et al. A Randomized Phase III Trial Comparing Trabectedin to Best Supportive Care in Patients with Pre-Treated Soft Tissue Sarcoma: T-SAR, a French Sarcoma Group Trial. Ann. Oncol. 2021, 32, 1034–1044. [Google Scholar] [CrossRef]
- Ng, V.Y.; Scharschmidt, T.J.; Mayerson, J.L.; Fisher, J.L. Incidence and survival in sarcoma in the United States: A focus on musculoskeletal lesions. Anticancer. Res. 2013, 33, 2597–2604. [Google Scholar]
- Nummi, A.; Partanen, T.A.; Setala, L.; Soini, Y.; Berg, L.; Mustonen, P. The Influence of National Guidelines on Soft Tissue Sarcoma Patient Outcome: A Single Center Experience. Plast. Aesthetic Res. 2014, 1, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.L.; Yuan, L.; Li, W.; Li, J.Y. Ferroptosis in Parkinson’s Disease: Glia–Neuron Crosstalk. Trends Mol. Med. 2022, 28, 258–269. [Google Scholar] [CrossRef]
- Han, C.; Liu, Y.; Dai, R.; Ismail, N.; Su, W.; Li, B. Ferroptosis and Its Potential Role in Human Diseases. Front. Pharmacol. 2020, 11, 239. [Google Scholar] [CrossRef]
- Zhang, C. Essential Functions of Iron-Requiring Proteins in DNA Replication, Repair and Cell Cycle Control. Protein Cell 2014, 5, 750–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Qi, H.; Liu, Y.; Duan, C.; Liu, X.; Xia, T.; Chen, D.; Piao, H.-L.; Liu, H.-X. The Double-Edged Roles of ROS in Cancer Prevention and Therapy. Theranostics 2021, 11, 4839–4857. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chloupková, M. Abnormal iron uptake and liver cancer. Cancer Biol. Ther. 2009, 8, 1699–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torti, S.V.; Torti, F.M. Iron and Cancer: More Ore to Be Mined. Nat. Rev. Cancer 2013, 13, 342–355. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Nam, M.; Son, H.Y.; Hyun, K.; Jang, S.Y.; Kim, J.W.; Kim, M.W.; Jung, Y.; Jang, E.; Yoon, S.-J.; et al. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 32433–32442. [Google Scholar] [CrossRef]
- Wang, T.-X.; Liang, J.-Y.; Zhang, C.; Xiong, Y.; Guan, K.-L.; Yuan, H.-X. The Oncometabolite 2-Hydroxyglutarate Produced by Mutant IDH1 Sensitizes Cells to Ferroptosis. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Poursaitidis, I.; Wang, X.; Crighton, T.; Labuschagne, C.; Mason, D.; Cramer, S.L.; Triplett, K.; Roy, R.; Pardo, O.E.; Seckl, M.J.; et al. Oncogene-Selective Sensitivity to Synchronous Cell Death following Modulation of the Amino Acid Nutrient Cystine. Cell Rep. 2017, 18, 2547–2556. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, V.S.; Ryan, M.J.; Dhruv, H.D.; Gill, S.; Eichhoff, O.M.; Seashore-Ludlow, B.; Kaffenberger, S.D.; Eaton, J.K.; Shimada, K.; Aguirre, A.J.; et al. Dependency of a Therapy-Resistant State of Cancer Cells on a Lipid Peroxidase Pathway. Nature 2017, 547, 453–457. [Google Scholar] [CrossRef]
- Kim, H.; Ronai, Z.A. PRMT5 Function and Targeting in Cancer. Cell Stress 2020, 4, 199–215. [Google Scholar] [CrossRef]
- Tokunaga, F.; Nakagawa, T.; Nakahara, M.; Saeki, Y.; Taniguchi, M.; Sakata, S.-I.; Tanaka, K.; Nakano, H.; Iwai, K. SHARPIN is a Component of the NF-κB-activating Linear Ubiquitin Chain Assembly Complex. Nature 2011, 471, 633–636. [Google Scholar] [CrossRef]
- Tamiya, H.; Kim, H.; Klymenko, O.; Kim, H.; Feng, Y.; Zhang, T.; Han, J.Y.; Murao, A.; Snipas, S.J.; Jilaveanu, L.; et al. SHARPIN-Mediated Regulation of Protein Arginine Methyltransferase 5 Controls Melanoma Growth. J. Clin. Investig. 2018, 128, 517–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, T.; Lv, X.; Kong, Q.; Yuan, C. A Novel SHARPIN-PRMT5-H3R2me1 Axis is Essential for Lung Cancer Cell Invasion. Oncotarget 2017, 8, 54809–54820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Huang, H.; Zhou, H.; Du, T.; Zeng, L.; Cao, Y.; Chen, J.; Lai, Y.; Li, J.; Wang, G.; et al. Activation of Nuclear Factor κB Pathway and Downstream Targets Survivin and Livin by SHARPIN Contributes to the Progression and Metastasis of Prostate Cancer. Cancer 2014, 120, 3208–3218. [Google Scholar] [CrossRef] [PubMed]
- Gullà, A.; Hideshima, T.; Bianchi, G.; Fulciniti, M.; Samur, M.K.; Qi, J.; Tai, Y.-T.; Harada, T.; Morelli, E.; Amodio, N.; et al. Protein Arginine Methyltransferase 5 has Prognostic Relevance and is a Druggable Target in Multiple Myeloma. Leukemia 2017, 32, 996–1002. [Google Scholar] [CrossRef]
- Li, S.; He, Y.; Chen, K.; Sun, J.; Zhang, L.; He, Y.; Yu, H.; Li, Q. RSL3 Drives Ferroptosis through NF-κB Pathway Activation and GPX4 Depletion in Glioblastoma. Oxidative Med. Cell. Longev. 2021, 2021, 1–10. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the Freely Available Easy-to-Use Software ‘EZR’ for Medical Statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, F.; Li, D.; Yan, Y.; Wang, H. Transferrin Receptor-Mediated Reactive Oxygen Species Promotes Ferroptosis of KGN Cells Via Regulating NADPH Oxidase 1/PTEN Induced Kinase 1/acyl-CoA Synthetase Long Chain Family Member 4 Signaling. Bioengineered 2021, 12, 4983–4994. [Google Scholar] [CrossRef]
- Feng, H.; Schorpp, K.; Jin, J.; Yozwiak, C.E.; Hoffstrom, B.G.; Decker, A.M.; Rajbhandari, P.; Stokes, M.E.; Bender, H.G.; Csuka, J.M.; et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020, 30, 3411–3423. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Chung, S.W. ROS-Mediated Autophagy Increases Intracellular Iron Levels and Ferroptosis by Ferritin and Transferrin Receptor Regulation. Cell Death Dis. 2019, 10, 822. [Google Scholar] [CrossRef] [Green Version]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.J.; Patel, D.N.; Welsch, M.; Skouta, R.; Lee, E.D.; Hayano, M.; Thomas, A.G.; Gleason, C.E.; Tatonetti, N.P.; Slusher, B.S.; et al. Pharmacological Inhibition of Cystine–Glutamate Exchange Induces Endoplasmic Reticulum Stress and Ferroptosis. eLife 2014, 2014, e02523. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine Transporter SLC7A11/xCT in Cancer: Ferroptosis, Nutrient Dependency, and Cancer Therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Lu, J.; Hao, X.; Li, H.; Zhang, G.; Liu, X.; Li, X.; Zhao, C.; Kuang, W.; Chen, D.; et al. FTH1 Inhibits Ferroptosis Through Ferritinophagy in the 6-OHDA Model of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1796–1812. [Google Scholar] [CrossRef]
- Dempster, J.M.; Boyle, I.; Vazquez, F.; Root, D.; Boehm, J.S.; Hahn, W.C.; Tsherniak, A.; McFarland, J.M. Chronos: A CRISPR Cell Population Dynamics Model. bioRxiv 2021, 432728. [Google Scholar] [CrossRef]
- Dempster, J.M.; Rossen, J.; Kazachkova, M.; Pan, J.; Root, D.E.; Sherika, A. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. BioRxiv 2019, 720243. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational Correction of Copy Number Effect Improves Specificity of CRISPR–Cas9 Essentiality Screens in Cancer Cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yu, S.; Wang, W.; Li, X.; Hou, Y.; Liu, Z.; Shi, Y.; Mu, K.; Niu, G.; Xu, J.; et al. SHARPIN Facilitates p53 Degradation in Breast Cancer Cells. Neoplasia 2017, 19, 84–92. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Q.; Liu, K.-W.; Qin, Z.-Y.; Zhu, G.-X.; Shen, L.-T.; Zhang, N.; Liu, B.-Y.; Che, L.-R.; Li, J.-Y.; et al. SHARPIN stabilizes β-catenin through a linear ubiquitination-independent manner to support gastric tumorigenesis. Gastric Cancer 2020, 24, 402–416. [Google Scholar] [CrossRef]
- Deng, X.; Lin, N.; Fu, J.; Xu, L.; Luo, H.; Jin, Y.; Liu, Y.; Sun, L.; Su, J. The Nrf2/PGC1α Pathway Regulates Antioxidant and Proteasomal Activity to Alter Cisplatin Sensitivity in Ovarian Cancer. Oxidative Med. Cell. Longev. 2020, 2020, 4830418. [Google Scholar] [CrossRef]
- Feng, L.; Zhao, K.; Sun, L.; Yin, X.; Zhang, J.; Liu, C.; Li, B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021, 19, 1–16. [Google Scholar] [CrossRef]
- Tan, Z.; Luo, X.; Xiao, L.; Tang, M.; Bode, A.M.; Dong, Z.; Cao, Y. The Role of PGC1α in Cancer Metabolism and its Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 774–782. [Google Scholar] [CrossRef] [Green Version]
- Marinković, M.; Novak, I. A brief overview of BNIP3L/NIX receptor-mediated mitophagy. FEBS Open Bio 2021, 11, 3230–3236. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.H.; Rathore, M.G.; Allende-Vega, N.; Vo, D.-N.; Belkhala, S.; Orecchioni, S.; Talarico, G.; Bertolini, F.; Cartron, G.; Lecellier, C.-H.; et al. Human Leukemic Cells performing Oxidative Phosphorylation (OXPHOS) Generate an Antioxidant Response Independently of Reactive Oxygen species (ROS) Production. Ebiomedicine 2016, 3, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Liao, C.; Quan, J.; Cheng, C.; Zhao, X.; Bode, A.M.; Cao, Y. Posttranslational regulation of PGC-1α and its implication in cancer metabolism. Int. J. Cancer 2019, 145, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Soyal, S.M.; Felder, T.K.; Auer, S.; Hahne, P.; Oberkofler, H.; Witting, A.; Paulmichl, M.; Landwehrmeyer, G.B.; Weydt, P.; Patsch, W.; et al. A greatly extended PPARGC1A genomic locus encodes several new brain-specific isoforms and influences Huntington disease age of onset. Hum. Mol. Genet. 2012, 21, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013, 23, 302–315. [Google Scholar] [CrossRef] [Green Version]
- Davis, I.J.; Kim, J.J.; Ozsolak, F.; Widlund, H.R.; Rozenblatt-Rosen, O.; Granter, S.R.; Du, J.; Fletcher, J.A.; Denny, C.T.; Lessnick, S.L.; et al. Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell 2006, 9, 473–484. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Van, T.-M.; Preukschat, D.; Schuenke, H.; Basic, M.; Bleich, A.; Klein, U.; Pasparakis, M. NF-κB inhibition in keratinocytes causes RIPK1-mediated necroptosis and skin inflammation. Life Sci. Alliance 2021, 4, e202000956. [Google Scholar] [CrossRef]
- Xia, T.; Liu, M.; Zhao, Q.; Ouyang, J.; Xu, P.; Chen, B. PRMT5 regulates cell pyroptosis by silencing CASP1 in multiple myeloma. Cell Death Dis. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Rantala, J.K.; Pouwels, J.; Pellinen, T.; Veltel, S.; Laasola, P.; Mattila, E.; Potter, C.S.; Duffy, T.; Sundberg, J.P.; Kallioniemi, O.; et al. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat. Cell Biol. 2011, 13, 1315–1324. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Fu, X.; Wang, Y.; Liu, H.; Jiang, Y.; Zhao, Z.; You, F. Transferrin receptor regulates malignancies and the stemness of hepatocellular carcinoma-derived cancer stem-like cells by affecting iron accumulation. PLoS ONE 2020, 15, e0243812. [Google Scholar] [CrossRef]
- Shen, Y.; Li, X.; Dong, D.; Zhang, B.; Xue, Y.; Shang, P. Transferrin Receptor 1 in Cancer: A New Sight for Cancer Therapy. Am. J. Cancer Res. 2018, 8, 916–931. [Google Scholar] [PubMed]
- Wu, H.; Zhang, J.; Dai, R.; Xu, J.; Feng, H. Transferrin receptor-1 and VEGF are prognostic factors for osteosarcoma. J. Orthop. Surg. Res. 2019, 14, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, C.; Lin, J.; Zhang, K.; Ou, H.; Shen, K.; Liu, Q.; Wei, Z.; Dong, X.; Zeng, X.; Zeng, L.; et al. SHARPIN promotes cell proliferation of cholangiocarcinoma and inhibits ferroptosis via p53/SLC7A11/GPX4 signaling. Cancer Sci. 2022, 113, 3766–3775. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.; Wu, J.; Ding, C.-K.C.; Lin, C.-C.; Pan, S.; Bossa, N.; Xu, Y.; Yang, W.-H.; Mathey-Prevot, B.; Chi, J.-T. Kinome screen of ferroptosis reveals a novel role of ATM in regulating iron metabolism. Cell Death Differ. 2020, 27, 1008–1022. [Google Scholar] [CrossRef]
- Guo, X.; Hong, S.; He, H.; Zeng, Y.; Chen, Y.; Mo, X.; Li, J.; Li, L.; Steinmetz, R.; Liu, Q. NFκB promotes oxidative stress-induced necrosis and ischemia/reperfusion injury by inhibiting Nrf2-ARE pathway. Free. Radic. Biol. Med. 2020, 159, 125–135. [Google Scholar] [CrossRef]
- Huang, L.; Liu, J.; Zhang, X.-O.; Sibley, K.; Najjar, S.M.; Lee, M.M.; Wu, Q. Inhibition of protein arginine methyltransferase 5 enhances hepatic mitochondrial biogenesis. J. Biol. Chem. 2018, 293, 10884–10894. [Google Scholar] [CrossRef] [Green Version]
- Hartley, A.-V.; Wang, B.; Jiang, G.; Wei, H.; Sun, M.; Prabhu, L.; Martin, M.; Safa, A.; Sun, S.; Liu, Y.; et al. Regulation of a PRMT5/NF-κB Axis by Phosphorylation of PRMT5 at Serine 15 in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3684. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Cho, Y.; Bae, G.-U.; Kim, S.-N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Stotland, A.; Gottlieb, R.A. Mitochondrial quality control: Easy come, easy go. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 2802–2811. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Wirth, A.-K.; Chen, D.; Wruck, C.J.; Rauh, M.; Buchfelder, M.; Savaskan, N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017, 6, e371. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Yi, J.; Zhu, J.; Minikes, A.M.; Monian, P.; Thompson, C.B.; Jiang, X. Role of Mitochondria in Ferroptosis. Mol. Cell 2018, 73, 354–363.e3. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ney, P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009, 16, 939–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zheng, W.; Lu, Y.; Zheng, Y.; Pan, L.; Wu, X.; Yuan, Y.; Shen, Z.; Ma, S.; Zhang, X.; et al. BNIP3L/NIX-mediated mitophagy: Molecular mechanisms and implications for human disease. Cell Death Dis. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Liang, Y.; Sundberg, J.P. SHARPIN regulates mitochondria-dependent apoptosis in keratinocytes. J. Dermatol. Sci. 2011, 63, 148–153. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamiya, H.; Urushihara, N.; Shizuma, K.; Ogawa, H.; Nakai, S.; Wakamatsu, T.; Takenaka, S.; Kakunaga, S. SHARPIN Enhances Ferroptosis in Synovial Sarcoma Cells via NF-κB- and PRMT5-Mediated PGC1α Reduction. Cancers 2023, 15, 3484. https://doi.org/10.3390/cancers15133484
Tamiya H, Urushihara N, Shizuma K, Ogawa H, Nakai S, Wakamatsu T, Takenaka S, Kakunaga S. SHARPIN Enhances Ferroptosis in Synovial Sarcoma Cells via NF-κB- and PRMT5-Mediated PGC1α Reduction. Cancers. 2023; 15(13):3484. https://doi.org/10.3390/cancers15133484
Chicago/Turabian StyleTamiya, Hironari, Naoko Urushihara, Kazuko Shizuma, Hisataka Ogawa, Sho Nakai, Toru Wakamatsu, Satoshi Takenaka, and Shigeki Kakunaga. 2023. "SHARPIN Enhances Ferroptosis in Synovial Sarcoma Cells via NF-κB- and PRMT5-Mediated PGC1α Reduction" Cancers 15, no. 13: 3484. https://doi.org/10.3390/cancers15133484
APA StyleTamiya, H., Urushihara, N., Shizuma, K., Ogawa, H., Nakai, S., Wakamatsu, T., Takenaka, S., & Kakunaga, S. (2023). SHARPIN Enhances Ferroptosis in Synovial Sarcoma Cells via NF-κB- and PRMT5-Mediated PGC1α Reduction. Cancers, 15(13), 3484. https://doi.org/10.3390/cancers15133484




