Cell-Free DNA Extracted from CSF for the Molecular Diagnosis of Pediatric Embryonal Brain Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Patients
2.2. Sample Collection and Processing
2.3. Molecular Analysis of Primary Tumors
2.4. cfDNA Extraction, Library Construction and Whole-Exome Sequencing of cfDNA
2.5. Targeted Sequencing Panel Design
2.6. Bioinformatics Analysis
2.6.1. Copy Number and SNV Analysis
2.6.2. Sequencing across the TSS to Infer Expression Profiles
2.6.3. Sequencing Coverage, Quality Statistics and Data Availability
3. Results
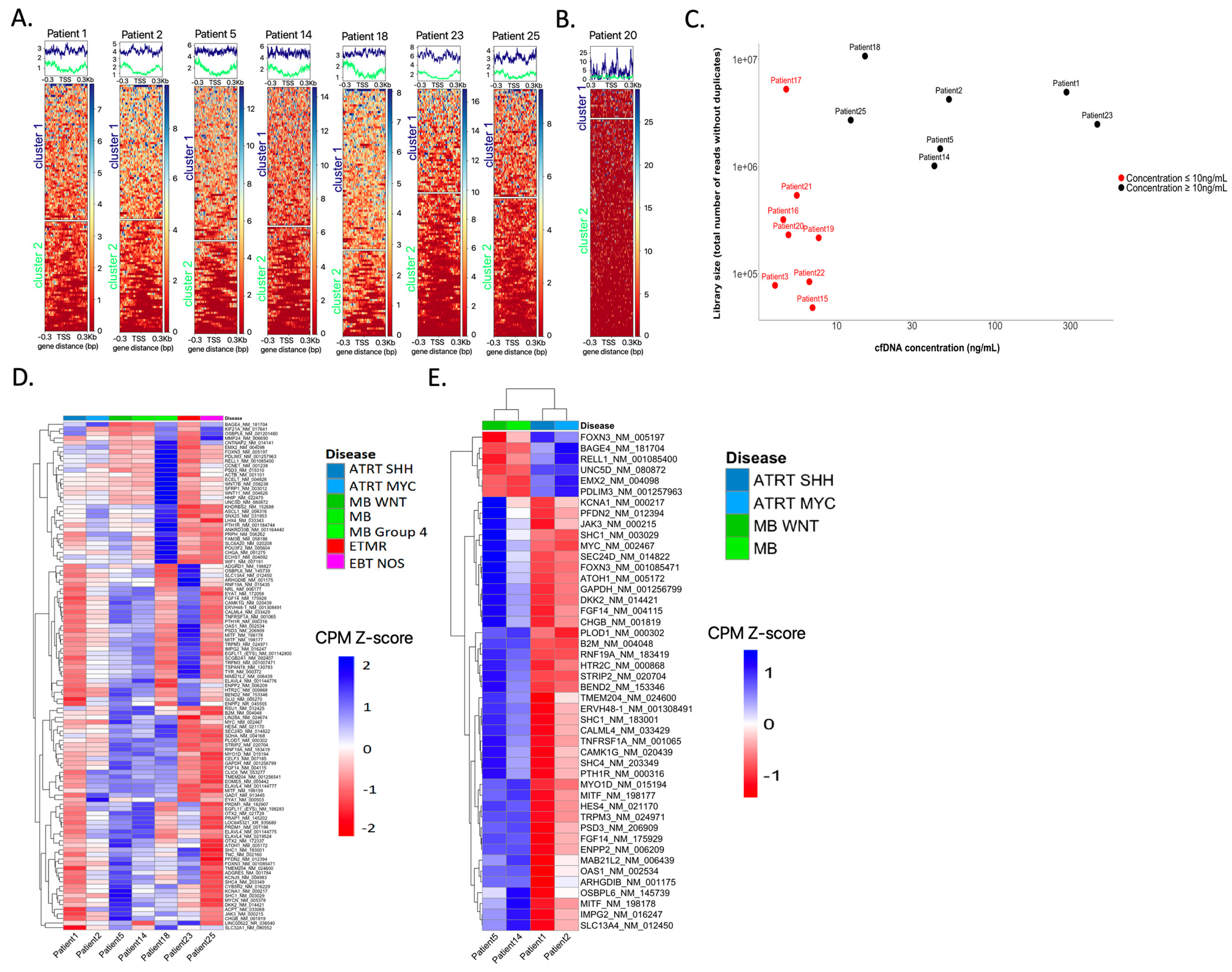
3.1. cfDNA Concentrations
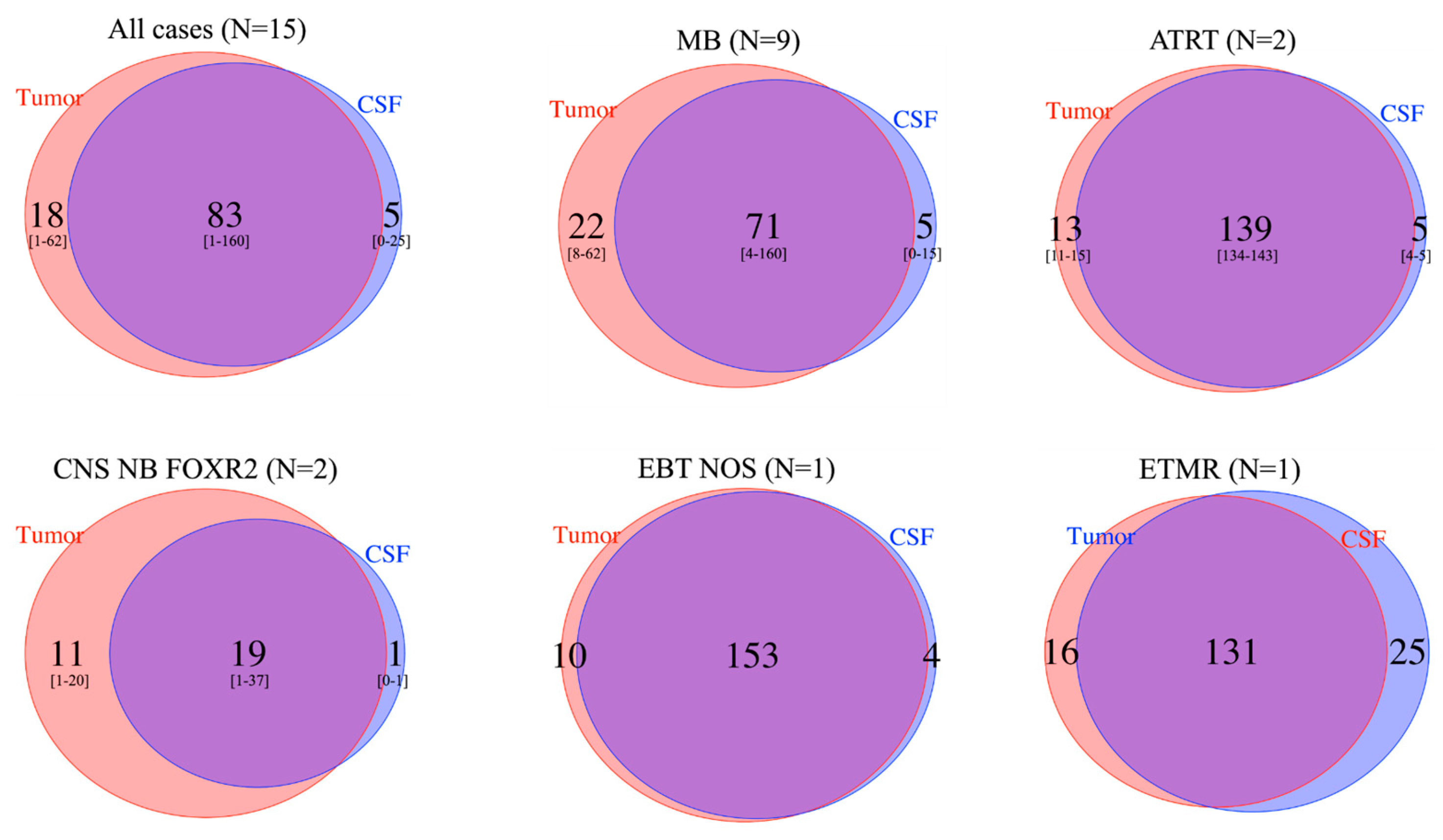
3.2. Informative Sequencing Approaches and Determining the ctDNA Fraction in CSF cfDNA
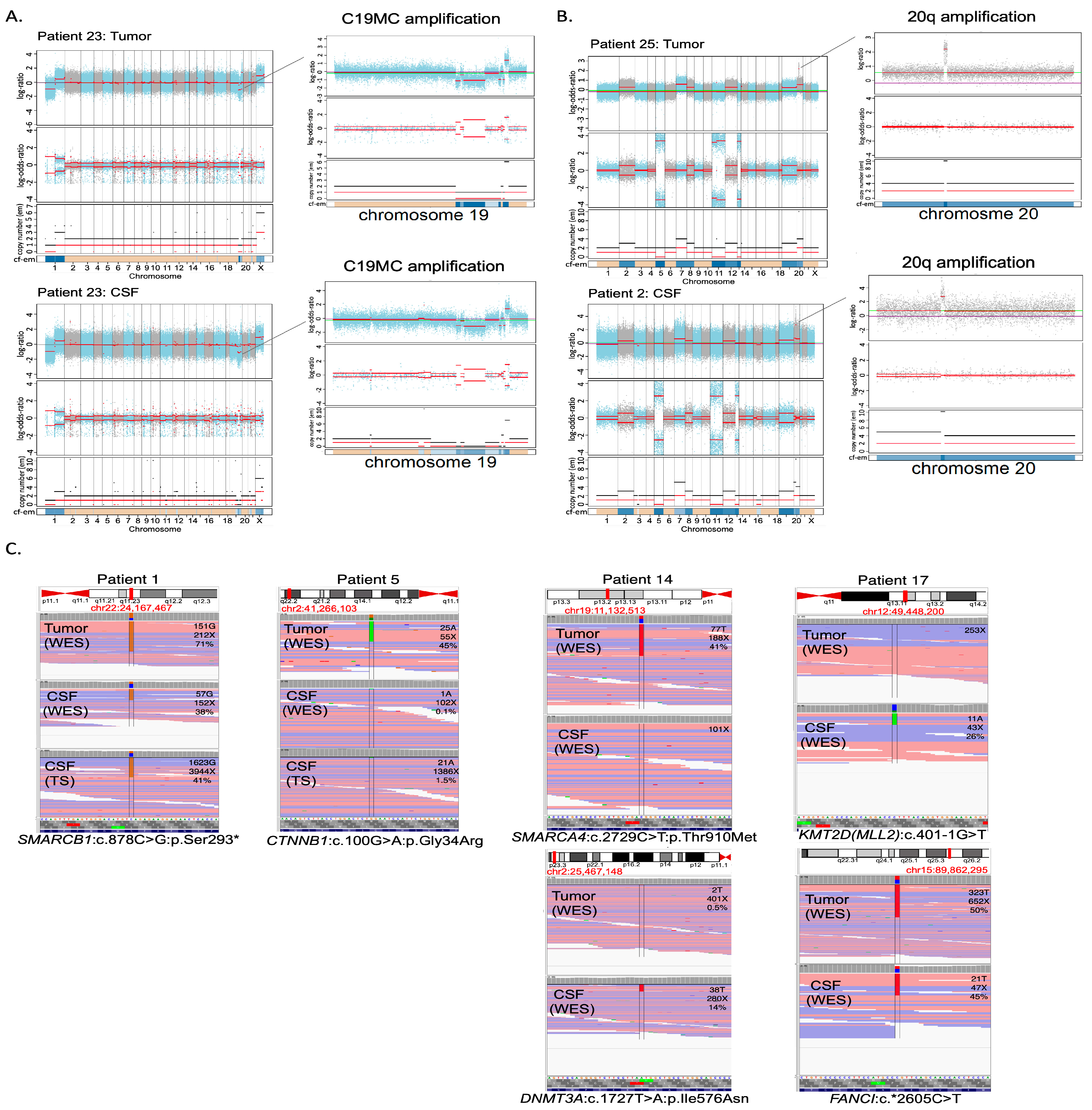
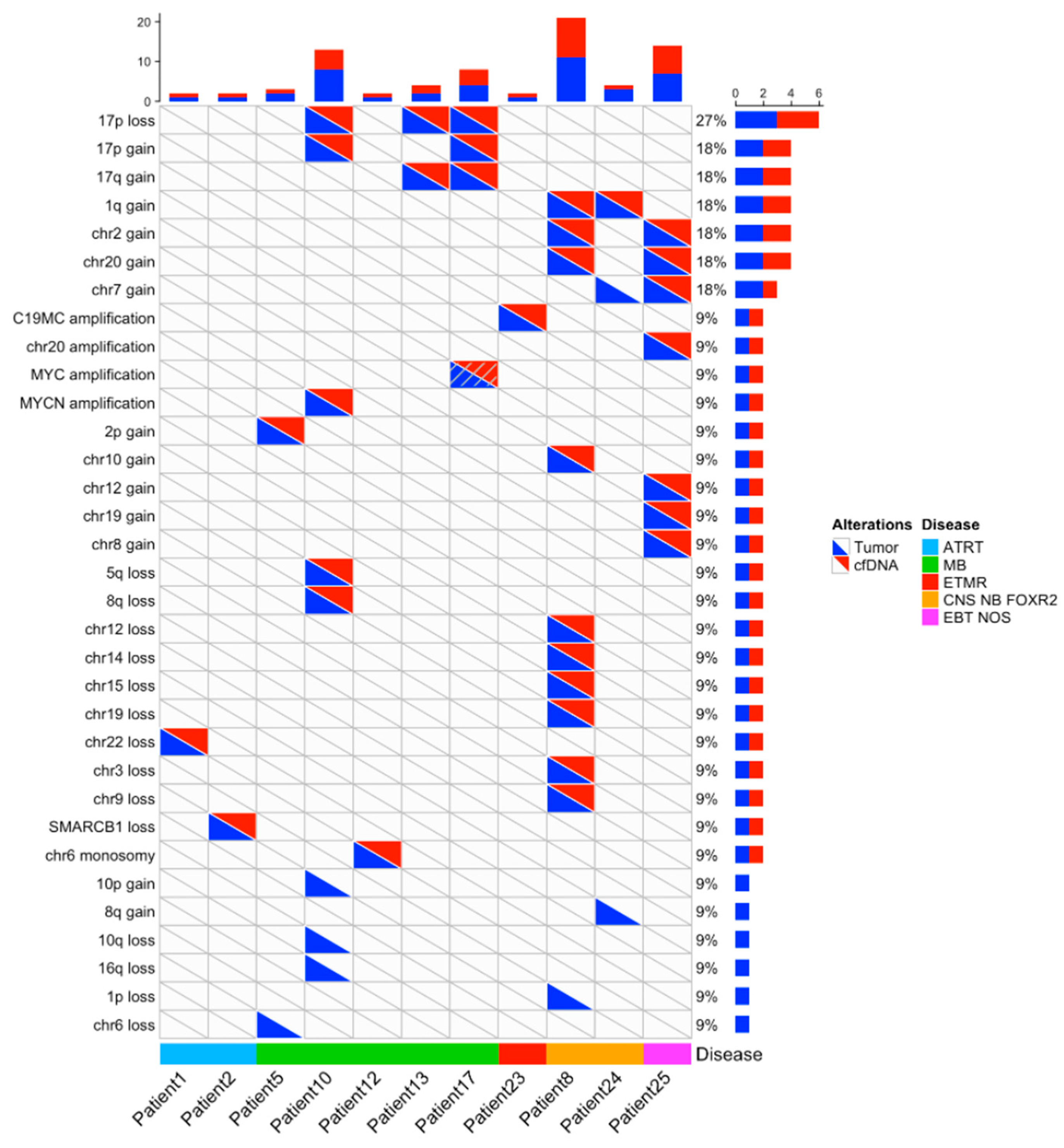
3.3. Molecular Characterization: Copy Number Profiling
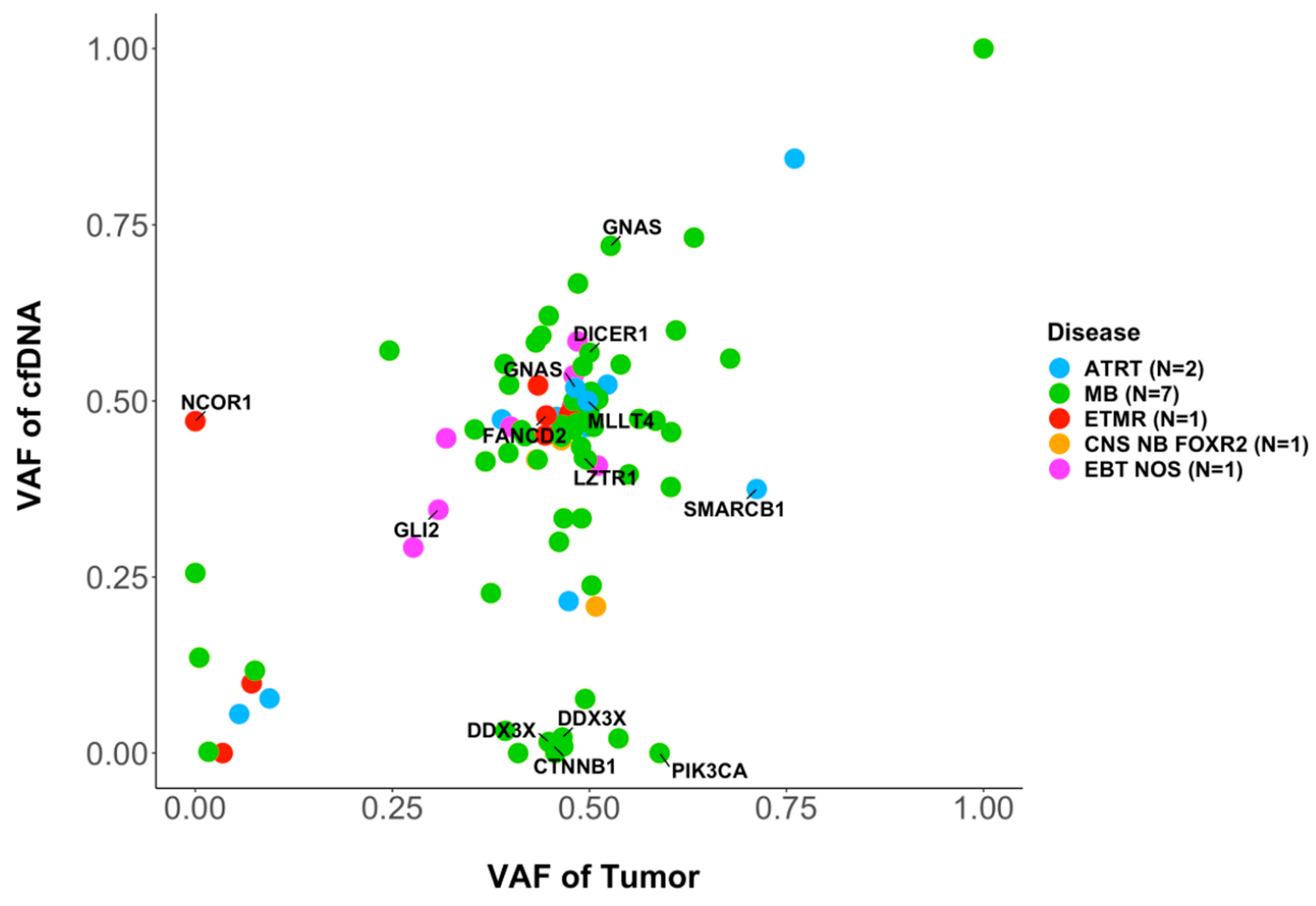
3.4. Molecular Characterization: SNVs
3.5. Diagnostic Genetic Alterations Can Be Identified in CSF cfDNA
3.6. Intratumor Heterogeneity
3.7. Inferring Expression Patterns for Tumor Classification Based on the Coverage around the tSS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grobner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Li, B.K.; Al-Karmi, S.; Huang, A.; Bouffet, E. Pediatric embryonal brain tumors in the molecular era. Expert Rev. Mol. Diagn. 2020, 20, 293–303. [Google Scholar] [CrossRef]
- Pfister, S.M.; Reyes-Mugica, M.; Chan, J.K.C.; Hasle, H.; Lazar, A.J.; Rossi, S.; Ferrari, A.; Jarzembowski, J.A.; Pritchard-Jones, K.; Hill, D.A.; et al. A Summary of the Inaugural WHO Classification of Pediatric Tumors: Transitioning from the Optical into the Molecular Era. Cancer Discov. 2022, 12, 331–355. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.6. [Google Scholar] [CrossRef]
- Ho, B.; Johann, P.D.; Grabovska, Y.; De Dieu Andrianteranagna, M.J.; Yao, F.; Fruhwald, M.; Hasselblatt, M.; Bourdeaut, F.; Williamson, D.; Huang, A.; et al. Molecular subgrouping of atypical teratoid/rhabdoid tumors-a reinvestigation and current consensus. Neuro Oncol. 2020, 22, 613–624. [Google Scholar] [CrossRef]
- Sturm, D.; Orr, B.A.; Toprak, U.H.; Hovestadt, V.; Jones, D.T.W.; Capper, D.; Sill, M.; Buchhalter, I.; Northcott, P.A.; Leis, I.; et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 2016, 164, 1060–1072. [Google Scholar] [CrossRef]
- Berlanga, P.; Pierron, G.; Lacroix, L.; Chicard, M.; Adam de Beaumais, T.; Marchais, A.; Harttrampf, A.C.; Iddir, Y.; Larive, A.; Soriano Fernandez, A.; et al. The European MAPPYACTS Trial: Precision Medicine Program in Pediatric and Adolescent Patients with Recurrent Malignancies. Cancer Discov. 2022, 12, 1266–1281. [Google Scholar] [CrossRef]
- Van Tilburg, C.M.; Pfaff, E.; Pajtler, K.W.; Langenberg, K.P.S.; Fiesel, P.; Jones, B.C.; Balasubramanian, G.P.; Stark, S.; Johann, P.D.; Blattner-Johnson, M.; et al. The Pediatric Precision Oncology INFORM Registry: Clinical Outcome and Benefit for Patients with Very High-Evidence Targets. Cancer Discov. 2021, 11, 2764–2779. [Google Scholar] [CrossRef]
- Seoane, J.; De Mattos-Arruda, L.; Le Rhun, E.; Bardelli, A.; Weller, M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann. Oncol. 2019, 30, 211–218. [Google Scholar] [CrossRef]
- Bounajem, M.T.; Karsy, M.; Jensen, R.L. Liquid biopsies for the diagnosis and surveillance of primary pediatric central nervous system tumors: A review for practicing neurosurgeons. Neurosurg. Focus 2020, 48, E8. [Google Scholar] [CrossRef]
- Li, J.; Zhao, S.; Lee, M.; Yin, Y.; Zhou, Y.; Ballester, L.Y.; Esquenazi, Y.; Dashwood, R.H.; Davies, P.J.A.; Parsons, D.W.; et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci. Adv. 2020, 6, eabb5427. [Google Scholar] [CrossRef]
- Sun, Y.; Li, M.; Ren, S.; Liu, Y.; Zhang, J.; Li, S.; Gao, W.; Gong, X.; Liu, J.; Wang, Y.; et al. Exploring genetic alterations in circulating tumor DNA from cerebrospinal fluid of pediatric medulloblastoma. Sci. Rep. 2021, 11, 5638. [Google Scholar] [CrossRef]
- Stankunaite, R.; Marshall, L.V.; Carceller, F.; Chesler, L.; Hubank, M.; George, S.L. Liquid biopsy for children with central nervous system tumours: Clinical integration and technical considerations. Front. Pediatr. 2022, 10, 957944. [Google Scholar] [CrossRef]
- Liu, A.P.Y.; Smith, K.S.; Kumar, R.; Paul, L.; Bihannic, L.; Lin, T.; Maass, K.K.; Pajtler, K.W.; Chintagumpala, M.; Su, J.M.; et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell 2021, 39, 1519–1530.e4. [Google Scholar] [CrossRef]
- Zuccato, J.A.; Patil, V.; Mansouri, S.; Voisin, M.; Chakravarthy, A.; Shen, S.Y.; Nassiri, F.; Mikolajewicz, N.; Trifoi, M.; Skakodub, A.; et al. Cerebrospinal fluid methylome-based liquid biopsies for accurate malignant brain neoplasm classification. Neuro Oncol. 2022, noac264. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Ulz, P.; Perakis, S.; Zhou, Q.; Moser, T.; Belic, J.; Lazzeri, I.; Wolfler, A.; Zebisch, A.; Gerger, A.; Pristauz, G.; et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat. Commun. 2019, 10, 4666. [Google Scholar] [CrossRef]
- Ulz, P.; Thallinger, G.G.; Auer, M.; Graf, R.; Kashofer, K.; Jahn, S.W.; Abete, L.; Pristauz, G.; Petru, E.; Geigl, J.B.; et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 2016, 48, 1273–1278. [Google Scholar] [CrossRef]
- Vanderstichele, A.; Busschaert, P.; Landolfo, C.; Olbrecht, S.; Coosemans, A.; Froyman, W.; Loverix, L.; Concin, N.; Braicu, E.I.; Wimberger, P.; et al. Nucleosome footprinting in plasma cell-free DNA for the pre-surgical diagnosis of ovarian cancer. NPJ Genom. Med. 2022, 7, 30. [Google Scholar] [CrossRef]
- Oh, S.; Geistlinger, L.; Ramos, M.; Morgan, M.; Waldron, L.; Riester, M. Reliable Analysis of Clinical Tumor-Only Whole-Exome Sequencing Data. JCO Clin. Cancer Inform. 2020, 4, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Euskirchen, P.; Bielle, F.; Labreche, K.; Kloosterman, W.P.; Rosenberg, S.; Daniau, M.; Schmitt, C.; Masliah-Planchon, J.; Bourdeaut, F.; Dehais, C.; et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017, 134, 691–703. [Google Scholar] [CrossRef]
- Passeri, T.; Dahmani, A.; Masliah-Planchon, J.; Naguez, A.; Michou, M.; El Botty, R.; Vacher, S.; Bouarich, R.; Nicolas, A.; Polivka, M.; et al. Dramatic In Vivo Efficacy of the EZH2-Inhibitor Tazemetostat in PBRM1-Mutated Human Chordoma Xenograft. Cancers 2022, 14, 1486. [Google Scholar] [CrossRef] [PubMed]
- Richer, W.; Masliah-Planchon, J.; Clement, N.; Jimenez, I.; Maillot, L.; Gentien, D.; Albaud, B.; Chemlali, W.; Galant, C.; Larousserie, F.; et al. Embryonic signature distinguishes pediatric and adult rhabdoid tumors from other SMARCB1-deficient cancers. Oncotarget 2017, 8, 34245–34257. [Google Scholar] [CrossRef] [PubMed]
- Chicard, M.; Colmet-Daage, L.; Clement, N.; Danzon, A.; Bohec, M.; Bernard, V.; Baulande, S.; Bellini, A.; Deveau, P.; Pierron, G.; et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin. Cancer Res. 2018, 24, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Johann, P.D.; Erkek, S.; Zapatka, M.; Kerl, K.; Buchhalter, I.; Hovestadt, V.; Jones, D.T.W.; Sturm, D.; Hermann, C.; Segura Wang, M.; et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 2016, 29, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Remke, M.; Bouffet, E.; Faria, C.C.; Perreault, S.; Cho, Y.J.; Shih, D.J.; Luu, B.; Dubuc, A.M.; Northcott, P.A.; et al. Recurrence patterns across medulloblastoma subgroups: An integrated clinical and molecular analysis. Lancet Oncol. 2013, 14, 1200–1207. [Google Scholar] [CrossRef]
- Behling, F.; Schittenhelm, J. Oncogenic BRAF Alterations and Their Role in Brain Tumors. Cancers 2019, 11, 794. [Google Scholar] [CrossRef]
- Wu, G.; Diaz, A.K.; Paugh, B.S.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Lizio, M.; Harshbarger, J.; Shimoji, H.; Severin, J.; Kasukawa, T.; Sahin, S.; Abugessaisa, I.; Fukuda, S.; Hori, F.; Ishikawa-Kato, S.; et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.Y.; Smith, K.S.; Kumar, R.; Robinson, G.W.; Northcott, P.A. Low-coverage whole-genome sequencing of cerebrospinal-fluid-derived cell-free DNA in brain tumor patients. STAR Protoc. 2022, 3, 101292. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.W.; Jäger, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.-J.; Pugh, T.J.; Hovestadt, V.; Stütz, A.M.; et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef]
- Escudero, L.; Llort, A.; Arias, A.; Diaz-Navarro, A.; Martinez-Ricarte, F.; Rubio-Perez, C.; Mayor, R.; Caratu, G.; Martinez-Saez, E.; Vazquez-Mendez, E.; et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat. Commun. 2020, 11, 5376. [Google Scholar] [CrossRef]
- Hill, R.M.; Kuijper, S.; Lindsey, J.C.; Petrie, K.; Schwalbe, E.C.; Barker, K.; Boult, J.K.; Williamson, D.; Ahmad, Z.; Hallsworth, A.; et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015, 27, 72–84. [Google Scholar] [CrossRef]
- Izquierdo, E.; Proszek, P.; Pericoli, G.; Temelso, S.; Clarke, M.; Carvalho, D.M.; Mackay, A.; Marshall, L.V.; Carceller, F.; Hargrave, D.; et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neuro-Oncol. Adv. 2021, 3, vdab013. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium; Hu, T.; Kumar, Y.; Ma, E.Z.; Wu, Z.; Xue, H. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Lambo, S.; Grobner, S.N.; Rausch, T.; Waszak, S.M.; Schmidt, C.; Gorthi, A.; Romero, J.C.; Mauermann, M.; Brabetz, S.; Krausert, S.; et al. The molecular landscape of ETMR at diagnosis and relapse. Nature 2019, 576, 274–280. [Google Scholar] [CrossRef]
- Hesson, L.B.; Sloane, M.A.; Wong, J.W.; Nunez, A.C.; Srivastava, S.; Ng, B.; Hawkins, N.J.; Bourke, M.J.; Ward, R.L. Altered promoter nucleosome positioning is an early event in gene silencing. Epigenetics 2014, 9, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Seshan, V.E. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016, 44, e131. [Google Scholar] [CrossRef] [PubMed]
| Patient Number | Primary Tumor Molecular Diagnosis: SNV | SNV in CSF cfDNA | Primary Tumor Molecular Diagnosis: CNA | CNA in CSF cfDNA |
|---|---|---|---|---|
| 1 | SMARCB1: c.851C>G/(p.Ser284*) | SMARCB1: c.851C>G/(p.Ser284*) | SMARCB1 loss | SMARCB1 loss |
| 2 | SMARCB1 loss | SMARCB1 loss | ||
| 3 | SMARCB1: c.601C>T/p.(Arg201*) | not found/CSF cfDNA non-contributive | ||
| 4 | Gains: 2pter-p22.3 (subclone), 7, 17q12.2-qter Losses: 11pter-p11.12, 17q12.2-qter | not found/CSF cfDNA non-contributive | ||
| 5 | CTNNB1: Gly34Arg (c. 100G>A) | CTNNB1: Gly34Arg (c. 100G>A) | chr6 loss chr2pter-p13.2 gain | chr2pter-p13.2 gain |
| 6 | 17q gain, 17p loss | not found/CSF cfDNA non-contributive | ||
| 7 | Losses: chr10, 2, 11, 13, 16, et 20 | not found/CSF cfDNA non-contributive | ||
| 8 | Gains: 1q, 2, 10, 20 Losses: 1p, 3, 9, 12, 14, 15, 19, X | Gains: 1q, 2, 10, 20 Losses: 3, 9, 12, 14, 15, 19 | ||
| 9 | 17p loss, 17q gain | not found/CSF cfDNA non-contributive | ||
| 10 | Amplicon 2p24.3-2 containing MYCN. Losses: 5q31.2-ter 8q12.3-ter, 10q, 16q, 17pter-p11.2 Gains: 10p, 17p11.2-qter | Amplicon 2p24.3-2 containing MYCN. Losses: 5q31,2-ter, 8q12.3-ter, 17pter-p11.2 Gains: 17p11.2-qter | ||
| 11 | PTCH1: c.1359_1360insG (p.cys454valfs*43) | not found/CSF cfDNA non-contributive | 9q loss 3q gain | not found/CSF cfDNA non-contributive |
| 12 | APC: c.3183_3187del/p.Gln1062* germline APC: c.3758del/p.Ser1253Leufs*12 | APC: c.3183_3187del/p.Gln1062* germline APC: c.3758del/p.Ser1253Leufs*12 | chr6 monosomy | chr6 monosomy |
| 13 | 17p loss 17pq gain | 17p loss 17pq gain | ||
| 14 | ||||
| 15 | CCND3 c.774_775delCTinsTG p.(Ser259Ala) COL3A1 c.946G>A p.(Ala316Thr) FANCD2 (c.1588C>T p.(Arg530*) NCKIPSD c.1650_1651delGCinsTT p.(Pro551Ser) PTPRC c.3452A>G p.(Lys1151Arg) | altered genes not included in targeted sequencing CSF cfDNA panel | ||
| 16 | Gains: 1q(210.34 Mb-tel), 3p(tel-5.17 Mb) | not found/CSF cfDNA non-contributive | ||
| 17 | Losses: 17p(tel-18.91 Mb) harboring TP53 Gains: 17pq(19.14 Mb-tel) | Losses: 17p(tel-18.91 Mb) harboring TP53 Gains: 17pq(19.14 Mb-tel) | ||
| 18 | Losses: 11p-, 17p(tel-22.22 Mb) Gains: 4q(142.07 Mb-tel), 13q(90.81–96.80 Mb), 15q(54.53 Mb-tel), 17pq(25.28 Mb-tel) Amplicons: 7q(92.20–96.80 Mb) harboring CDK6 | not found/CSF cfDNA non-contributive | ||
| 19 | Losses: 2q(213.21 Mb-tel), 8p(tel-10,18 Mb), 16q(57.83 Mb-tel), 17p(tel-18.15 Mb), X(78.64-Tel), X(16.68–17.87 Mb) Subclonal segmental losses: 4q(170.70 Mb-tel), 5q(170.87 Mb-tel), 8q(55.00–70.23 Mb), 9p(tel-11.78 Mb), 10q(107.71 Mb-tel), 12q(40.77–57.69 Mb) | No CN analysis for target seq data | ||
| 20 | CTNNB1: c.98C>G/p.(Ser33Cys) PIK3CA: c.311C>G/p.(Pro104Arg) (50, 3%; 193X) | CTNNB1: c.98C>G/p.(Ser33Cys) | chr6 monosomy | No CN analysis for target seq data |
| 21 | Losses: 17p(tel-18.91 Mb) Gains: 17pq(19.14 Mb-tel) | No CN analysis for target seq data | ||
| 22 | Losses: 16q(66.57 Mb-tel) Subclonal segmental losses: 8q(52.57 Mb-tel), 11q(75.97 Mb-tel), 13q(25.30–31.39 Mb), 13q(50.05–81.10 Mb) Gains: 7p(tel-4,88 Mb), 13q(20,28–25,27 Mb), 13q(31.45–49.99 Mb), 13q(81.14 Mb-tel), 17q(46.78 Mb-tel) | No CN analysis for target seq data | ||
| 23 | Amplification 19q13.41 | Amplification 19q13.41 | ||
| 24 | PTPRK p.(Thr395AspfsTer6) | not found/CSF cfDNA non-contributive | Gains: 1q, 7, 8q | Gains: 1q |
| 25 | KDR: Hist1144Asp TP53: Arg175His KRAS: Gly12Asp | not found/CSF cfDNA non-contributive | Gains: 2, 7, 12, 19. Amplification chr 20(30.5–30.8 Mb) | Gains: 2, 7, 12, 19. Amplification chr 20(30.5–30.8 Mb) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicard, M.; Iddir, Y.; Masliah Planchon, J.; Combaret, V.; Attignon, V.; Saint-Charles, A.; Frappaz, D.; Faure-Conter, C.; Beccaria, K.; Varlet, P.; et al. Cell-Free DNA Extracted from CSF for the Molecular Diagnosis of Pediatric Embryonal Brain Tumors. Cancers 2023, 15, 3532. https://doi.org/10.3390/cancers15133532
Chicard M, Iddir Y, Masliah Planchon J, Combaret V, Attignon V, Saint-Charles A, Frappaz D, Faure-Conter C, Beccaria K, Varlet P, et al. Cell-Free DNA Extracted from CSF for the Molecular Diagnosis of Pediatric Embryonal Brain Tumors. Cancers. 2023; 15(13):3532. https://doi.org/10.3390/cancers15133532
Chicago/Turabian StyleChicard, Mathieu, Yasmine Iddir, Julien Masliah Planchon, Valérie Combaret, Valéry Attignon, Alexandra Saint-Charles, Didier Frappaz, Cécile Faure-Conter, Kévin Beccaria, Pascale Varlet, and et al. 2023. "Cell-Free DNA Extracted from CSF for the Molecular Diagnosis of Pediatric Embryonal Brain Tumors" Cancers 15, no. 13: 3532. https://doi.org/10.3390/cancers15133532
APA StyleChicard, M., Iddir, Y., Masliah Planchon, J., Combaret, V., Attignon, V., Saint-Charles, A., Frappaz, D., Faure-Conter, C., Beccaria, K., Varlet, P., Geoerger, B., Baulande, S., Pierron, G., Bouchoucha, Y., Doz, F., Delattre, O., Waterfall, J. J., Bourdeaut, F., & Schleiermacher, G. (2023). Cell-Free DNA Extracted from CSF for the Molecular Diagnosis of Pediatric Embryonal Brain Tumors. Cancers, 15(13), 3532. https://doi.org/10.3390/cancers15133532







