An Assessment of the Penile Squamous Cell Carcinoma Surfaceome for Biomarker and Therapeutic Target Discovery
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. PSCC Cell Line Translatomic Data
2.1.2. MCC3651 Gene Expression Data
2.1.3. Johnstone Gene Expression Data
2.1.4. GSE57955 Gene Expression Data
2.2. RNA Sequencing
2.3. Surfaceome Inference
2.4. Immunohistochemistry
2.5. Statistical Analysis
3. Results
3.1. PSCC Cell Line Surfaceome Characterization
3.2. Patient Tumor Surfaceome and Druggability Potential
3.3. Validation of PSCC Surfaceome Protein Expression
3.4. Prognostic Association of Select Surfaceome Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jakobsen, J.K.; Sørensen, C.M.; Krarup, K.P.; Jensen, J.B. Quality of life, voiding and sexual function of penile cancer patients: DaPeCa-10-a cross-sectional questionnaire survey. BJUI Compass 2022, 3, 354–362. [Google Scholar] [CrossRef]
- Thomas, A.; Necchi, A.; Muneer, A.; Tobias-Machado, M.; Tran, A.T.H.; Van Rompuy, A.-S.; Spiess, P.E.; Albersen, M. Penile cancer. Nat. Rev. Dis. Prim. 2021, 7, 11. [Google Scholar] [CrossRef]
- Olesen, T.B.; Sand, F.L.; Rasmussen, C.L.; Albieri, V.; Toft, B.G.; Norrild, B.; Munk, C.; Kjær, S.K. Prevalence of human papillomavirus DNA and p16INK4a in penile cancer and penile intraepithelial neoplasia: A systematic review and meta-analysis. Lancet Oncol. 2019, 20, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Alencar, A.M.; Sonpavde, G. Emerging Therapies in Penile Cancer. Front. Oncol. 2022, 12, 910335. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Alvim, L.M.D.C.; Rainho, C.A.; Juengel, E.; Blaheta, R.A.; Spiess, P.E.; Rogatto, S.R.; Tsaur, I. Systemic treatment of penile squamous cell carcinoma—Hurdles and hopes of preclinical models and clinical regimens: A narrative review. Transl. Androl. Urol. 2021, 10, 4085–4098. [Google Scholar] [CrossRef] [PubMed]
- Medeiros-Fonseca, B.; Cubilla, A.; Brito, H.; Martins, T.; Medeiros, R.; Oliveira, P.; Gil da Costa, R.M. Experimental Models for Studying HPV-Positive and HPV-Negative Penile Cancer: New Tools for An Old Disease. Cancers 2021, 13, 460. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Milani, E.S.; Wollscheid, B. Surfaceome nanoscale organization and extracellular interaction networks. Curr. Opin. Chem. Biol. 2019, 48, 26–33. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. MemLoci: Predicting subcellular localization of membrane proteins in eukaryotes. Bioinformatics 2011, 27, 1224–1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.-C.; Shen, H.-B. MemType-2L: A web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biophys. Res. Commun. 2007, 360, 339–345. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Tian, Y.; Yan, Y. Subcellular localization prediction for human internal and organelle membrane proteins with projected gene ontology scores. J. Theor. Biol. 2012, 313, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Briesemeister, S.; Rahnenführer, J.; Kohlbacher, O. YLoc--an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010, 38, W497–W502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bausch-Fluck, D.; Hofmann, A.; Bock, T.K.C.; Frei, A.P.; Cerciello, F.; Jacobs, A.; Moest, H.; Omasits, U.; Gundry, R.L.; Yoon, C.; et al. A Mass Spectrometric-Derived Cell Surface Protein Atlas. PLoS ONE 2015, 10, e0121314. [Google Scholar] [CrossRef] [Green Version]
- Bausch-Fluck, D.; Goldmann, U.; Müller, S.; van Oostrum, M.; Müller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, A.L.; da Silva, V.L.; da Fonsêca, M.M.; Meira, I.T.J.; da Silva, T.E.; Kroll, J.E.; Ribeiro-Dos-Santos, A.M.; Freitas, C.R.; Furtado, R.; de Souza, J.E.; et al. Bioinformatics Analysis of the Human Surfaceome Reveals New Targets for a Variety of Tumor Types. Int. J. Genom. 2016, 2016, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Syafruddin, S.E.; Nazarie, W.F.W.M.; Moidu, N.A.; Soon, B.H.; Mohtar, M.A. Integration of RNA-Seq and proteomics data identifies glioblastoma multiforme surfaceome signature. BMC Cancer 2021, 21, 850. [Google Scholar] [CrossRef]
- Hu, Z.; Yuan, J.; Long, M.; Jiang, J.; Zhang, Y.; Zhang, T.; Xu, M.; Fan, Y.; Tanyi, J.L.; Montone, K.T.; et al. The Cancer Surfaceome Atlas integrates genomic, functional and drug response data to identify actionable targets. Nat. Cancer 2021, 2, 1406–1422. [Google Scholar] [CrossRef]
- Kuasne, H.; Canto, L.M.D.; Aagaard, M.M.; Muñoz, J.J.M.; De Jamblinne, C.; Marchi, F.A.; Scapulatempo-Neto, C.; Faria, E.F.; Lopes, A.; Carréno, S.; et al. Penile Cancer-Derived Cells Molecularly Characterized as Models to Guide Targeted Therapies. Cells 2021, 10, 814. [Google Scholar] [CrossRef]
- Azizi, M.; Tang, D.H.; Verduzco, D.; Peyton, C.C.; Chipollini, J.; Yuan, Z.; Schaible, B.J.; Zhou, J.-M.; Johnstone, P.A.; Giuliano, A.; et al. Impact of PI3K-AKT-mTOR Signaling Pathway Up-regulation on Prognosis of Penile Squamous-Cell Carcinoma: Results From a Tissue Microarray Study and Review of the Literature. Cancer 2019, 17, e80–e91. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.; Chahoud, J.; Lopez, A.; Dhillon, J.; Eschrich, S.A.; Johnstone, P.A.; Spiess, P.E. An Analysis of Nectin-4 (PVRL4) in Penile Squamous Cell Carcinoma. Eur. Urol. Open Sci. 2023, 49, 1–5. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Grass, G.; Azizi, M.; Ahmed, K.A.; Yoder, G.S.J.; Welsh, E.A.; Fulp, W.J.; Dhillon, J.; Torres-Roca, J.F.; Giuliano, A.R.; et al. Intrinsic radiosensitivity, genomic-based radiation dose and patterns of failure of penile cancer in response to adjuvant radiation therapy. Rep. Pract. Oncol. Radiother. 2019, 24, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Kuasne, H.; Cólus, I.M.d.S.; Busso, A.F.; Hernandez-Vargas, H.; Barros-Filho, M.C.; Marchi, F.A.; Scapulatempo-Neto, C.; Faria, E.F.; Lopes, A.; Guimarães, G.C.; et al. Genome-wide methylation and transcriptome analysis in penile carcinoma: Uncovering new molecular markers. Clin. Epigenetics 2015, 7, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almén, M.S.; Nordström, K.J.; Fredriksson, R.; Schiöth, H.B. Mapping the human membrane proteome: A majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Li, C.; Wang, M.; Webb, G.I.; Zhang, Y.; Whisstock, J.C.; Song, J. GlycoMine: A machine learning-based approach for predicting N-, C- and O-linked glycosylation in the human proteome. Bioinformatics 2015, 31, 1411–1419. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Buccitelli, C.; Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Jogalekar, M.P.; Rajendran, R.L.; Khan, F.; Dmello, C.; Gangadaran, P.; Ahn, B.-C. CAR T-Cell-Based gene therapy for cancers: New perspectives, challenges, and clinical developments. Front. Immunol. 2022, 13, 925985. [Google Scholar] [CrossRef]
- Bald, T.; Krummel, M.F.; Smyth, M.J.; Barry, K.C. The NK cell-cancer cycle: Advances and new challenges in NK cell-based immunotherapies. Nat. Immunol. 2020, 21, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Drago, J.Z.; Modi, S.; Chandarlapaty, S. Unlocking the potential of antibody-drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Wollscheid, B.; Bausch-Fluck, D.; Henderson, C.; O’Brien, R.; Bibel, M.; Schiess, R.; Aebersold, R.; Watts, J.D. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009, 27, 378–386. [Google Scholar] [CrossRef] [Green Version]
- Governa, V.; Talbot, H.; de Oliveira, K.G.; Cerezo-Magaña, M.; Bång-Rudenstam, A.; Johansson, M.C.; Månsson, A.-S.; Forsberg-Nilsson, K.; Marko-Varga, G.; Pérez, J.E.; et al. Landscape of surfaceome and endocytome in human glioma is divergent and depends on cellular spatial organization. Proc. Natl. Acad. Sci. USA 2022, 119, e2114456119. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, G.; Tuveson, D.A. Activated fibroblasts in cancer: Perspectives and challenges. Cancer Cell 2023, 41, 434–449. [Google Scholar] [CrossRef]
- Alquraini, A.; El Khoury, J. Scavenger receptors. Curr. Biol. 2020, 30, R790–R795. [Google Scholar] [CrossRef] [PubMed]
- Taban, Q.; Mumtaz, P.T.; Masoodi, K.Z.; Haq, E.; Ahmad, S.M. Scavenger receptors in host defense: From functional aspects to mode of action. Cell Commun. Signal. 2022, 20, 2. [Google Scholar] [CrossRef]
- Kanai, Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharmacol. Ther. 2022, 230, 107964. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Kudelka, M.R.; Antonopoulos, A.; Wang, Y.; Duong, D.M.; Song, X.; Seyfried, N.T.; Dell, A.; Haslam, S.M.; Cummings, R.D.; Ju, T. Cellular O-Glycome Reporter/Amplification to explore O-glycans of living cells. Nat. Methods 2016, 13, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhang, Y.; Ocansey, D.K.W.; Wang, B.; Mao, F. Glycosylation in Cervical Cancer: New Insights and Clinical Implications. Front. Oncol. 2021, 11, 706862. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhao, R.-X.; Chen, J.; Li, Y.; Li, X.-D.; Liu, X.-L.; Zhang, W.-M.; Quan, C.-S.; Wang, Y.-S.; Zhai, Y.-X.; et al. O-linked GlcNAcylation elevated by HPV E6 mediates viral oncogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 9333–9338. [Google Scholar] [CrossRef]
- Jing, W.; Zhang, R.; Chen, X.; Zhang, X.; Qiu, J. Association of Glycosylation-Related Genes with Different Patterns of Immune Profiles and Prognosis in Cervical Cancer. J. Pers. Med. 2023, 13, 529. [Google Scholar] [CrossRef]
- Rasheduzzaman, M.; Murugan, A.V.M.; Zhang, X.; Oliveira, T.; Dolcetti, R.; Kenny, L.; Johnson, N.W.; Kolarich, D.; Punyadeera, C. Head and neck cancer N-glycome traits are cell line and HPV status-dependent. Anal. Bioanal. Chem. 2022, 414, 8401–8411. [Google Scholar] [CrossRef]
- Joshi, V.B.; Spiess, P.E.; Necchi, A.; Pettaway, C.A.; Chahoud, J. Immune-based therapies in penile cancer. Nat. Rev. Urol. 2022, 19, 457–474. [Google Scholar] [CrossRef] [PubMed]
- McGregor, B.A.; Campbell, M.T.; Xie, W.; Farah, S.; Bilen, M.A.; Schmidt, A.L.; Sonpavde, G.P.; Kilbridge, K.L.; Choudhury, A.D.; Mortazavi, A.; et al. Results of a multicenter, phase 2 study of nivolumab and ipilimumab for patients with advanced rare genitourinary malignancies. Cancer 2021, 127, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Araldi, R.P.; Sant’Ana, T.A.; Módolo, D.G.; de Melo, T.C.; Spadacci-Morena, D.D.; de Cassia Stocco, R.; Cerutti, J.M.; de Souza, E.B. The human papillomavirus (HPV)-related cancer biology: An overview. Biomed. Pharmacother. 2018, 106, 1537–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosper, P.F.; Bradley, S.; Luo, Q.; Kimple, R.J. Biology of HPV Mediated Carcinogenesis and Tumor Progression. Semin. Radiat. Oncol. 2021, 31, 265–273. [Google Scholar] [CrossRef]
- Medda, A.; Duca, D.; Chiocca, S. Human Papillomavirus and Cellular Pathways: Hits and Targets. Pathogens 2021, 10, 262. [Google Scholar] [CrossRef]
- Macedo, J.; Silva, E.; Nogueira, L.; Coelho, R.; Silva, J.; Santos, A.; Teixeira-Júnior, A.A.; Belfort, M.; Silva, G.; Khayat, A.; et al. Genomic profiling reveals the pivotal role of hrHPV driving copy number and gene expression alterations, including mRNA downregulation of TP53 and RB1 in penile cancer. Mol. Carcinog. 2020, 59, 604–617. [Google Scholar] [CrossRef]
- Chahoud, J.; Gleber-Netto, F.O.; McCormick, B.Z.; Rao, P.; Lu, X.; Guo, M.; Morgan, M.B.; Chu, R.A.; Martinez-Ferrer, M.; Eterovic, A.K.; et al. Whole-exome Sequencing in Penile Squamous Cell Carcinoma Uncovers Novel Prognostic Categorization and Drug Targets Similar to Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2021, 27, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.W.; Mirabello, L. Human papillomavirus genomics: Understanding carcinogenicity. Tumour Virus Res. 2023, 15, 200258. [Google Scholar] [CrossRef] [PubMed]
- Kahlhofer, J.; Teis, D. The human LAT1-4F2hc (SLC7A5-SLC3A2) transporter complex: Physiological and pathophysiological implications. Basic Clin. Pharmacol. Toxicol. 2022. [Google Scholar] [CrossRef]
- Kobayashi, M.; Narumi, K.; Furugen, A.; Iseki, K. Transport function, regulation, and biology of human monocarboxylate transporter 1 (hMCT1) and 4 (hMCT4). Pharmacol. Ther. 2021, 226, 107862. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, A.; Schafer, J.; Dzieciatkowska, M.; Nemkov, T.; D’Alessandro, A.; Neelakantan, D.; Ford, H.L.; Pearson, C.G.; Weekes, C.D.; Hansen, K.C.; et al. CD147: A small molecule transporter ancillary protein at the crossroad of multiple hallmarks of cancer and metabolic reprogramming. Oncotarget 2017, 8, 6742–6762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Huang, Q.; Long, X.; Zhang, J.; Huang, X.; Aa, J.; Yang, H.; Chen, Z.; Xing, J. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARα pathways. J. Hepatol. 2015, 63, 1378–1389. [Google Scholar] [CrossRef]
- Xu, D.; Hemler, M.E. Metabolic activation-related CD147-CD98 complex. Mol. Cell. Proteom. 2005, 4, 1061–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, X.; Zeng, X.; Gu, H.; Li, M.; Tan, H.; Jin, Z.; Hua, T.; Shi, R.; Wang, H. CD147/EMMPRIN overexpression and prognosis in cancer: A systematic review and meta-analysis. Sci. Rep. 2016, 6, 32804. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xing, J.; Yang, Y.; Liu, J.; Wang, W.; Xia, Y.; Yan, Z.; Wang, K.; Wu, D.; Wu, L.; et al. Adjuvant 131I-metuximab for hepatocellular carcinoma after liver resection: A randomised, controlled, multicentre, open-label, phase 2 trial. Lancet Gastroenterol. Hepatol. 2020, 5, 548–560. [Google Scholar] [CrossRef]
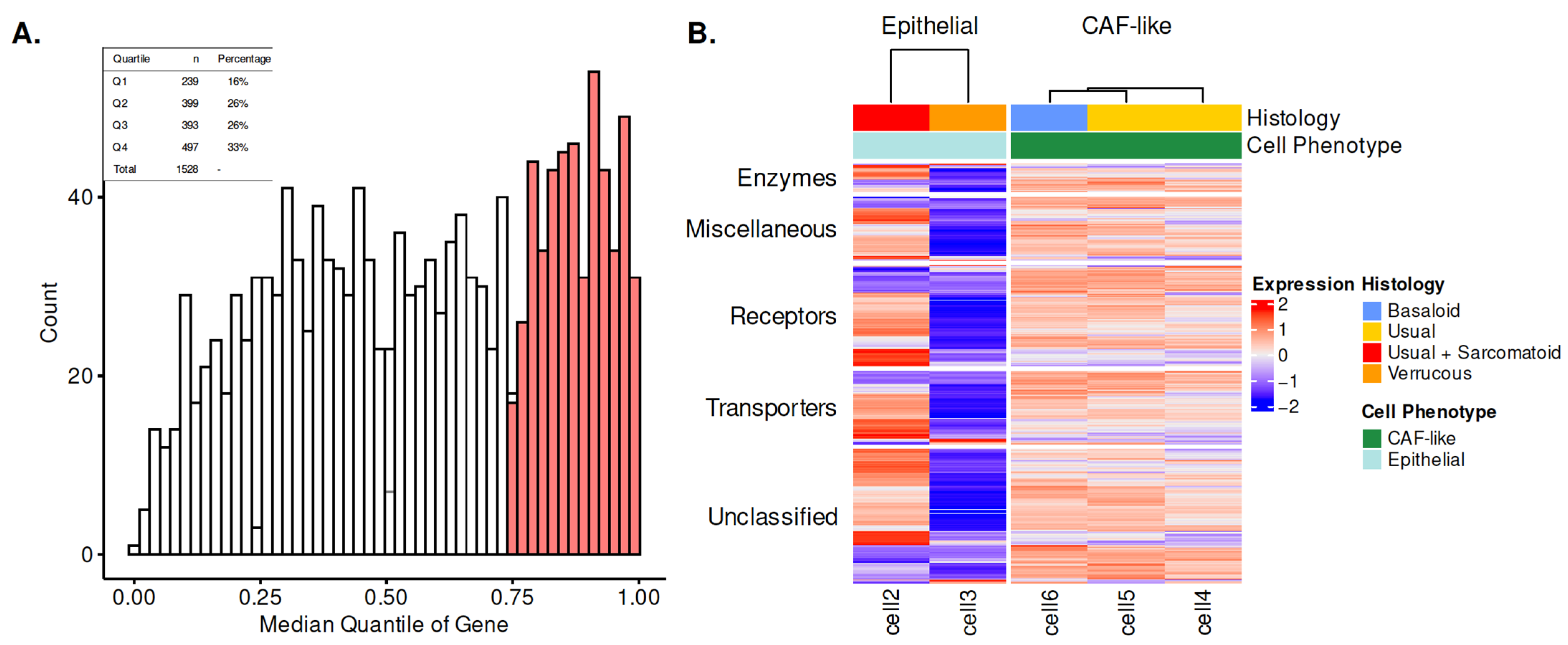

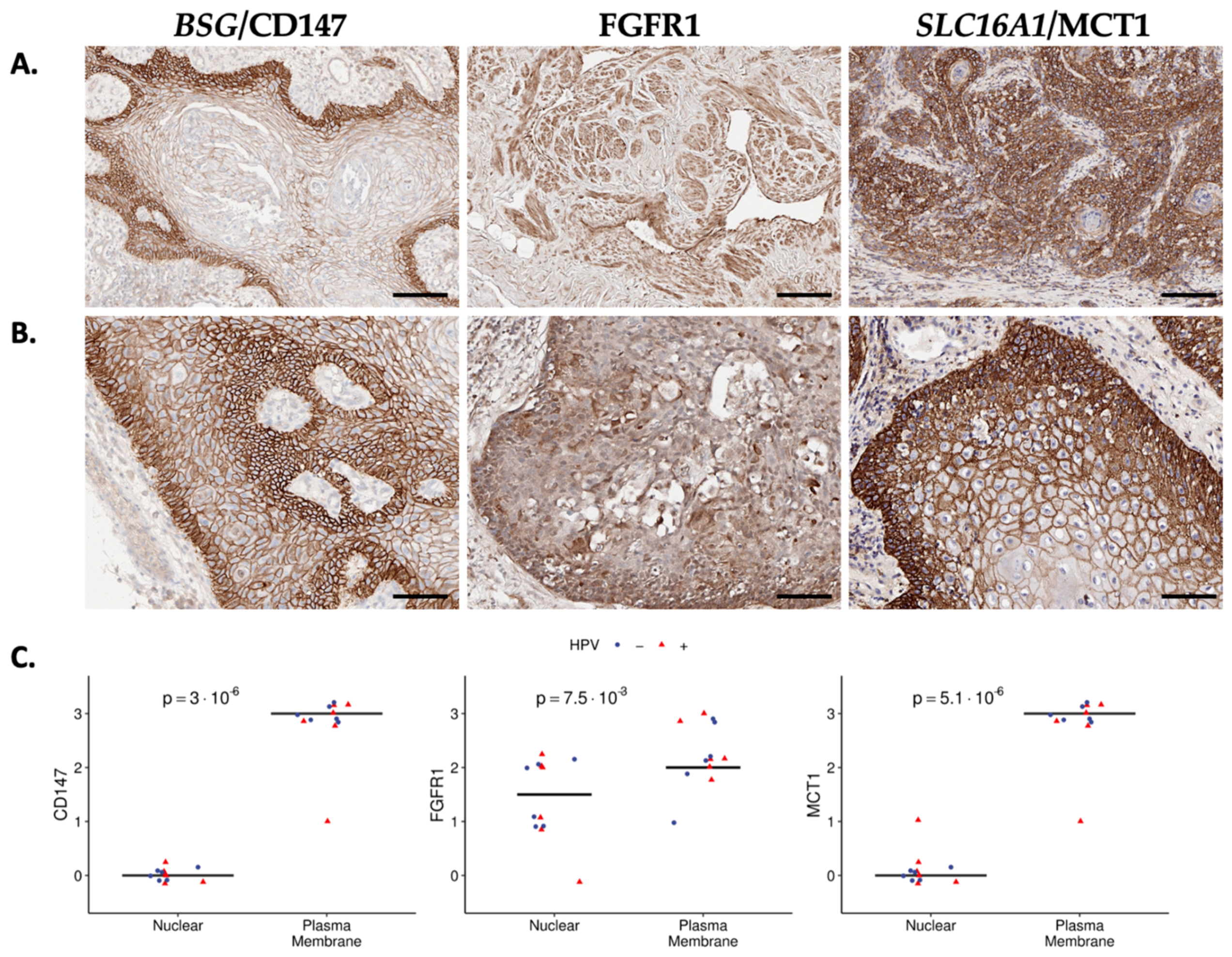
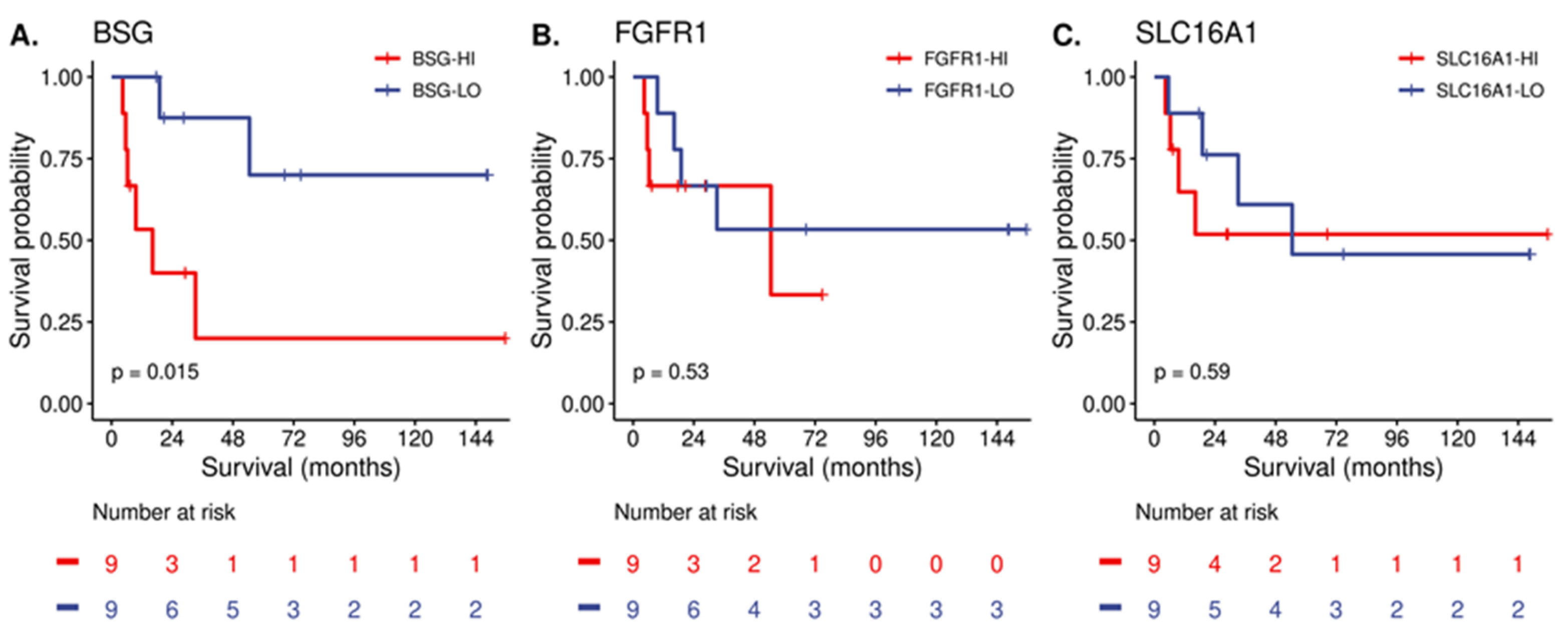
| Characteristic | N = 497 1 |
|---|---|
| Almén Category | |
| Enzymes | 35 (7%) |
| Miscellaneous | 79 (16%) |
| Receptors | 125 (25%) |
| Transporters | 91 (18%) |
| Unclassified | 167 (34%) |
| Characteristic | Q1, N = 239 1 | Q4, N = 497 1 | p-Value 2 |
|---|---|---|---|
| glycomineO_present | 24 (10%) | 111 (22%) | <0.001 |
| glycomineC_present | 45 (19%) | 97 (20%) | 0.5 |
| noncyt. nxst present | 224 (94%) | 483 (97%) | 0.024 |
| Variable | HPV Negative n = 8 | HPV Positive n = 10 | p-Value |
|---|---|---|---|
| Tissue Source | |||
| Penis | 8 (100%) | 10 (100%) | |
| Age at Surgery | 58 (50, 67) | 58 (52, 65) | >0.9 |
| Race | 0.11 | ||
| Asian | 2 (25%) | 0 (0%) | |
| Black | 0 (0%) | 2 (20%) | |
| Hispanic | 1 (12%) | 0 (0%) | |
| White | 5 (62%) | 8 (80%) | |
| Histology | 0.086 | ||
| Basaloid | 0 (0%) | 2 (20%) | |
| Mixed | 0 (0%) | 2 (20%) | |
| Other | 1 (12%) | 0 (0%) | |
| Usual | 5 (62%) | 4 (40%) | |
| Verrucous | 2 (25%) | 0 (0%) | |
| Warty | 0 (0%) | 2 (20%) | |
| LVI | 0.6 | ||
| No | 3 (38%) | 2 (20%) | |
| Yes | 5 (62%) | 8 (80%) | |
| p16 IHC | <0.001 | ||
| Negative | 6 (75%) | 0 (0%) | |
| Positive | 1 (12%) | 10 (100%) | |
| Unknown | 1 (12%) | 0 (0%) | |
| pT | 0.2 | ||
| 1 | 2 (25%) | 3 (30%) | |
| 2 | 4 (50%) | 1 (10%) | |
| 3 | 2 (25%) | 6 (60%) | |
| pN | 0.6 | ||
| 0 | 3 (38%) | 1 (10%) | |
| 1 | 0 (0%) | 1 (10%) | |
| 2 | 4 (50%) | 5 (50%) | |
| 3 | 1 (12%) | 3 (30%) | |
| n (%); median (IQR). | |||
| Wilcoxon rank sum exact test; Fisher’s exact test. | |||
| Almén Category | MCC 3651 (% Difference by HPV Status) |
|---|---|
| Enzymes | 7/33 (21%) |
| Miscellaneous | 13/68 (19%) |
| Receptors | 17/115 (15%) |
| Transporters | 20/80 (25%) |
| Unclassified | 29/156 (19%) |
| Almén Category | N | Total # Drugs | Mean # Drugs | Targets with Drug(s) | Percent Targets with Drug(s) |
|---|---|---|---|---|---|
| Receptors | 115 | 85 | 0.74 | 27 | 23.47 |
| Transporters | 80 | 56 | 0.70 | 18 | 22.50 |
| Unclassified | 222 | 25 | 0.11 | 14 | 6.31 |
| Enzymes | 33 | 12 | 0.36 | 7 | 21.21 |
| Miscellaneous | 69 | 1 | 0.01 | 1 | 1.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grass, G.D.; Ercan, D.; Obermayer, A.N.; Shaw, T.; Stewart, P.A.; Chahoud, J.; Dhillon, J.; Lopez, A.; Johnstone, P.A.S.; Rogatto, S.R.; et al. An Assessment of the Penile Squamous Cell Carcinoma Surfaceome for Biomarker and Therapeutic Target Discovery. Cancers 2023, 15, 3636. https://doi.org/10.3390/cancers15143636
Grass GD, Ercan D, Obermayer AN, Shaw T, Stewart PA, Chahoud J, Dhillon J, Lopez A, Johnstone PAS, Rogatto SR, et al. An Assessment of the Penile Squamous Cell Carcinoma Surfaceome for Biomarker and Therapeutic Target Discovery. Cancers. 2023; 15(14):3636. https://doi.org/10.3390/cancers15143636
Chicago/Turabian StyleGrass, George Daniel, Dalia Ercan, Alyssa N. Obermayer, Timothy Shaw, Paul A. Stewart, Jad Chahoud, Jasreman Dhillon, Alex Lopez, Peter A. S. Johnstone, Silvia Regina Rogatto, and et al. 2023. "An Assessment of the Penile Squamous Cell Carcinoma Surfaceome for Biomarker and Therapeutic Target Discovery" Cancers 15, no. 14: 3636. https://doi.org/10.3390/cancers15143636
APA StyleGrass, G. D., Ercan, D., Obermayer, A. N., Shaw, T., Stewart, P. A., Chahoud, J., Dhillon, J., Lopez, A., Johnstone, P. A. S., Rogatto, S. R., Spiess, P. E., & Eschrich, S. A. (2023). An Assessment of the Penile Squamous Cell Carcinoma Surfaceome for Biomarker and Therapeutic Target Discovery. Cancers, 15(14), 3636. https://doi.org/10.3390/cancers15143636








