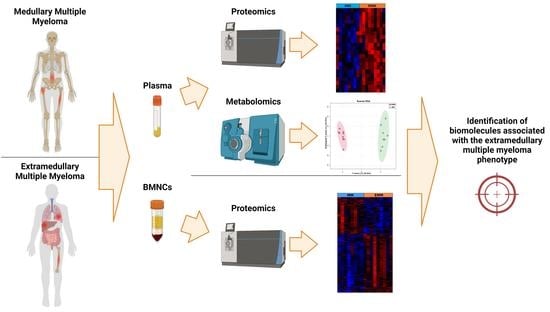
Proteomic and Metabolomic Analysis of Bone Marrow and Plasma from Patients with Extramedullary Multiple Myeloma Identifies Distinct Protein and Metabolite Signatures
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Information and Sample Collection
2.2. Bone Marrow Mononuclear Cells Sample Preparation
2.3. Label-Free Liquid Chromatography—Tandem Mass Spectrometry Analysis of BMNCs
2.4. Data Analysis of BMNCs Mass Spectrometry Results
2.5. Gene Expression Analysis Using the CoMMpass Dataset
2.6. Blood Plasma Sample Preparation
2.7. Label-Free Liquid Chromatography-Tandem Mass Spectrometry Analysis of Plasma
2.8. Data Analysis of Plasma Mass Spectrometry Results
2.9. Enzyme-Linked Immunosorbent Assays (ELISAs)
2.10. Statistical Analysis
2.11. Targeted Metabolomic Analysis
3. Results
3.1. Clinical Information
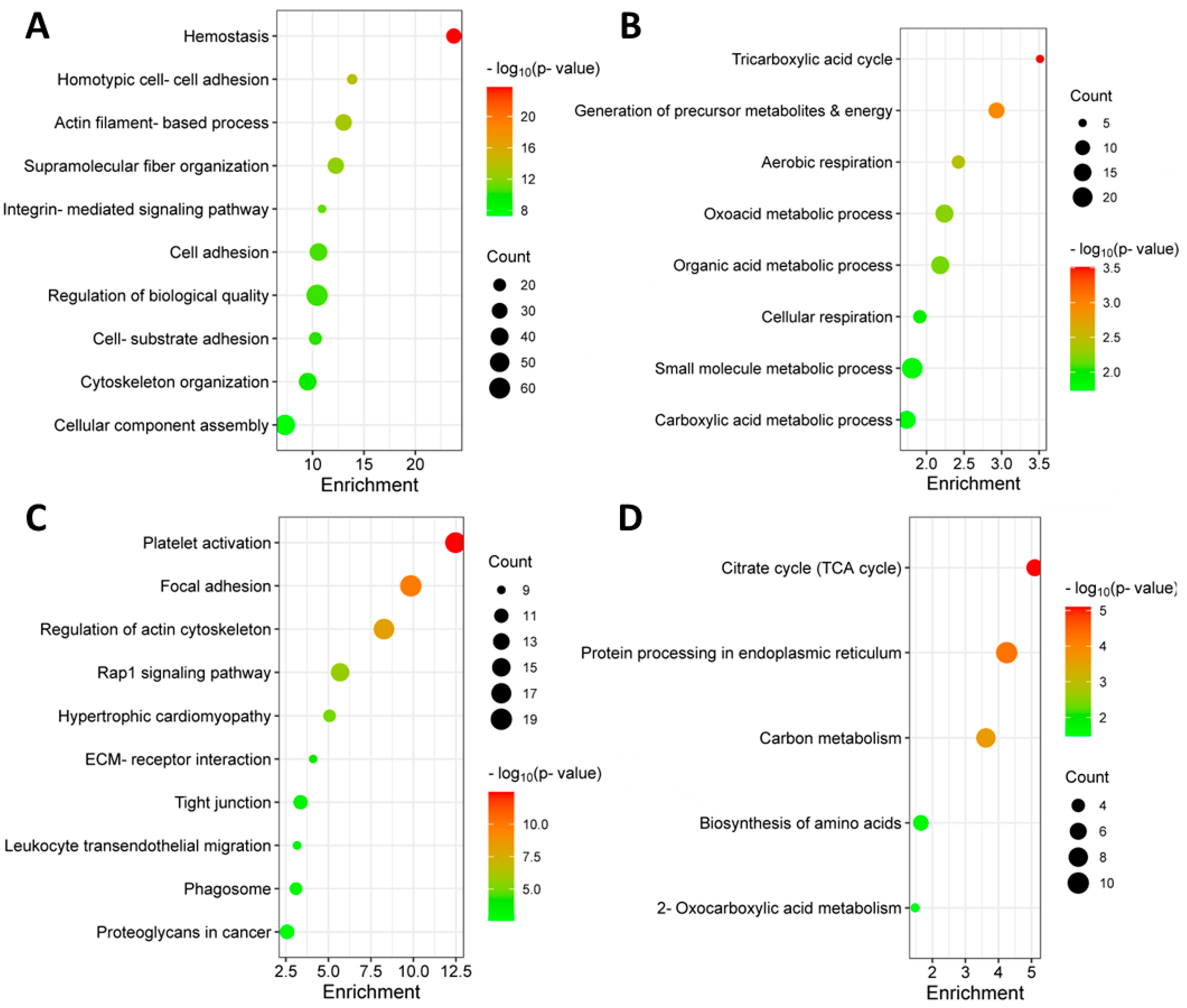
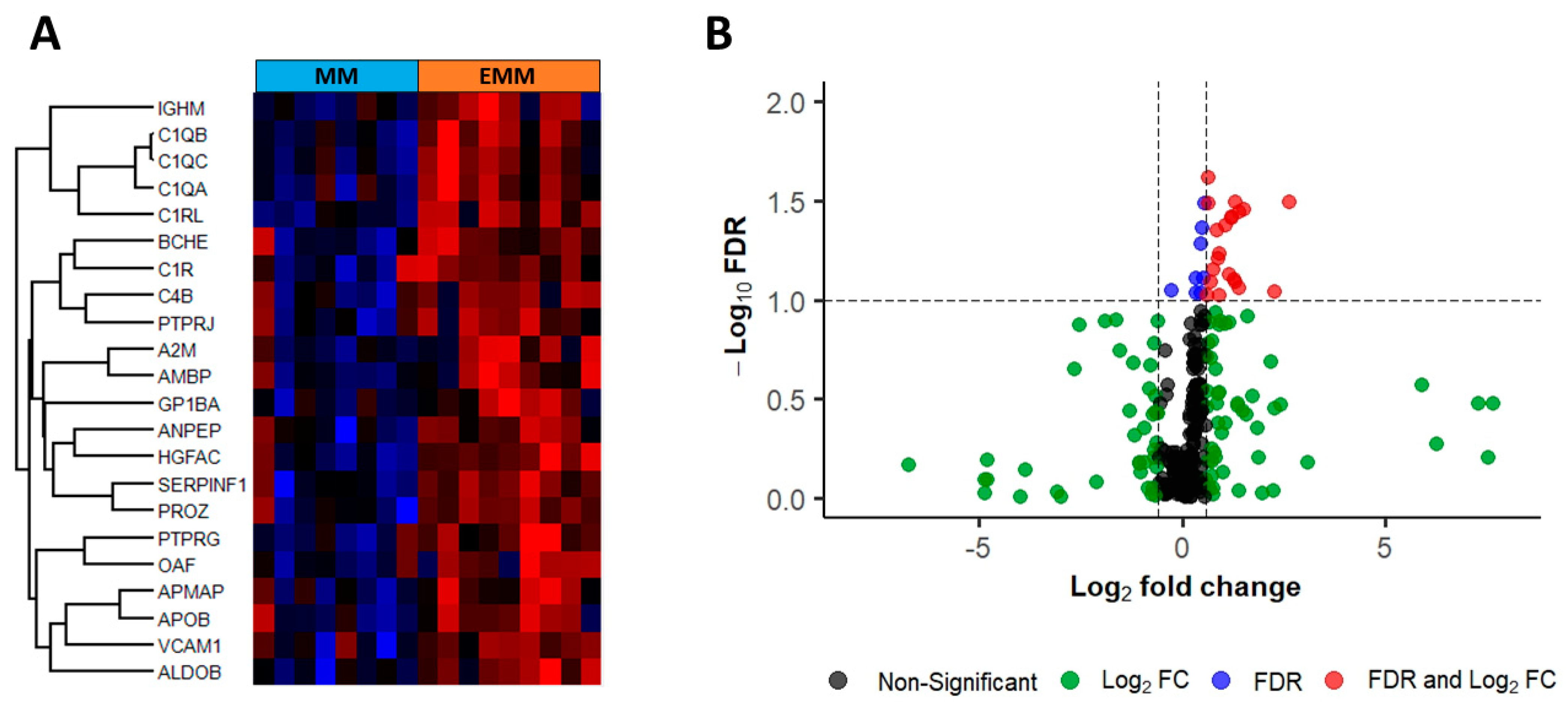
3.2. Identification of Differentially Abundant Proteins in the Bone Marrow of MM Patients with and without Extramedullary Spread
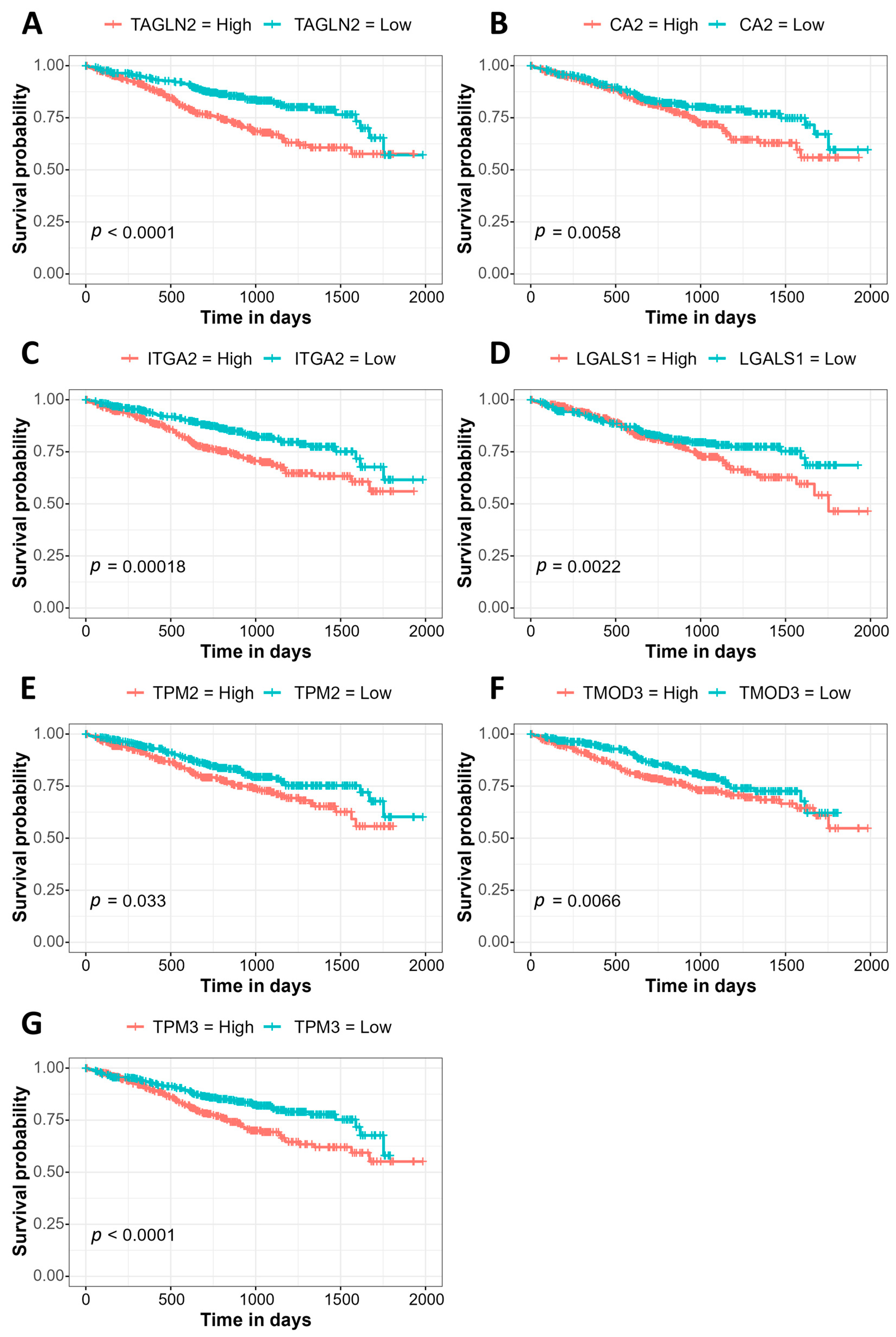
3.3. Association of Gene Expression with Prognosis Using the MMRF CoMMpass Study Data
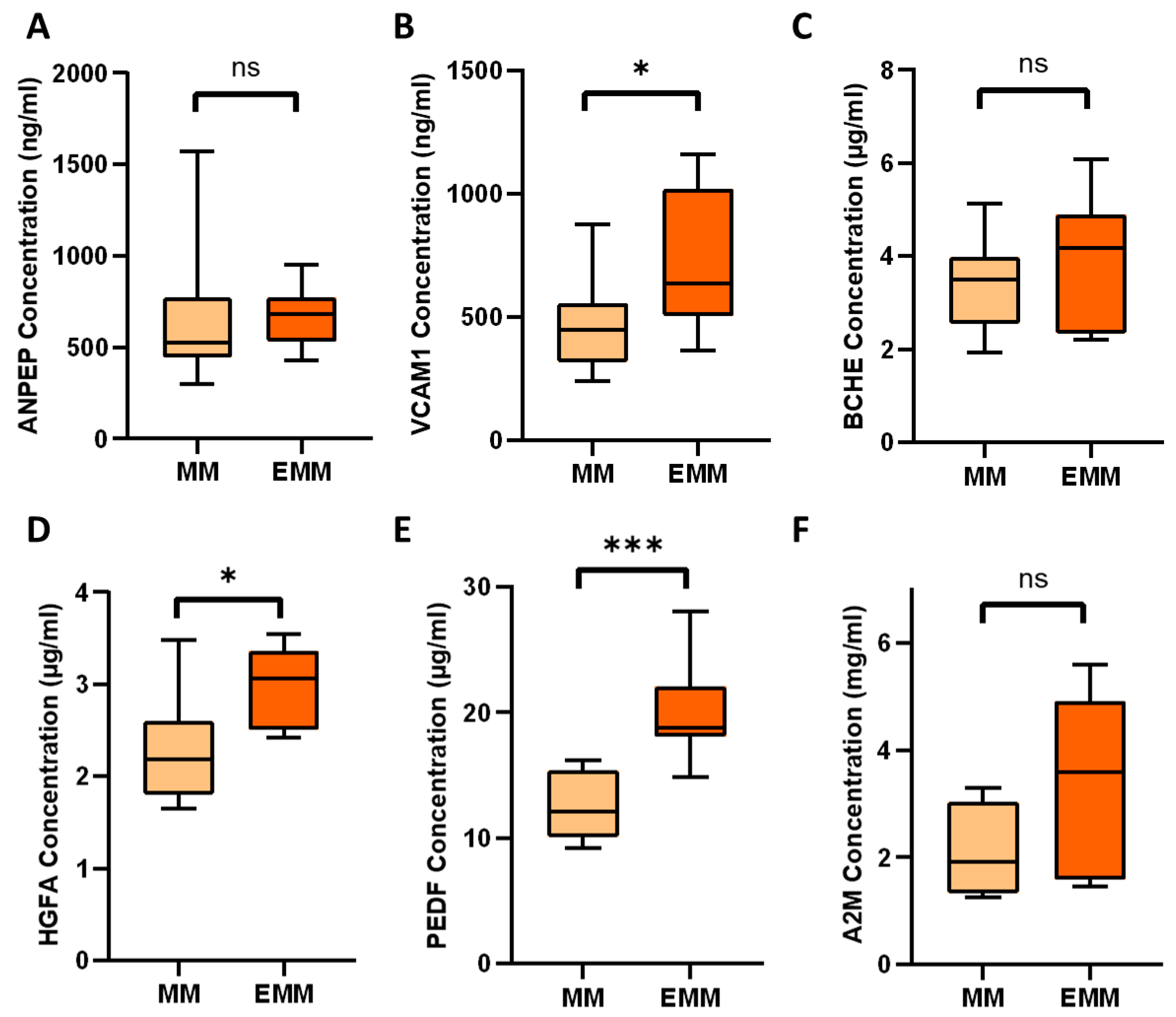
3.4. Identification of Significantly Differentially Abundant Proteins in the Plasma of MM Patients with and without Extramedullary Spread
3.5. Verification of Differentially Expressed Plasma Proteins Identified by LC-MS/MS
3.6. VCAM1 Plasma Concentrations Are Increased in Patients Most Sensitive to the BCL-2 Inhibitors, Venetoclax and Navitoclax
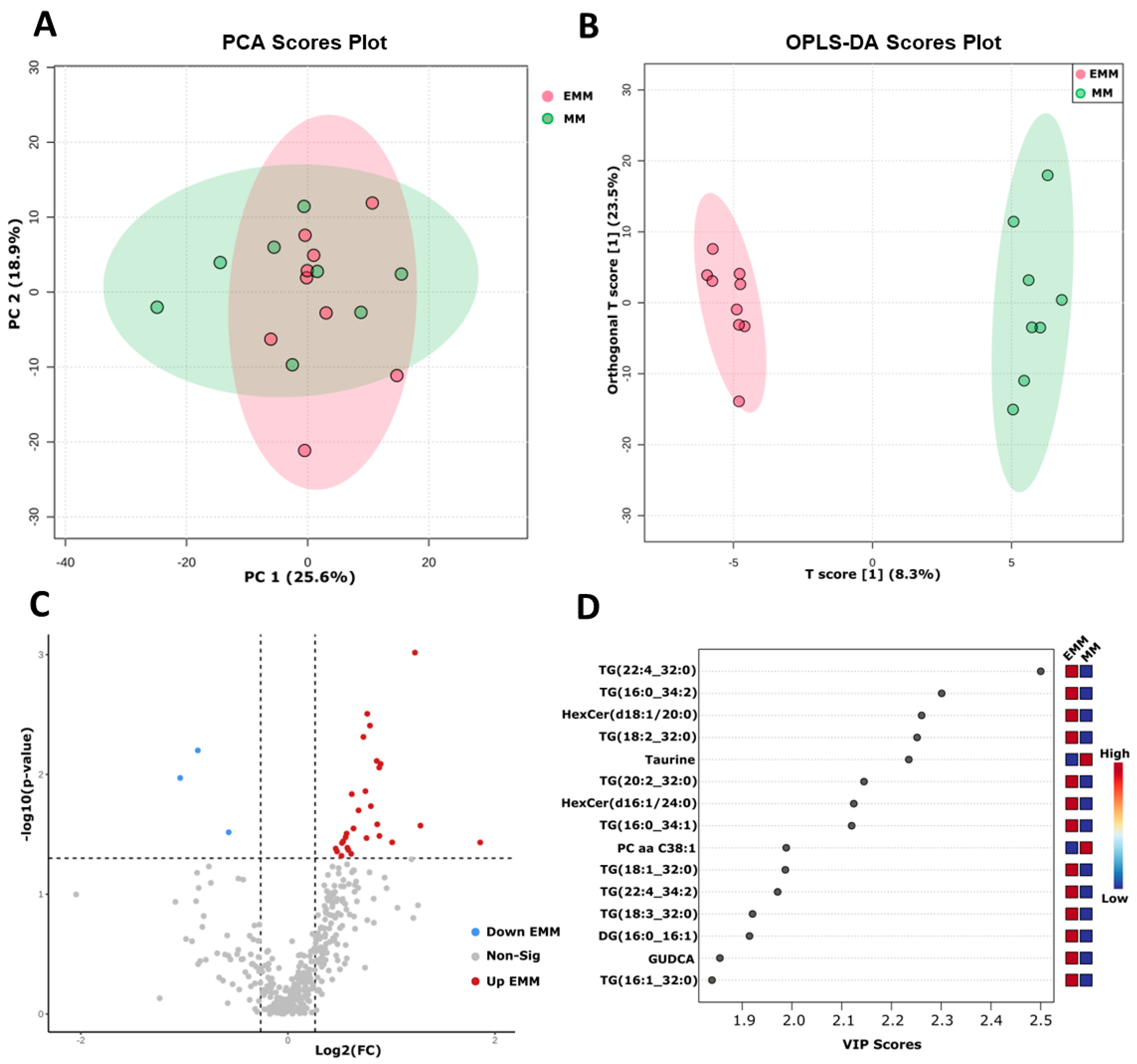
3.7. Targeted Metabolomic Analysis of Blood Plasma from MM Patients with and without Extramedullary Spread
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple Myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef] [PubMed]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimarães, J.E.; Vasconcelos, M.H. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple Myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V. Updated Diagnostic Criteria and Staging System for Multiple Myeloma. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e418–e423. [Google Scholar] [CrossRef]
- Bhutani, M.; Foureau, D.M.; Atrash, S.; Voorhees, P.M.; Usmani, S.Z. Extramedullary Multiple Myeloma. Leukemia 2020, 34, 1–20. [Google Scholar] [CrossRef]
- Gagelmann, N.; Eikema, D.-J.; Iacobelli, S.; Koster, L.; Nahi, H.; Stoppa, A.-M.; Masszi, T.; Caillot, D.; Lenhoff, S.; Udvardy, M.; et al. Impact of Extramedullary Disease in Patients with Newly Diagnosed Multiple Myeloma Undergoing Autologous Stem Cell Transplantation: A Study from the Chronic Malignancies Working Party of the EBMT. Haematologica 2018, 103, 890–897. [Google Scholar] [CrossRef] [Green Version]
- Beksac, M.; Seval, G.C.; Kanellias, N.; Coriu, D.; Rosiñol, L.; Ozet, G.; Goranova-Marinova, V.; Unal, A.; Bila, J.; Ozsan, H.; et al. A Real World Multicenter Retrospective Study on Extramedullary Disease from Balkan Myeloma Study Group and Barcelona University: Analysis of Parameters That Improve Outcome. Haematologica 2020, 105, 201–208. [Google Scholar] [CrossRef]
- Bladé, J.; Beksac, M.; Caers, J.; Jurczyszyn, A.; von Lilienfeld-Toal, M.; Moreau, P.; Rasche, L.; Rosiñol, L.; Usmani, S.Z.; Zamagni, E.; et al. Extramedullary Disease in Multiple Myeloma: A Systematic Literature Review. Blood Cancer J. 2022, 12, 45. [Google Scholar] [CrossRef]
- Moreau, P.; Attal, M.; Caillot, D.; Macro, M.; Karlin, L.; Garderet, L.; Facon, T.; Benboubker, L.; Escoffre-Barbe, M.; Stoppa, A.-M.; et al. Prospective Evaluation of Magnetic Resonance Imaging and [18F]Fluorodeoxyglucose Positron Emission Tomography-Computed Tomography at Diagnosis and Before Maintenance Therapy in Symptomatic Patients with Multiple Myeloma Included in the IFM/DFCI 2009 Trial: Results of the IMAJEM Study. J. Clin. Oncol. 2017, 35, 2911–2918. [Google Scholar] [CrossRef]
- Badar, T.; Srour, S.; Bashir, Q.; Shah, N.; Al-Atrash, G.; Hosing, C.; Popat, U.; Nieto, Y.; Orlowski, R.Z.; Champlin, R.; et al. Predictors of Inferior Clinical Outcome in Patients with Standard-Risk Multiple Myeloma. Eur. J. Haematol. 2017, 98, 263–268. [Google Scholar] [CrossRef]
- Touzeau, C.; Moreau, P. How I Treat Extramedullary Myeloma. Blood 2016, 127, 971–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billecke, L.; Murga Penas, E.M.; May, A.M.; Engelhardt, M.; Nagler, A.; Leiba, M.; Schiby, G.; Kröger, N.; Zustin, J.; Marx, A.; et al. Cytogenetics of Extramedullary Manifestations in Multiple Myeloma. Br. J. Haematol. 2013, 161, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Ortiz, A.; Rodríguez-García, Y.; Encinas, J.; Maroto-Martín, E.; Castellano, E.; Teixidó, J.; Martínez-López, J. The Role of Tumor Microenvironment in Multiple Myeloma Development and Progression. Cancers 2021, 13, 217. [Google Scholar] [CrossRef]
- Solimando, A.; Vià, M.D.; Croci, G.; Borrelli, P.; Tabares, P.; Brandl, A.; Munawar, U.; Steinbrunn, T.; Balduini, A.; Rauert-Wunderlich, H.; et al. OAB-041: Epithelial-Mesenchymal-Transition Regulated by Junctional Adhesion Molecule-A (JAM-A) Associates with Aggressive Extramedullary Multiple Myeloma Disease. Clin. Lymphoma Myeloma Leuk. 2021, 21, S26–S27. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Mishima, Y.; Sacco, A.; Moschetta, M.; Tai, Y.-T.; Shi, J.; Zhang, Y.; Reagan, M.R.; Huynh, D.; Kawano, Y.; et al. CXCR4 Regulates Extra-Medullary Myeloma through Epithelial-Mesenchymal-Transition-like Transcriptional Activation. Cell Rep. 2015, 12, 622–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greipp, P.R.; Leong, T.; Bennett, J.M.; Gaillard, J.P.; Klein, B.; Stewart, J.A.; Oken, M.M.; Kay, N.E.; Van Ness, B.; Kyle, R.A. Plasmablastic Morphology—An Independent Prognostic Factor with Clinical and Laboratory Correlates: Eastern Cooperative Oncology Group (ECOG) Myeloma Trial E9486 Report by the ECOG Myeloma Laboratory Group. Blood 1998, 91, 2501–2507. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jelloul, F.; Zhang, Y.; Bhavsar, T.; Ho, C.; Rao, M.; Lewis, N.E.; Cimera, R.; Baik, J.; Sigler, A.; et al. Genetic Basis of Extramedullary Plasmablastic Transformation of Multiple Myeloma. Am. J. Surg. Pathol. 2020, 44, 838–848. [Google Scholar] [CrossRef]
- Weinstock, M.; Ghobrial, I.M. Extramedullary Multiple Myeloma. Leuk. Lymphoma 2013, 54, 1135–1141. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Forster, S.; Radpour, R. Molecular Impact of the Tumor Microenvironment on Multiple Myeloma Dissemination and Extramedullary Disease. Front. Oncol. 2022, 12, 941437. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.M.; Silvennoinen, R.; Anttila, P.; Tamborero, D.; Eldfors, S.; Yadav, B.; Karjalainen, R.; Kuusanmäki, H.; Lievonen, J.; Parsons, A.; et al. Identification of Precision Treatment Strategies for Relapsed/Refractory Multiple Myeloma by Functional Drug Sensitivity Testing. Oncotarget 2017, 8, 56338–56350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tierney, C.; Bazou, D.; Majumder, M.M.; Anttila, P.; Silvennoinen, R.; Heckman, C.A.; Dowling, P.; O’Gorman, P. Next Generation Proteomics with Drug Sensitivity Screening Identifies Sub-Clones Informing Therapeutic and Drug Development Strategies for Multiple Myeloma Patients. Sci. Rep. 2021, 11, 12866. [Google Scholar] [CrossRef]
- Godzien, J.; Ciborowski, M.; Angulo, S.; Barbas, C. From Numbers to a Biological Sense: How the Strategy Chosen for Metabolomics Data Treatment May Affect Final Results. A Practical Example Based on Urine Fingerprints Obtained by LC-MS. Electrophoresis 2013, 34, 2812–2826. [Google Scholar] [CrossRef]
- An, R.; Yu, H.; Wang, Y.; Lu, J.; Gao, Y.; Xie, X.; Zhang, J. Integrative Analysis of Plasma Metabolomics and Proteomics Reveals the Metabolic Landscape of Breast Cancer. Cancer Metab. 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, S.; Minarik, J.; Stork, M.; Jelinek, T.; Pour, L.; Hajek, R. Extramedullary Disease in Multiple Myeloma—Controversies and Future Directions. Blood Rev. 2019, 36, 32–39. [Google Scholar] [CrossRef]
- Ormond Filho, A.G.; Carneiro, B.C.; Pastore, D.; Silva, I.P.; Yamashita, S.R.; Consolo, F.D.; Hungria, V.T.M.; Sandes, A.F.; Rizzatti, E.G.; Nico, M.A.C. Whole-Body Imaging of Multiple Myeloma: Diagnostic Criteria. RadioGraphics 2019, 39, 1077–1097. [Google Scholar] [CrossRef] [PubMed]
- Gozzetti, A.; Kok, C.H.; Li, C.-F. Editorial: Molecular Mechanisms of Multiple Myeloma. Front. Oncol. 2022, 12, 870123. [Google Scholar] [CrossRef] [PubMed]
- Kriegova, E.; Fillerova, R.; Minarik, J.; Savara, J.; Manakova, J.; Petrackova, A.; Dihel, M.; Balcarkova, J.; Krhovska, P.; Pika, T.; et al. Whole-Genome Optical Mapping of Bone-Marrow Myeloma Cells Reveals Association of Extramedullary Multiple Myeloma with Chromosome 1 Abnormalities. Sci. Rep. 2021, 11, 14671. [Google Scholar] [CrossRef]
- Ryu, D.; Kim, S.J.; Hong, Y.; Jo, A.; Kim, N.; Kim, H.-J.; Lee, H.-O.; Kim, K.; Park, W.-Y. Alterations in the Transcriptional Programs of Myeloma Cells and the Microenvironment during Extramedullary Progression Affect Proliferation and Immune Evasion. Clin. Cancer Res. 2020, 26, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Gregorova, J.; Vychytilova-Faltejskova, P.; Kramarova, T.; Knechtova, Z.; Almasi, M.; Stork, M.; Pour, L.; Kohoutek, J.; Sevcikova, S. Proteomic Analysis of the Bone Marrow Microenvironment in Extramedullary Multiple Myeloma Patients. Neoplasma 2022, 69, 412–424. [Google Scholar] [CrossRef]
- Bou Zerdan, M.; Nasr, L.; Kassab, J.; Saba, L.; Ghossein, M.; Yaghi, M.; Dominguez, B.; Chaulagain, C.P. Adhesion Molecules in Multiple Myeloma Oncogenesis and Targeted Therapy. Int. J. Hematol. Oncol. 2022, 11, IJH39. [Google Scholar] [CrossRef] [PubMed]
- Hathi, D.; Chanswangphuwana, C.; Cho, N.; Fontana, F.; Maji, D.; Ritchey, J.; O’Neal, J.; Ghai, A.; Duncan, K.; Akers, W.J.; et al. Ablation of VLA4 in Multiple Myeloma Cells Redirects Tumor Spread and Prolongs Survival. Sci. Rep. 2022, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Hedvat, C.V.; Comenzo, R.L.; Teruya-Feldstein, J.; Olshen, A.B.; Ely, S.A.; Osman, K.; Zhang, Y.; Kalakonda, N.; Nimer, S.D. Insights into Extramedullary Tumour Cell Growth Revealed by Expression Profiling of Human Plasmacytomas and Multiple Myeloma. Br. J. Haematol. 2003, 122, 728–744. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.; Hoshino, D.; Koshikawa, N.; Akizawa, T.; Seiki, M. Turnover of Focal Adhesions and Cancer Cell Migration. Int. J. Cell Biol. 2012, 2012, e310616. [Google Scholar] [CrossRef] [Green Version]
- Górska, A.; Mazur, A.J. Integrin-Linked Kinase (ILK): The Known vs. the Unknown and Perspectives. Cell. Mol. Life Sci. 2022, 79, 100. [Google Scholar] [CrossRef]
- McDonald, P.C.; Dedhar, S. New Perspectives on the Role of Integrin-Linked Kinase (ILK) Signaling in Cancer Metastasis. Cancers 2022, 14, 3209. [Google Scholar] [CrossRef]
- Nikou, S.; Arbi, M.; Dimitrakopoulos, F.-I.D.; Sirinian, C.; Chadla, P.; Pappa, I.; Ntaliarda, G.; Stathopoulos, G.T.; Papadaki, H.; Zolota, V.; et al. Integrin-Linked Kinase (ILK) Regulates KRAS, IPP Complex and Ras Suppressor-1 (RSU1) Promoting Lung Adenocarcinoma Progression and Poor Survival. J. Mol. Hist. 2020, 51, 385–400. [Google Scholar] [CrossRef]
- Tsoumas, D.; Nikou, S.; Giannopoulou, E.; Tsaniras, S.C.; Sirinian, C.; Maroulis, I.; Taraviras, S.; Zolota, V.; Kalofonos, H.P.; Bravou, V. ILK Expression in Colorectal Cancer Is Associated with EMT, Cancer Stem Cell Markers and Chemoresistance. Cancer Genom. Proteom. 2018, 15, 127–141. [Google Scholar]
- Chen, D.; Zhang, Y.; Zhang, X.; Li, J.; Han, B.; Liu, S.; Wang, L.; Ling, Y.; Mao, S.; Wang, X. Overexpression of Integrin-Linked Kinase Correlates with Malignant Phenotype in Non-Small Cell Lung Cancer and Promotes Lung Cancer Cell Invasion and Migration via Regulating Epithelial–Mesenchymal Transition (EMT)-Related Genes. Acta Histochem. 2013, 115, 128–136. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Z.; Yao, C. Targeting Integrin-Linked Kinase Increases Apoptosis and Decreases Invasion of Myeloma Cell Lines and Inhibits IL-6 and VEGF Secretion from BMSCs. Med. Oncol. 2011, 28, 1596–1600. [Google Scholar] [CrossRef] [PubMed]
- Steinbrunn, T.; Siegmund, D.; Andrulis, M.; Grella, E.; Kortüm, M.; Einsele, H.; Wajant, H.; Bargou, R.C.; Stühmer, T. Integrin-Linked Kinase Is Dispensable for Multiple Myeloma Cell Survival. Leuk. Res. 2012, 36, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Y.; Liu, H.; Xu, X.; He, S.; Tang, J.; Huang, Y.; Miao, X.; Wu, Y.; Wang, Q.; et al. Pyruvate Kinase Isoform M2 (PKM2) Participates in Multiple Myeloma Cell Proliferation, Adhesion and Chemoresistance. Leuk. Res. 2015, 39, 1428–1436. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, S.; Liu, J.; Tian, Y.; Ma, B.; Xu, S.; Fu, Y.; Luo, Y. Secreted Pyruvate Kinase M2 Promotes Lung Cancer Metastasis through Activating the Integrin Beta1/FAK Signaling Pathway. Cell Rep. 2020, 30, 1780–1797.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dah, K.; Lavezo, J.L.; Dihowm, F. Aggressive Plasmablastic Myeloma with Extramedullary Cord Compression and Hyperammonemic Encephalopathy: Case Report and Literature Review. Anticancer Res. 2021, 41, 5839–5845. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Luderer, M.; Azab, A.K. Hypoxia Promotes Stem Cell-like Phenotype in Multiple Myeloma Cells. Blood Cancer J. 2014, 4, e262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, K.; Ramani, V.C.; Bandari, S.K.; Amin, R.; Brown, E.E.; Ritchie, J.P.; Stewart, M.D.; Sanderson, R.D. Heparanase Promotes Myeloma Stemness and in Vivo Tumorigenesis. Matrix Biol. 2020, 88, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Jung, O.; Trapp-Stamborski, V.; Purushothaman, A.; Jin, H.; Wang, H.; Sanderson, R.D.; Rapraeger, A.C. Heparanase-Induced Shedding of Syndecan-1/CD138 in Myeloma and Endothelial Cells Activates VEGFR2 and an Invasive Phenotype: Prevention by Novel Synstatins. Oncogenesis 2016, 5, e202. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Bi, J.; Liang, Q.; Wang, S.; Zhang, L.; Han, F.; Li, S.; Qiu, B.; Fan, X.; Chen, W.; et al. VCAM1 Promotes Tumor Cell Invasion and Metastasis by Inducing EMT and Transendothelial Migration in Colorectal Cancer. Front. Oncol. 2020, 10, 1066. [Google Scholar] [CrossRef]
- Hao, P.; Zhang, C.; Wang, R.; Yan, P.; Peng, R. Expression and Pathogenesis of VCAM-1 and VLA-4 Cytokines in Multiple Myeloma. Saudi J. Biol. Sci. 2020, 27, 1674–1678. [Google Scholar] [CrossRef]
- Terpos, E.; Migkou, M.; Christoulas, D.; Gavriatopoulou, M.; Eleutherakis-Papaiakovou, E.; Kanellias, N.; Iakovaki, M.; Panagiotidis, I.; Ziogas, D.C.; Fotiou, D.; et al. Increased Circulating VCAM-1 Correlates with Advanced Disease and Poor Survival in Patients with Multiple Myeloma: Reduction by Post-Bortezomib and Lenalidomide Treatment. Blood Cancer J. 2016, 6, e428. [Google Scholar] [CrossRef] [Green Version]
- Okugawa, Y.; Miki, C.; Toiyama, Y.; Koike, Y.; Yokoe, T.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Kusunoki, M. Soluble VCAM-1 and Its Relation to Disease Progression in Colorectal Carcinoma. Exp. Ther. Med. 2010, 1, 463–469. [Google Scholar] [CrossRef] [Green Version]
- Liao, T.; Chen, W.; Sun, J.; Zhang, Y.; Hu, X.; Yang, S.; Qiu, H.; Li, S.; Chu, T. CXCR4 Accelerates Osteoclastogenesis Induced by Non-Small Cell Lung Carcinoma Cells Through Self-Potentiation and VCAM1 Secretion. CPB 2018, 50, 1084–1099. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, D.; Karayiannakis, A.J.; Syrigos, K.N.; Zbar, A.; Kremmyda, A.; Bramis, I.; Tsigris, C. Serum Levels of E-Selectin, ICAM-1 and VCAM-1 in Colorectal Cancer Patients: Correlations with Clinicopathological Features, Patient Survival and Tumour Surgery. Eur. J. Cancer 2001, 37, 2392–2397. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.-B.; Chen, G.-Y.; Xia, J.-G.; Zang, X.-W.; Yang, H.-Y.; Yang, L. Association of VCAM-1 Overexpression with Oncogenesis, Tumor Angiogenesis and Metastasis of Gastric Carcinoma. World J. Gastroenterol. 2003, 9, 1409–1414. [Google Scholar] [CrossRef]
- Kitani, A.; Nakashima, N.; Izumihara, T.; Inagaki, M.; Baoui, X.; Yu, S.; Matsuda, T.; Matsuyama, T. Soluble VCAM-1 Induces Chemotaxis of Jurkat and Synovial Fluid T Cells Bearing High Affinity Very Late Antigen-4. J. Immunol. 1998, 161, 4931–4938. [Google Scholar] [CrossRef]
- Roy, P.; Sarkar, U.A.; Basak, S. The NF-ΚB Activating Pathways in Multiple Myeloma. Biomedicines 2018, 6, 59. [Google Scholar] [CrossRef] [Green Version]
- Demchenko, Y.N.; Glebov, O.K.; Zingone, A.; Keats, J.J.; Bergsagel, P.L.; Kuehl, W.M. Classical and/or Alternative NF-ΚB Pathway Activation in Multiple Myeloma. Blood 2010, 115, 3541–3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becerra, S.P.; Notario, V. The Effects of PEDF on Cancer Biology: Mechanisms of Action and Therapeutic Potential. Nat. Rev. Cancer 2013, 13, 258–271. [Google Scholar] [CrossRef] [Green Version]
- Seki, R.; Yoshida, T.; Nakamura, K.; Yamagishi, S.; Imaizumi, T.; Okamura, T.; Sata, M. Pigment Epithelium-Derived Factor (PEDF) Inhibits Multiple Myeloma through Suppressing NADPH Oxidase ROS Generation. Blood 2005, 106, 3389. [Google Scholar] [CrossRef]
- Seki, R.; Yamagishi, S.; Matsui, T.; Yoshida, T.; Torimura, T.; Ueno, T.; Sata, M.; Okamura, T. Pigment Epithelium-Derived Factor (PEDF) Inhibits Survival and Proliferation of VEGF-Exposed Multiple Myeloma Cells through Its Anti-Oxidative Properties. Biochem. Biophys. Res. Commun. 2013, 431, 693–697. [Google Scholar] [CrossRef]
- Chen, Z.; Che, D.; Gu, X.; Lin, J.; Deng, J.; Jiang, P.; Xu, K.; Xu, B.; Zhang, T. Upregulation of PEDF Predicts a Poor Prognosis and Promotes Esophageal Squamous Cell Carcinoma Progression by Modulating the MAPK/ERK Signaling Pathway. Front. Oncol. 2021, 11, 625612. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Ge, C.; Cui, M.; Liu, T.; Liu, X.; Tian, H.; Zhao, F.; Chen, T.; Cui, Y.; Yao, M.; et al. Pigment Epithelium-Derived Factor Promotes Tumor Metastasis through an Interaction with Laminin Receptor in Hepatocellular Carcinomas. Cell Death Dis. 2017, 8, e2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abooshahab, R.; Al-Salami, H.; Dass, C.R. The Increasing Role of Pigment Epithelium-Derived Factor in Metastasis: From Biological Importance to a Promising Target. Biochem. Pharmacol. 2021, 193, 114787. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, S.; Tanaka, G.; Takagane, K.; Itoh, G.; Tanaka, M. Pigment Epithelium Derived Factor Is Involved in the Late Phase of Osteosarcoma Metastasis by Increasing Extravasation and Cell-Cell Adhesion. Front. Oncol. 2022, 12, 818182. [Google Scholar] [CrossRef]
- Tjin, E.P.M.; Derksen, P.W.B.; Kataoka, H.; Spaargaren, M.; Pals, S.T. Multiple Myeloma Cells Catalyze Hepatocyte Growth Factor (HGF) Activation by Secreting the Serine Protease HGF-Activator. Blood 2004, 104, 2172–2175. [Google Scholar] [CrossRef]
- Wader, K.F.; Fagerli, U.M.; Holt, R.U.; Stordal, B.; Børset, M.; Sundan, A.; Waage, A. Elevated Serum Concentrations of Activated Hepatocyte Growth Factor Activator in Patients with Multiple Myeloma. Eur. J. Haematol. 2008, 81, 380–383. [Google Scholar] [CrossRef] [Green Version]
- Giannoni, P.; de Totero, D. The HGF/c-MET Axis as a Potential Target to Overcome Survival Signals and Improve Therapeutic Efficacy in Multiple Myeloma. Cancer Drug Resist. 2021, 4, 923–933. [Google Scholar] [CrossRef]
- Ramani, V.C.; Yang, Y.; Ren, Y.; Nan, L.; Sanderson, R.D. Heparanase Plays a Dual Role in Driving Hepatocyte Growth Factor (HGF) Signaling by Enhancing HGF Expression and Activity. J. Biol. Chem. 2011, 286, 6490–6499. [Google Scholar] [CrossRef] [Green Version]
- Misselwitz, B.; Goede, J.S.; Pestalozzi, B.C.; Schanz, U.; Seebach, J.D. Hyperlipidemic Myeloma: Review of 53 Cases. Ann. Hematol. 2010, 89, 569–577. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, U.; Umar, Z.; Pansuriya, A.M.; Mahmood, A.; Lopez, R. Multiple Myeloma with Retroperitoneal Extramedullary Plasmacytoma Causing Renal Failure and Obstructive Shock From Inferior Vena Cava Compression: A Case Report. Cureus 2022, 14, e31056. [Google Scholar] [CrossRef]
- Shimokihara, K.; Kawahara, T.; Chiba, S.; Takamoto, D.; Yao, M.; Uemura, H. Extramedullary Plasmacytoma of the Testis: A Case Report. Urol. Case Rep. 2018, 16, 101–103. [Google Scholar] [CrossRef]
- Stork, M.; Sevcikova, S.; Minarik, J.; Krhovska, P.; Radocha, J.; Pospisilova, L.; Brozova, L.; Jarkovsky, J.; Spicka, I.; Straub, J.; et al. Identification of Patients at High Risk of Secondary Extramedullary Multiple Myeloma Development. Br. J. Haematol. 2022, 196, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Lazaris, V.; Hatziri, A.; Symeonidis, A.; Kypreos, K.E. The Lipoprotein Transport System in the Pathogenesis of Multiple Myeloma: Advances and Challenges. Front. Oncol. 2021, 11, 638288. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.; Fairfield, H.; Karam, M.; D’Amico, A.; Murphy, C.S.; Falank, C.; Pistofidi, R.S.; Cao, A.; Marinac, C.R.; Dragon, J.A.; et al. Targeting the Fatty Acid Binding Proteins Disrupts Multiple Myeloma Cell Cycle Progression and MYC Signaling. eLife 2023, 12, e81184. [Google Scholar] [CrossRef] [PubMed]
- Kobari, L.; Auclair, M.; Piau, O.; Ferrand, N.; Zaoui, M.; Delhommeau, F.; Fève, B.; Sabbah, M.; Garderet, L. Circulating Cytokines Present in Multiple Myeloma Patients Inhibit the Osteoblastic Differentiation of Adipose Stem Cells. Leukemia 2022, 36, 540–548. [Google Scholar] [CrossRef]
- Ponvilawan, B.; Charoenngam, N.; Rittiphairoj, T.; Ungprasert, P. Receipt of Statins Is Associated with Lower Risk of Multiple Myeloma: Systematic Review and Meta-Analysis. Clin. Lymphoma Myeloma Leuk. 2020, 20, e399–e413. [Google Scholar] [CrossRef] [PubMed]
- Brånvall, E.; Ekberg, S.; Eloranta, S.; Wästerlid, T.; Birmann, B.M.; Smedby, K.E. Statin Use Is Associated with Improved Survival in Multiple Myeloma: A Swedish Population-Based Study of 4315 Patients. Am. J. Hematol. 2020, 95, 652–661. [Google Scholar] [CrossRef]
- Gohlke, B.; Zincke, F.; Eckert, A.; Kobelt, D.; Preissner, S.; Liebeskind, J.M.; Gunkel, N.; Putzker, K.; Lewis, J.; Preissner, S.; et al. Real-world Evidence for Preventive Effects of Statins on Cancer Incidence: A Trans-Atlantic Analysis. Clin. Transl. Med. 2022, 12, e726. [Google Scholar] [CrossRef]
- Juneja, M.; Kobelt, D.; Walther, W.; Voss, C.; Smith, J.; Specker, E.; Neuenschwander, M.; Gohlke, B.-O.; Dahlmann, M.; Radetzki, S.; et al. Statin and Rottlerin Small-Molecule Inhibitors Restrict Colon Cancer Progression and Metastasis via MACC1. PLoS Biol. 2017, 15, e2000784. [Google Scholar] [CrossRef] [Green Version]
- Lonial, S.; Lee, H.C.; Badros, A.; Trudel, S.; Nooka, A.K.; Chari, A.; Abdallah, A.; Callander, N.; Sborov, D.; Suvannasankha, A.; et al. Longer Term Outcomes with Single-agent Belantamab Mafodotin in Patients with Relapsed or Refractory Multiple Myeloma: 13-month Follow-up from the Pivotal DREAMM-2 Study. Cancer 2021, 127, 4198–4212. [Google Scholar] [CrossRef]
- Rosiñol, L.; Beksac, M.; Zamagni, E.; Van de Donk, N.W.C.J.; Anderson, K.C.; Badros, A.; Caers, J.; Cavo, M.; Dimopoulos, M.-A.; Dispenzieri, A.; et al. Expert Review on Soft-Tissue Plasmacytomas in Multiple Myeloma: Definition, Disease Assessment and Treatment Considerations. Br. J. Haematol. 2021, 194, 496–507. [Google Scholar] [CrossRef]
- Jelinek, T.; Sevcikova, T.; Zihala, D.; Popkova, T.; Kapustova, V.; Broskevicova, L.; Capkova, L.; Rihova, L.; Bezdekova, R.; Sevcikova, S.; et al. Limited Efficacy of Daratumumab in Multiple Myeloma with Extramedullary Disease. Leukemia 2022, 36, 288–291. [Google Scholar] [CrossRef]
- Li, W.; Liu, M.; Yuan, T.; Yan, L.; Cui, R.; Deng, Q. Efficacy and Follow-up of Humanized Anti-BCMA CAR-T Cell Therapy in Relapsed/Refractory Multiple Myeloma Patients with Extramedullary-Extraosseous, Extramedullary-Bone Related, and without Extramedullary Disease. Hematol. Oncol. 2022, 40, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.; Xiao, A.; Yi, D.; Zanwar, S.; Bianchi, G. Treating Multiple Myeloma in the Context of the Bone Marrow Microenvironment. Curr. Oncol. 2022, 29, 8975–9005. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Chatterjee, M.; Grasso, M.; Specchia, G.; Magen, H.; Einsele, H.; Celeghini, I.; Barbieri, P.; Paoletti, D.; Pace, S.; et al. Phase I Study of the Heparanase Inhibitor Roneparstat: An Innovative Approach for Ultiple Myeloma Therapy. Haematologica 2018, 103, e469–e472. [Google Scholar] [CrossRef]
- Federico, C.; Alhallak, K.; Sun, J.; Duncan, K.; Azab, F.; Sudlow, G.P.; de la Puente, P.; Muz, B.; Kapoor, V.; Zhang, L.; et al. Tumor Microenvironment-Targeted Nanoparticles Loaded with Bortezomib and ROCK Inhibitor Improve Efficacy in Multiple Myeloma. Nat. Commun. 2020, 11, 6037. [Google Scholar] [CrossRef]
- Sidiqi, M.H.; Al Saleh, A.S.; Kumar, S.K.; Leung, N.; Jevremovic, D.; Muchtar, E.; Gonsalves, W.I.; Kourelis, T.V.; Warsame, R.; Buadi, F.K.; et al. Venetoclax for the Treatment of Multiple Myeloma: Outcomes Outside of Clinical Trials. Am. J. Hematol. 2021, 96, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, L.M.; Maxcy, K.L.; LaBelle, J.L. Flow Cytometry-Based Detection and Analysis of BCL-2 Family Proteins and Mitochondrial Outer Membrane Permeabilization (MOMP). Methods Mol. Biol. 2019, 1877, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Ackler, S.; Mitten, M.J.; Foster, K.; Oleksijew, A.; Refici, M.; Tahir, S.K.; Xiao, Y.; Tse, C.; Frost, D.J.; Fesik, S.W.; et al. The Bcl-2 Inhibitor ABT-263 Enhances the Response of Multiple Chemotherapeutic Regimens in Hematologic Tumors in Vivo. Cancer Chemother. Pharmacol. 2010, 66, 869–880. [Google Scholar] [CrossRef]
- Yee, A.J.; Huff, C.A.; Chari, A.; Vogl, D.T.; Gavriatopoulou, M.; Nooka, A.K.; Moreau, P.; Dingli, D.; Cole, C.E.; Lonial, S.; et al. Response to Therapy and the Effectiveness of Treatment with Selinexor and Dexamethasone in Patients with Penta-Exposed Triple-Class Refractory Myeloma Who Had Plasmacytomas. Blood 2019, 134, 3140. [Google Scholar] [CrossRef]
- Bahlis, N.J.; Sutherland, H.; White, D.; Sebag, M.; Lentzsch, S.; Kotb, R.; Venner, C.P.; Gasparetto, C.; Del Col, A.; Neri, P.; et al. Selinexor plus Low-Dose Bortezomib and Dexamethasone for Patients with Relapsed or Refractory Multiple Myeloma. Blood 2018, 132, 2546–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, P.G.; Oriol, A.; Larocca, A.; Bladé, J.; Cavo, M.; Rodriguez-Otero, P.; Leleu, X.; Nadeem, O.; Hiemenz, J.W.; Hassoun, H.; et al. Melflufen and Dexamethasone in Heavily Pretreated Relapsed and Refractory Multiple Myeloma. J. Clin. Oncol. 2021, 39, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, J.J.; Kumari, R.; Traustadottir, G.A.; Huppunen, M.-E.; Sergeev, P.; Majumder, M.M.; Schepsky, A.; Gudjonsson, T.; Lievonen, J.; Bazou, D.; et al. Aminopeptidase Expression in Multiple Myeloma Associates with Disease Progression and Sensitivity to Melflufen. Cancers 2021, 13, 1527. [Google Scholar] [CrossRef]
- Sriskandarajah, P.; De Haven Brandon, A.; MacLeod, K.; Carragher, N.O.; Kirkin, V.; Kaiser, M.; Whittaker, S.R. Combined Targeting of MEK and the Glucocorticoid Receptor for the Treatment of RAS-Mutant Multiple Myeloma. BMC Cancer 2020, 20, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Master, S.; Graham, C. Extramedullary Multiple Myeloma: A Patient-Focused Review of the Pathogenesis of Bone Marrow Escape. World J. Oncol. 2022, 13, 311–319. [Google Scholar] [CrossRef]
- Dahl, I.M.S.; Rasmussen, T.; Kauric, G.; Husebekk, A. Differential Expression of CD56 and CD44 in the Evolution of Extramedullary Myeloma. Br. J. Haematol. 2002, 116, 273–277. [Google Scholar] [CrossRef]
- Mey, U.J.M.; Renner, C.; von Moos, R. Vemurafenib in Combination with Cobimetinib in Relapsed and Refractory Extramedullary Multiple Myeloma Harboring the BRAF V600E Mutation. Hematol. Oncol. 2017, 35, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Giesen, N.; Chatterjee, M.; Scheid, C.; Poos, A.M.; Besemer, B.; Miah, K.; Benner, A.; Becker, N.; Moehler, T.; Metzler, I.; et al. A Phase 2 Clinical Trial of Combined BRAF/MEK Inhibition for BRAFV600E-Mutated Multiple Myeloma. Blood 2023, 141, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Mariño, K.V.; Cagnoni, A.J.; Croci, D.O.; Rabinovich, G.A. Targeting Galectin-Driven Regulatory Circuits in Cancer and Fibrosis. Nat. Rev. Drug Discov. 2023, 22, 295–316. [Google Scholar] [CrossRef]
- Storti, P.; Marchica, V.; Giuliani, N. Role of Galectins in Multiple Myeloma. Int. J. Mol. Sci. 2017, 18, 2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storti, P.; Marchica, V.; Airoldi, I.; Donofrio, G.; Fiorini, E.; Ferri, V.; Guasco, D.; Todoerti, K.; Silbermann, R.; Anderson, J.L.; et al. Galectin-1 Suppression Delineates a New Strategy to Inhibit Myeloma-Induced Angiogenesis and Tumoral Growth in Vivo. Leukemia 2016, 30, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.; Lee, S.J.; Arai, S.; Rotta, M.; Zoghi, B.; Lazaryan, A.; Ramakrishnan, A.; DeFilipp, Z.; Salhotra, A.; Chai-Ho, W.; et al. Belumosudil for Chronic Graft-versus-Host Disease after 2 or More Prior Lines of Therapy: The ROCKstar Study. Blood 2021, 138, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Kalra, J.; Warburton, C.; Fang, K.; Edwards, L.; Daynard, T.; Waterhouse, D.; Dragowska, W.; Sutherland, B.W.; Dedhar, S.; Gelmon, K.; et al. QLT0267, a Small Molecule Inhibitor Targeting Integrin-Linked Kinase (ILK), and Docetaxel Can Combine to Produce Synergistic Interactions Linked to Enhanced Cytotoxicity, Reductions in P-AKT Levels, Altered F-Actin Architecture and Improved Treatment Outcomes in an Orthotopic Breast Cancer Model. Breast Cancer Res. 2009, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- García-Marín, J.; Rodríguez-Puyol, D.; Vaquero, J.J. Insight into the Mechanism of Molecular Recognition between Human Integrin-Linked Kinase and Cpd22 and Its Implication at Atomic Level. J. Comput. Aided Mol. Des. 2022, 36, 575–589. [Google Scholar] [CrossRef] [PubMed]



| Sample ID | Diagnosis | Status | Sex | Age | OS (mo) from Diagnosis |
|---|---|---|---|---|---|
| D_EMM_2689 | Myeloma, extramedullary | Diagnostic | Male | 65 | 80 |
| D_EMM_3497 | Myeloma, extramedullary | Diagnostic | Male | 65 | 87 * |
| D_EMM_3674 | Myeloma, extramedullary | Diagnostic | Male | 58 | 8 |
| D_EMM_4296 | Myeloma, extramedullary | Diagnostic | Male | 65 | 22 |
| D_EMM_1994 | Myeloma, extramedullary | Diagnostic | Female | 67 | 16 |
| D_EMM_40725 | Myeloma, extramedullary | Diagnostic | Male | 49 | 2 |
| PD_EMM_874 | Myeloma, extramedullary | Progressive disease | Male | 72 | 31 |
| PD_EMM_1994 † | Myeloma, extramedullary | Progressive disease | Female | 68 | 16 |
| PD_EMM_40795 | Myeloma, extramedullary | Progressive disease | Female | 69 | 7 |
| D_MM_5215 | Myeloma, no extramedullary | Diagnostic | Male | 65 | 61 * |
| D_MM_4314 | Myeloma, no extramedullary | Diagnostic | Male | 65 | 53 |
| D_MM_5187 | Myeloma, no extramedullary | Diagnostic | Male | 59 | 62 * |
| D_MM_4317 | Myeloma, no extramedullary | Diagnostic | Male | 65 | 65 |
| D_MM_40141 | Myeloma, no extramedullary | Diagnostic | Male | 49 | 43 * |
| PD_MM_899 | Myeloma, no extramedullary | Progressive disease | Male | 72 | 124 |
| PD_MM_1579 | Myeloma, no extramedullary | Progressive disease | Female | 68 | 83 |
| PD_MM_40301 | Myeloma, no extramedullary | Progressive disease | Female | 70 | 129 * |
| Uniprot ID | Description | Gene Name | Fold Change | FDR-Adjusted p-Value |
|---|---|---|---|---|
| P00918 | Carbonic anhydrase 2 | CA2 | 4.42 | 0.0001 |
| Q8NBJ5 | Procollagen galactosyltransferase 1 | COLGALT1 | 1.77 | 0.0003 |
| P09382 | Galectin-1 | LGALS1 | 1.94 | 0.0005 |
| Q5JRX3 | Presequence protease, mitochondrial | PITRM1 | 3.05 | 0.0006 |
| P37802 | Transgelin-2 | TAGLN2 | 4.65 | 0.0006 |
| P17301 | Integrin alpha-2 | ITGA2 | 34.90 | 0.0009 |
| Q86WV6 | Stimulator of interferon genes protein | TMEM173 | 3.02 | 0.0009 |
| Q32MZ4 | Leucine-rich repeat flightless-interacting protein 1 | LRRFIP1 | 2.13 | 0.0009 |
| Q15833 | Syntaxin-binding protein 2 | STXBP2 | 2.26 | 0.0011 |
| P62328 | Thymosin beta-4 | TMSB4X | 8.64 | 0.0011 |
| P08567 | Pleckstrin | PLEK | 5.45 | 0.0015 |
| Q9UGT4 | Sushi domain-containing protein 2 | SUSD2 | 53.35 | 0.0017 |
| O60610 | Protein diaphanous homolog 1 | DIAPH1 | 2.21 | 0.0018 |
| P08758 | Annexin A5 | ANXA5 | 7.68 | 0.0018 |
| P07951 | Tropomyosin beta chain | TPM2 | 3.45 | 0.0019 |
| Q7LDG7 | RAS guanyl-releasing protein 2 | RASGRP2 | 3.54 | 0.0019 |
| Q14019 | Coactosin-like protein | COTL1 | 2.38 | 0.0020 |
| P18054 | Polyunsaturated fatty acid lipoxygenase ALOX12 | ALOX12 | 26.46 | 0.0020 |
| Q9NYL9 | Tropomodulin-3 | TMOD3 | 2.90 | 0.0020 |
| P63000 | Ras-related C3 botulinum toxin substrate 1 | RAC1 | 2.73 | 0.0025 |
| P37840 | Alpha-synuclein | SNCA | 18.57 | 0.0025 |
| Q9HBI1 | Beta-parvin | PARVB | 12.09 | 0.0026 |
| P18206 | Vinculin | VCL | 5.78 | 0.0028 |
| Q15942 | Zyxin | ZYX | 4.93 | 0.0029 |
| P06753 | Tropomyosin alpha-3 chain | TPM3 | 2.07 | 0.0030 |
| Uniprot ID | Description | Gene Name | Fold Change | FDR Adjusted p-Value |
|---|---|---|---|---|
| P22087 | rRNA 2′-O-methyltransferase fibrillarin | FBL | 1.65 | 0.0003 |
| P16402 | Histone H1.3 | HIST1H1D | 2.89 | 0.0007 |
| Q8NBS9 | Thioredoxin domain-containing protein 5 | TXNDC5 | 4.33 | 0.0008 |
| Q99798 | Aconitate hydratase, mitochondrial | ACO2 | 1.61 | 0.0012 |
| Q9NSE4 | Isoleucine--tRNA ligase, mitochondrial | IARS2 | 2.22 | 0.0014 |
| Q9Y320 | Thioredoxin-related transmembrane protein 2 | TMX2 | 5.92 | 0.0014 |
| Q13263 | Transcription intermediary factor 1-beta | TRIM28 | 2.19 | 0.0015 |
| P30837 | Aldehyde dehydrogenase X, mitochondrial | ALDH1B1 | 8.19 | 0.0015 |
| Q9BY50 | Signal peptidase complex catalytic subunit SEC11C | SEC11C | 5.19 | 0.0016 |
| Q13813 | Spectrin alpha chain, non-erythrocytic 1 | SPTAN1 | 2.10 | 0.0023 |
| Q3SY69 | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | ALDH1L2 | 7.33 | 0.0033 |
| P08240 | Signal recognition particle receptor subunit alpha | SRPR | 2.48 | 0.0035 |
| P30044 | Peroxiredoxin-5, mitochondrial | PRDX5 | 1.77 | 0.0037 |
| Q7KZF4 | Staphylococcal nuclease domain-containing protein 1 | SND1 | 2.91 | 0.0042 |
| P49257 | Protein ERGIC-53 | LMAN1 | 2.44 | 0.0043 |
| Q9Y4P3 | Transducin beta-like protein 2 | TBL2 | 4.53 | 0.0045 |
| P09874 | Poly [ADP-ribose] polymerase 1 | PARP1 | 2.67 | 0.0047 |
| Q01105 | Protein SET | SET | 3.29 | 0.0054 |
| Q92506 | Estradiol 17-beta-dehydrogenase 8 | HSD17B8 | 3.37 | 0.0054 |
| P12235 | ADP/ATP translocase 1 | SLC25A4 | 4.43 | 0.0055 |
| Q13310 | Polyadenylate-binding protein 4 | PABPC4 | 4.47 | 0.0056 |
| P53992 | Protein transport protein Sec24C | SEC24C | 38.68 | 0.0057 |
| Q16706 | Alpha-mannosidase 2 | MAN2A1 | 6.43 | 0.0058 |
| Q01082 | Spectrin beta chain, non-erythrocytic 1 | SPTBN1 | 1.96 | 0.0060 |
| P54886 | Delta-1-pyrroline-5-carboxylate synthase | ALDH18A1 | 10.26 | 0.0072 |
| Uniprot ID | Description | Gene Name | Fold Change | FDR-Adjusted p-Value |
|---|---|---|---|---|
| Q9NZP8 | Complement C1r subcomponent-like protein | C1RL | 1.58 | 0.012 |
| P02747 | Complement C1q subcomponent subunit C | C1QC | 2.45 | 0.030 |
| P36955 | Pigment epithelium-derived factor | SERPINF1 | 1.56 | 0.031 |
| P23470 | Receptor-type tyrosine-protein phosphatase gamma | PTPRG | 2.87 | 0.035 |
| P02745 | Complement C1q subcomponent subunit A | C1QA | 2.33 | 0.038 |
| Q04756 | Hepatocyte growth factor activator | HGFA | 2.09 | 0.038 |
| P05062 | Fructose-bisphosphate aldolase B | ALDOB | 2.31 | 0.039 |
| P02746 | Complement C1q subcomponent subunit B | C1QB | 2.66 | 0.041 |
| P22891 | Vitamin K-dependent protein Z | PROZ | 1.83 | 0.042 |
| P06276 | Cholinesterase | BCHE | 1.88 | 0.056 |
| Q9HDC9 | Adipocyte plasma membrane-associated protein | APMAP | 1.85 | 0.058 |
| P02760 | Protein AMBP | AMBP | 1.69 | 0.062 |
| P07359 | Platelet glycoprotein Ib alpha chain | GP1BA | 2.40 | 0.070 |
| P19320 | Vascular cell adhesion protein 1 | VCAM1 | 2.23 | 0.078 |
| P01023 | Alpha-2-macroglobulin | A2M | 2.50 | 0.079 |
| P00736 | Complement C1r subcomponent | C1R | 1.63 | 0.079 |
| P15144 | Aminopeptidase N | ANPEP | 2.65 | 0.081 |
| P01871 | Ig mu chain C region | IGHM | 4.84 | 0.083 |
| P04114 | Apolipoprotein B-100 | APOB | 1.54 | 0.083 |
| Q86UD1 | Out at first protein homolog | OAF | 2.24 | 0.085 |
| P0C0L5 | Complement C4-B | C4B | 1.88 | 0.085 |
| Q12913 | Receptor-type tyrosine-protein phosphatase eta | PTPRJ | 1.79 | 0.095 |
| Protein Target/Marker | Potential Therapeutic | Method of Target Detection | FDA Approval | References |
|---|---|---|---|---|
| Potential protein targets in extramedullary multiple myeloma (identified from the literature) | ||||
| BCL2 | Venetoclax | Immunohistochemistry, qPCR, fl–w cytometry | Yes—Acute myeloid leukaemia, Chronic lymphocytic leukaemia | [88,89] |
| BCL2, BCL-XL | Navitoclax | Immunohistochemistry, qPCR, flow cytometry | No | [89,90] |
| XPO1 | Selinexor | Immunohistochemistry | Yes—Multiple myeloma | [91,92] |
| Aminopeptidase expression (Correlates with Melflufen sensitivity) | Melflufen | RNA sequencing | No | [93,94] |
| MEK | Trametinib | Targeted sequencing for RAS mutations | Yes (in combination with dabrafenib)–Various metastatic solid tumours with BRAF V600 E mutation | [11,95] |
| CD44v | 4SCAR-CD44v6 | Immunohistochemistry, flow cytometry | No | [96,97] |
| BRAF V600E | Vemurafenib, encorafenib, binimetinib | Allele-specific PCR | Yes–Metastatic melanoma with BRAF V600 E mutation | [98,99] |
| Potential protein targets in extramedullary multiple myeloma (identified in this study) | ||||
| LGALS1 | OTX008 | Immunohistochemistry | No | [100,101,102] |
| HPSE | Roneparstat | Immunohistochemistry | No | [86] |
| ROCK2 | Belumosudil | qPCR, immunohistochemistry | Yes–Chronic graft-versus-host disease | [103] |
| ILK | QLT0267, Compound 22 | Immunohistochemistry | No | [104,105] |
| Lipids | Statins | Unknown | Yes | [78,79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunphy, K.; Bazou, D.; Henry, M.; Meleady, P.; Miettinen, J.J.; Heckman, C.A.; Dowling, P.; O’Gorman, P. Proteomic and Metabolomic Analysis of Bone Marrow and Plasma from Patients with Extramedullary Multiple Myeloma Identifies Distinct Protein and Metabolite Signatures. Cancers 2023, 15, 3764. https://doi.org/10.3390/cancers15153764
Dunphy K, Bazou D, Henry M, Meleady P, Miettinen JJ, Heckman CA, Dowling P, O’Gorman P. Proteomic and Metabolomic Analysis of Bone Marrow and Plasma from Patients with Extramedullary Multiple Myeloma Identifies Distinct Protein and Metabolite Signatures. Cancers. 2023; 15(15):3764. https://doi.org/10.3390/cancers15153764
Chicago/Turabian StyleDunphy, Katie, Despina Bazou, Michael Henry, Paula Meleady, Juho J. Miettinen, Caroline A. Heckman, Paul Dowling, and Peter O’Gorman. 2023. "Proteomic and Metabolomic Analysis of Bone Marrow and Plasma from Patients with Extramedullary Multiple Myeloma Identifies Distinct Protein and Metabolite Signatures" Cancers 15, no. 15: 3764. https://doi.org/10.3390/cancers15153764
APA StyleDunphy, K., Bazou, D., Henry, M., Meleady, P., Miettinen, J. J., Heckman, C. A., Dowling, P., & O’Gorman, P. (2023). Proteomic and Metabolomic Analysis of Bone Marrow and Plasma from Patients with Extramedullary Multiple Myeloma Identifies Distinct Protein and Metabolite Signatures. Cancers, 15(15), 3764. https://doi.org/10.3390/cancers15153764