MicroRNAs Derived from Extracellular Vesicles: Keys to Understanding SARS-CoV-2 Vaccination Response in Cancer Patients?
Abstract
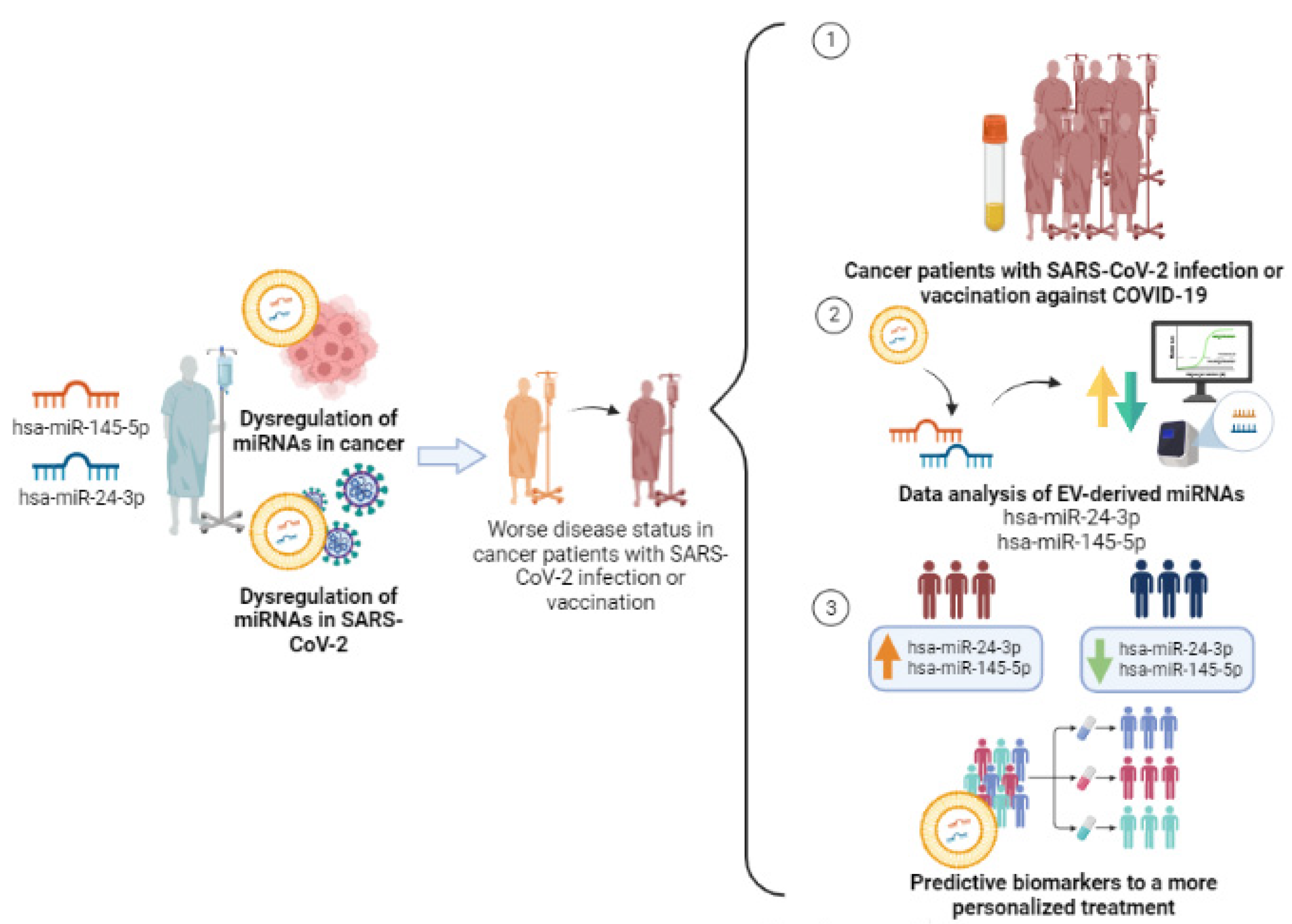
:Simple Summary
Abstract
1. Introduction
1.1. The Problematic behind COVID-19
1.2. Vaccines Development against COVID-19
1.3. Cancer Patients and mRNA Vaccines against COVID-19
1.4. Necessity of Biomarkers of Immune Response to COVID-19 Vaccines: Can EV-miRNAs Help in Cancer Patients’ Immune Response Stratification?
2. Evidence Acquisition
2.1. Literature Analysis and Evidence Synthesis
2.2. SARS-CoV-2-Related EV-miRNAs and Their Potential Impact in Cancer Patients’ Immune Response to Vaccination
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rito, T.; Fernandes, P.; Duarte, R.; Soares, P. Evaluating Data Sharing of SARS-CoV-2 Genomes for Molecular Epidemiology across the COVID-19 Pandemic. Viruses 2023, 15, 560. [Google Scholar] [CrossRef]
- Zhou, Z.; Qiu, Y.; Ge, X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim. Dis. 2021, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Park, B.K.; Kim, J.; Park, S.; Kim, D.; Kim, M.; Baek, K.; Bae, J.Y.; Park, M.S.; Kim, W.K.; Lee, Y.; et al. MERS-CoV and SARS-CoV-2 replication can be inhibited by targeting the interaction between the viral spike protein and the nucleocapsid protein. Theranostics 2021, 11, 3853–3867. [Google Scholar] [CrossRef] [PubMed]
- Jahirul Islam, M.; Nawal Islam, N.; Siddik Alom, M.; Kabir, M.; Halim, M.A. A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites. Immunobiology 2023, 228, 152302. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhong, C.; Tieleman, D.P. Supramolecular Organization of SARS-CoV and SARS-CoV-2 Virions Revealed by Coarse-Grained Models of Intact Virus Envelopes. J. Chem. Inf. Model. 2022, 62, 176–186. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Ching, K.L.; de Vries, M.; Gago, J.; Dancel-Manning, K.; Sall, J.; Rice, W.J.; Barnett, C.; Khodadadi-Jamayran, A.; Tsirigos, A.; Liang, F.X.; et al. ACE2-containing defensosomes serve as decoys to inhibit SARS-CoV-2 infection. PLoS Biol. 2022, 20, e3001754. [Google Scholar] [CrossRef]
- Berry, F.; Morin-Dewaele, M.; Majidipur, A.; Jamet, T.; Bartier, S.; Ignjatovic, E.; Toniutti, D.; Gaspar Lopes, J.; Soyeux-Porte, P.; Maillé, P.; et al. Proviral role of human respiratory epithelial cell-derived small extracellular vesicles in SARS-CoV-2 infection. J. Extracell. Vesicles 2022, 11, e12269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, F.; Xia, B.; Yuan, Y.; Yu, F.; Wang, G.; Chen, Q.; Wang, Q.; Li, Y.; Li, R.; et al. The interferon-stimulated exosomal hACE2 potently inhibits SARS-CoV-2 replication through competitively blocking the virus entry. Signal Transduct. Target. Ther. 2021, 6, 189. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e813. [Google Scholar] [CrossRef] [PubMed]
- Pontelli, M.C.; Castro, Í.A.; Martins, R.B.; La Serra, L.; Veras, F.P.; Nascimento, D.C.; Silva, C.M.; Cardoso, R.S.; Rosales, R.; Gomes, R.; et al. SARS-CoV-2 productively infects primary human immune system cells in vitro and in COVID-19 patients. J. Mol. Cell Biol. 2022, 14, mjac021. [Google Scholar] [CrossRef]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A.; et al. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J. Virol. 2022, 96, e0012822. [Google Scholar] [CrossRef]
- Marti, S.; Carsin, A.-E.; Sampol, J.; Pallero, M.; Aldas, I.; Marin, T.; Lujan, M.; Lalmolda, C.; Sabater, G.; Bonnin-Vilaplana, M.; et al. Higher mortality and intubation rate in COVID-19 patients treated with noninvasive ventilation compared with high-flow oxygen or CPAP. Sci. Rep. 2022, 12, 6527. [Google Scholar] [CrossRef]
- Silverstein, N.J.; Wang, Y.; Manickas-Hill, Z.; Carbone, C.; Dauphin, A.; Boribong, B.P.; Loiselle, M.; Davis, J.; Leonard, M.M.; Kuri-Cervantes, L.; et al. Innate lymphoid cells and COVID-19 severity in SARS-CoV-2 infection. Elife 2022, 11, e74681. [Google Scholar] [CrossRef]
- Olbei, M.; Hautefort, I.; Modos, D.; Treveil, A.; Poletti, M.; Gul, L.; Shannon-Lowe, C.D.; Korcsmaros, T. SARS-CoV-2 Causes a Different Cytokine Response Compared to Other Cytokine Storm-Causing Respiratory Viruses in Severely Ill Patients. Front. Immunol. 2021, 12, 629193. [Google Scholar] [CrossRef]
- Shen, W.X.; Luo, R.C.; Wang, J.Q.; Chen, Z.S. Features of Cytokine Storm Identified by Distinguishing Clinical Manifestations in COVID-19. Front. Public Health 2021, 9, 671788. [Google Scholar] [CrossRef]
- Hogan, M.J.; Pardi, N. mRNA Vaccines in the COVID-19 Pandemic and Beyond. Annu. Rev. Med. 2022, 73, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Perincheri, S.; Fleming, T.; Poulson, C.; Tiffany, B.; Bremner, R.M.; Mohanakumar, T. Cutting Edge: Circulating Exosomes with COVID Spike Protein Are Induced by BNT162b2 (Pfizer-BioNTech) Vaccination prior to Development of Antibodies: A Novel Mechanism for Immune Activation by mRNA Vaccines. J. Immunol. 2021, 207, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Defendi, H.G.T.; da Silva Madeira, L.; Borschiver, S. Analysis of the COVID-19 Vaccine Development Process: An Exploratory Study of Accelerating Factors and Innovative Environments. J. Pharm. Innov. 2022, 17, 555–571. [Google Scholar] [CrossRef]
- Zuo, F.; Abolhassani, H.; Du, L.; Piralla, A.; Bertoglio, F.; de Campos-Mata, L.; Wan, H.; Schubert, M.; Cassaniti, I.; Wang, Y.; et al. Heterologous immunization with inactivated vaccine followed by mRNA-booster elicits strong immunity against SARS-CoV-2 Omicron variant. Nat. Commun. 2022, 13, 2670. [Google Scholar] [CrossRef] [PubMed]
- Fanciullino, R.; Ciccolini, J.; Milano, G. COVID-19 vaccine race: Watch your step for cancer patients. Br. J. Cancer 2021, 124, 860–861. [Google Scholar] [CrossRef]
- Packer, M.; Gyawali, D.; Yerabolu, R.; Schariter, J.; White, P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 2021, 12, 6777. [Google Scholar] [CrossRef]
- Chi, W.-Y.; Li, Y.-D.; Huang, H.-C.; Chan, T.E.H.; Chow, S.-Y.; Su, J.-H.; Ferrall, L.; Hung, C.-F.; Wu, T.C. COVID-19 vaccine update: Vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. J. Biomed. Sci. 2022, 29, 82. [Google Scholar] [CrossRef]
- Vizcarra, P.; Haemmerle, J.; Velasco, H.; Velasco, T.; Fernández-Escribano, M.; Vallejo, A.; Casado, J.L. BNT162b2 mRNA COVID-19 vaccine Reactogenicity: The key role of immunity. Vaccine 2021, 39, 7367–7374. [Google Scholar] [CrossRef]
- Saadh, M.J.; Jaber, S.A. Efficacy of COVID-19 vaccines. Microb. Pathog. 2022, 171, 105729. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.; Cardoso, M.J.; Norton, P.; Sarmento, A.; Guimarães, J.T. Serological response to a single dose of a SARS-CoV-2 mRNA vaccine. J. Virol. Methods 2021, 296, 114223. [Google Scholar] [CrossRef] [PubMed]
- Lederer, K.; Castaño, D.; Gómez Atria, D.; Oguin, T.H., 3rd; Wang, S.; Manzoni, T.B.; Muramatsu, H.; Hogan, M.J.; Amanat, F.; Cherubin, P.; et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity 2020, 53, 1281–1295.e1285. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Zhang, C.; Walker, P.G.; Dong, Y. Formulation and Delivery Technologies for mRNA Vaccines. Curr. Top Microbiol. Immunol. 2022, 440, 71–110. [Google Scholar] [CrossRef]
- Brüssow, H. mRNA vaccines against COVID-19: A showcase for the importance of microbial biotechnology. Microb. Biotechnol. 2022, 15, 135–148. [Google Scholar] [CrossRef]
- Layan, M.; Gilboa, M.; Gonen, T.; Goldenfeld, M.; Meltzer, L.; Andronico, A.; Hozé, N.; Cauchemez, S.; Regev-Yochay, G. Impact of BNT162b2 Vaccination and Isolation on SARS-CoV-2 Transmission in Israeli Households: An Observational Study. Am. J. Epidemiol. 2022, 191, 1224–1234. [Google Scholar] [CrossRef]
- Castruita, J.A.S.; Schneider, U.V.; Mollerup, S.; Leineweber, T.D.; Weis, N.; Bukh, J.; Pedersen, M.S.; Westh, H. SARS-CoV-2 spike mRNA vaccine sequences circulate in blood up to 28 days after COVID-19 vaccination. Apmis 2023, 131, 128–132. [Google Scholar] [CrossRef]
- de Mey, W.; Locy, H.; De Ridder, K.; De Schrijver, P.; Autaers, D.; Lakdimi, A.; Esprit, A.; Franceschini, L.; Thielemans, K.; Verdonck, M.; et al. An mRNA mix redirects dendritic cells towards an antiviral program, inducing anticancer cytotoxic stem cell and central memory CD8(+) T cells. Front. Immunol. 2023, 14, 1111523. [Google Scholar] [CrossRef]
- Dagla, I.; Iliou, A.; Benaki, D.; Gikas, E.; Mikros, E.; Bagratuni, T.; Kastritis, E.; Dimopoulos, M.A.; Terpos, E.; Tsarbopoulos, A. Plasma Metabolomic Alterations Induced by COVID-19 Vaccination Reveal Putative Biomarkers Reflecting the Immune Response. Cells 2022, 11, 1241. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451.e2417. [Google Scholar] [CrossRef]
- Röltgen, K.; Nielsen, S.C.A.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A.; et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 2022, 185, 1025–1040.e1014. [Google Scholar] [CrossRef]
- Dobaño, C.; Jiménez, A.; Rubio, R.; Alonso, S.; Ramírez-Morros, A.; Vidal, M.; Vidal-Alaball, J.; Ruiz-Comellas, A.; García-Basteiro, A.L.; Izquierdo, L.; et al. Spike-based COVID-19 immunization increases antibodies to nucleocapsid antigen. Transl. Res. 2022, 240, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012, 9, 1319–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassett, K.J.; Higgins, J.; Woods, A.; Levy, B.; Xia, Y.; Hsiao, C.J.; Acosta, E.; Almarsson, Ö.; Moore, M.J.; Brito, L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release 2021, 335, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Di Noia, V.; Pimpinelli, F.; Renna, D.; Barberi, V.; Maccallini, M.T.; Gariazzo, L.; Pontone, M.; Monti, A.; Campo, F.; Taraborelli, E.; et al. Immunogenicity and Safety of COVID-19 Vaccine BNT162b2 for Patients with Solid Cancer: A Large Cohort Prospective Study from a Single Institution. Clin. Cancer Res. 2021, 27, 6815–6823. [Google Scholar] [CrossRef]
- Al Khames Aga, Q.A.; Alkhaffaf, W.H.; Hatem, T.H.; Nassir, K.F.; Batineh, Y.; Dahham, A.T.; Shaban, D.; Al Khames Aga, L.A.; Agha, M.Y.R.; Traqchi, M. Safety of COVID-19 vaccines. J. Med Virol. 2021, 93, 6588–6594. [Google Scholar] [CrossRef]
- Oberhardt, V.; Luxenburger, H.; Kemming, J.; Schulien, I.; Ciminski, K.; Giese, S.; Csernalabics, B.; Lang-Meli, J.; Janowska, I.; Staniek, J.; et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature 2021, 597, 268–273. [Google Scholar] [CrossRef]
- Sureshchandra, S.; Lewis, S.A.; Doratt, B.M.; Jankeel, A.; Coimbra Ibraim, I.; Messaoudi, I. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight 2021, 6, e153201. [Google Scholar] [CrossRef]
- Zhang, N.-N.; Li, X.-F.; Deng, Y.-Q.; Zhao, H.; Huang, Y.-J.; Yang, G.; Huang, W.-J.; Gao, P.; Zhou, C.; Zhang, R.-R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283.e1216. [Google Scholar] [CrossRef]
- Gobbi, F.; Buonfrate, D.; Moro, L.; Rodari, P.; Piubelli, C.; Caldrer, S.; Riccetti, S.; Sinigaglia, A.; Barzon, L. Antibody Response to the BNT162b2 mRNA COVID-19 Vaccine in Subjects with Prior SARS-CoV-2 Infection. Viruses 2021, 13, 422. [Google Scholar] [CrossRef]
- Goel, R.R.; Painter, M.M.; Apostolidis, S.A.; Mathew, D.; Meng, W.; Rosenfeld, A.M.; Lundgreen, K.A.; Reynaldi, A.; Khoury, D.S.; Pattekar, A.; et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 2021, 374, abm0829. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Morioka, T.; Natsuki, Y.; Sasaki, K.; Kakutani, Y.; Ochi, A.; Yamazaki, Y.; Shoji, T.; Emoto, M. New-onset Type 1 Diabetes after COVID-19 mRNA Vaccination. Intern. Med. 2022, 61, 1197–1200. [Google Scholar] [CrossRef]
- Watanabe, M.; Balena, A.; Tuccinardi, D.; Tozzi, R.; Risi, R.; Masi, D.; Caputi, A.; Rossetti, R.; Spoltore, M.E.; Filippi, V.; et al. Central obesity, smoking habit, and hypertension are associated with lower antibody titres in response to COVID-19 mRNA vaccine. Diabetes Metab. Res. Rev. 2022, 38, e3465. [Google Scholar] [CrossRef]
- Patone, M.; Mei, X.W.; Handunnetthi, L.; Dixon, S.; Zaccardi, F.; Shankar-Hari, M.; Watkinson, P.; Khunti, K.; Harnden, A.; Coupland, C.A.C.; et al. Risks of myocarditis, pericarditis, and cardiac arrhythmias associated with COVID-19 vaccination or SARS-CoV-2 infection. Nat. Med. 2022, 28, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Ma, W.; Sikavi, D.R.; Drew, D.A.; Nguyen, L.H.; Bowyer, R.C.E.; Cardoso, M.J.; Fall, T.; Freidin, M.B.; Gomez, M.; et al. Cancer and Risk of COVID-19 Through a General Community Survey. Oncologist 2021, 26, e182–e185. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Wong, S.Y.; Chai, L.Y.A.; Lee, S.C.; Lee, M.X.; Muthiah, M.D.; Tay, S.H.; Teo, C.B.; Tan, B.K.J.; Chan, Y.H.; et al. Efficacy of COVID-19 vaccines in immunocompromised patients: Systematic review and meta-analysis. BMJ 2022, 376, e068632. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Schmidt, A.L.; Labaki, C.; Hsu, C.Y.; Bakouny, Z.; Balanchivadze, N.; Berg, S.A.; Blau, S.; Daher, A.; El Zarif, T.; Friese, C.R.; et al. COVID-19 vaccination and breakthrough infections in patients with cancer. Ann. Oncol. 2022, 33, 340–346. [Google Scholar] [CrossRef]
- Shroff, R.T.; Chalasani, P.; Wei, R.; Pennington, D.; Quirk, G.; Schoenle, M.V.; Peyton, K.L.; Uhrlaub, J.L.; Ripperger, T.J.; Jergović, M.; et al. Immune responses to two and three doses of the BNT162b2 mRNA vaccine in adults with solid tumors. Nat. Med. 2021, 27, 2002–2011. [Google Scholar] [CrossRef]
- Obeid, M.; Suffiotti, M.; Pellaton, C.; Bouchaab, H.; Cairoli, A.; Salvadé, V.; Stevenel, C.; Hottinger, R.; Pythoud, C.; Coutechier, L.; et al. Humoral Responses Against Variants of Concern by COVID-19 mRNA Vaccines in Immunocompromised Patients. JAMA Oncol. 2022, 8, e220446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Graves-Deal, R.; Trinh, V.Q.; Ramirez, M.A.; Sohn, Y.; Neininger, A.C.; Taneja, N.; McKinley, E.T.; et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021, 23, 1240–1254. [Google Scholar] [CrossRef]
- Jia, E.; Ren, N.; Shi, X.; Zhang, R.; Yu, H.; Yu, F.; Qin, S.; Xue, J. Extracellular vesicle biomarkers for pancreatic cancer diagnosis: A systematic review and meta-analysis. BMC Cancer 2022, 22, 573. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Xu, Y.; Cai, S.; Li, X.; Li, D. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J. Extracell. Vesicles 2022, 11, e12186. [Google Scholar] [CrossRef]
- Adduri, R.S.R.; Vasireddy, R.; Mroz, M.M.; Bhakta, A.; Li, Y.; Chen, Z.; Miller, J.W.; Velasco-Alzate, K.Y.; Gopalakrishnan, V.; Maier, L.A.; et al. Realistic biomarkers from plasma extracellular vesicles for detection of beryllium exposure. Int. Arch. Occup. Environ. Health 2022, 95, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef] [PubMed]
- McKay, T.B.; Hutcheon, A.E.K.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Epithelial Cells Promote Myofibroblast Differentiation. Cells 2020, 9, 1080. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Plazas, X.; Altuna-Coy, A.; Alves-Santiago, M.; Vila-Barja, J.; García-Fontgivell, J.F.; Martínez-González, S.; Segarra-Tomás, J.; Chacón, M.R. Liquid Biopsy-Based Exo-oncomiRNAs Can Predict Prostate Cancer Aggressiveness. Cancers 2021, 13, 250. [Google Scholar] [CrossRef]
- Cocozza, F.; Névo, N.; Piovesana, E.; Lahaye, X.; Buchrieser, J.; Schwartz, O.; Manel, N.; Tkach, M.; Théry, C.; Martin-Jaular, L. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 Spike protein-containing virus. J. Extracell. Vesicles 2020, 10, e12050. [Google Scholar] [CrossRef]
- El-Shennawy, L.; Hoffmann, A.D.; Dashzeveg, N.K.; McAndrews, K.M.; Mehl, P.J.; Cornish, D.; Yu, Z.; Tokars, V.L.; Nicolaescu, V.; Tomatsidou, A.; et al. Circulating ACE2-expressing extracellular vesicles block broad strains of SARS-CoV-2. Nat. Commun. 2022, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Wang, W.; Jo, H.; Park, S.; Kim, H.; Kim, S.I.; Han, Y.; Lee, J.; Seol, A.; Kim, J.; Lee, M.; et al. Integrated analysis of ascites and plasma extracellular vesicles identifies a miRNA-based diagnostic signature in ovarian cancer. Cancer Lett. 2022, 542, 215735. [Google Scholar] [CrossRef]
- Fernández-Pato, A.; Virseda-Berdices, A.; Resino, S.; Ryan, P.; Martínez-González, O.; Pérez-García, F.; Martin-Vicente, M.; Valle-Millares, D.; Brochado-Kith, O.; Blancas, R.; et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg. Microbes Infect. 2022, 11, 676–688. [Google Scholar] [CrossRef]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, R.; Banerjea, A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal miR-148a to Activate Human Microglia. Front. Immunol. 2021, 12, 656700. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, X.; Jiang, X.-M.; Guo, J.; Fu, Z.; Zhou, Z.; Yang, P.; Guo, H.; Guo, X.; Liang, G.; et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target. Ther. 2021, 6, 300. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, Y.; Lim, C.W.; Park, J.M.; Yu, S.H.; Kim, Y.; Han, H.J.; Kim, C.H.; Song, Y.S.; Kim, C.; et al. Potential Therapeutic Effect of Micrornas in Extracellular Vesicles from Mesenchymal Stem Cells against SARS-CoV-2. Cells 2021, 10, 2393. [Google Scholar] [CrossRef]
- Meidert, A.S.; Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Billaud, J.N.; Klein, M.; Lindemann, A.; Aue, E.; Schelling, G.; et al. Extracellular Vesicle Associated miRNAs Regulate Signaling Pathways Involved in COVID-19 Pneumonia and the Progression to Severe Acute Respiratory Corona Virus-2 Syndrome. Front. Immunol. 2021, 12, 784028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.; Li, L.; Li, L.; Zhou, X.; Wan, M.; Lou, P.; Zhao, M.; Lv, K.; Yuan, Y.; et al. Peritoneal M2 macrophage-derived extracellular vesicles as natural multitarget nanotherapeutics to attenuate cytokine storms after severe infections. J. Control. Release 2022, 349, 118–132. [Google Scholar] [CrossRef]
- Gambardella, J.; Kansakar, U.; Sardu, C.; Messina, V.; Jankauskas, S.S.; Marfella, R.; Maggi, P.; Wang, X.; Mone, P.; Paolisso, G.; et al. Exosomal miR-145 and miR-885 Regulate Thrombosis in COVID-19. J. Pharmacol. Exp. Ther. 2023, 384, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Borrmann, M.; Brandes, F.; Kirchner, B.; Klein, M.; Billaud, J.N.; Reithmair, M.; Rehm, M.; Schelling, G.; Pfaffl, M.W.; Meidert, A.S. Extensive blood transcriptome analysis reveals cellular signaling networks activated by circulating glycocalyx components reflecting vascular injury in COVID-19. Front. Immunol. 2023, 14, 1129766. [Google Scholar] [CrossRef] [PubMed]
- Mimmi, S.; Zimbo, A.M.; Rotundo, S.; Cione, E.; Nisticò, N.; Aloisio, A.; Maisano, D.; Tolomeo, A.M.; Dattilo, V.; Lionello, R.; et al. SARS-CoV-2 spike protein-guided exosome isolation facilitates detection of potential miRNA biomarkers in COVID-19 infections. Clin. Chem. Lab. Med. (CCLM) 2023, 61, 1518–1524. [Google Scholar] [CrossRef]
- Latini, A.; Vancheri, C.; Amati, F.; Morini, E.; Grelli, S.; Matteucci, C.; Petrone, V.; Colona, V.L.; Murdocca, M.; Andreoni, M.; et al. Expression analysis of miRNA hsa-let7b-5p in naso-oropharyngeal swabs of COVID-19 patients supports its role in regulating ACE2 and DPP4 receptors. J. Cell. Mol. Med. 2022, 26, 4940–4948. [Google Scholar] [CrossRef]
- Mandolesi, G.; Rizzo, F.R.; Balletta, S.; Stampanoni Bassi, M.; Gilio, L.; Guadalupi, L.; Nencini, M.; Moscatelli, A.; Ryan, C.P.; Licursi, V.; et al. The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis. Cells 2021, 10, 330. [Google Scholar] [CrossRef]
- Yan, L.; Ma, J.; Zhu, Y.; Zan, J.; Wang, Z.; Ling, L.; Li, Q.; Lv, J.; Qi, S.; Cao, Y.; et al. miR-24-3p promotes cell migration and proliferation in lung cancer by targeting SOX7. J. Cell Biochem. 2018, 119, 3989–3998. [Google Scholar] [CrossRef]
- Gao, Z.; Zhou, L.; Hua, S.; Wu, H.; Luo, L.; Li, L.; Wang, S.; Liu, Y.; Zhou, Z.; Chen, X. miR-24-3p promotes colon cancer progression by targeting ING1. Signal Transduct. Target. Ther. 2020, 5, 171. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, D.; Huang, R.; Shi, Z. HMGB3 Targeted by miR-145-5p Impacts Proliferation, Migration, Invasion, and Apoptosis of Breast Cancer Cells. Comput. Math. Methods Med. 2022, 2022, 1954099. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, M.; Liu, Q.; Han, Z.; Zhao, Y.; Ji, S. miR-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol. Lett. 2018, 16, 6355–6360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. miR-24 targets SARS-CoV-2 co-factor Neuropilin-1 in human brain microvascular endothelial cells: Insights for COVID-19 neurological manifestations. Res. Sq. 2021, preprint. [Google Scholar] [CrossRef]
- Papi, C.; Gasparello, J.; Zurlo, M.; Cosenza, L.C.; Gambari, R.; Finotti, A. The Cystic Fibrosis Transmembrane Conductance Regulator Gene (CFTR) Is under Post-Transcriptional Control of microRNAs: Analysis of the Effects of agomiRNAs Mimicking miR-145-5p, miR-101-3p, and miR-335-5p. Non-Coding RNA 2023, 9, 29. [Google Scholar] [CrossRef]
- Dias, T.R.; Dias, F.; Teixeira, A.L.; Sousa, H.; Oliveira, J.; Medeiros, R. MicroRNAs as Potential Tools for Predicting Cancer Patients’ Susceptibility to SARS-CoV-2 Infection and Vaccination Response. Cells 2022, 11, 2279. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi-Jamayran, A.; Akgol-Oksuz, B.; Afanasyeva, Y.; Heguy, A.; Thompson, M.; Ray, K.; Giro-Perafita, A.; Sánchez, I.; Wu, X.; Tripathy, D.; et al. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 2018, 9, 12868–12878. [Google Scholar] [CrossRef]
- Garcia-Magallanes, N.; Beltran-Ontiveros, S.A.; Leal-León, E.A.; Luque-Ortega, F.; Romero-Quintana, J.G.; Osuna-Ramirez, I.; Barbosa-Jasso, M.; Arámbula-Meraz, E. Underexpression of circulating miR-145-5p and miR-133a-3p are associated with breast cancer and immunohistochemical markers. J. Cancer Res. Ther. 2020, 16, 1223–1228. [Google Scholar]
- Wang, H.; Chen, C.; Ding, K.; Zhang, W.; Hou, J. MiR-24-3p as a prognostic indicator for multiple cancers: From a meta-analysis view. Biosci. Rep. 2020, 40, BSR20202938. [Google Scholar] [CrossRef]
- Lu, K.; Wang, J.; Song, Y.; Zhao, S.; Liu, H.; Tang, D.; Pan, B.; Zhao, H.; Zhang, Q. miRNA-24-3p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting p27Kip1. Oncol. Rep. 2015, 34, 995–1002. [Google Scholar] [CrossRef] [Green Version]
- Thuringer, D.; Jego, G.; Berthenet, K.; Hammann, A.; Solary, E.; Garrido, C. Gap junction-mediated transfer of miR-145-5p from microvascular endothelial cells to colon cancer cells inhibits angiogenesis. Oncotarget 2016, 7, 28160–28168. [Google Scholar] [CrossRef]
- Wang, S.; Wang, T.; Gu, P. microRNA-145-5p Inhibits Migration, Invasion, and Metastasis in Hepatocellular Carcinoma by Inhibiting ARF6. Cancer Manag. Res. 2021, 13, 3473–3484. [Google Scholar] [CrossRef]
- Ji, S.; Shi, Y.; Yang, L.; Zhang, F.; Li, Y.; Xu, F. miR-145-5p Inhibits Neuroendocrine Differentiation and Tumor Growth by Regulating the SOX11/MYCN Axis in Prostate cancer. Front. Genet. 2022, 13, 790621. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Samples/EV Sources | Results | Reference |
|---|---|---|---|
| miR-148a and miR-590 | HEK-293T and human microglial cell line (CHME3) | Neuroinflammation induced by miR-148a and miR-590 derived by EVs after S protein stimulation. Hyperactivation of microglia, with a reduction in USP33 and IRF9 expression. | Mishra and Banerjea [78] |
| miR-7-5p, miR-24-3p, miR-223-3p, miR-145-5p, and miR-15b-5p | Two cohorts: 30 serum samples per group (4 groups) in cohort 1 and 20 serum samples per group (4 groups); HEK293T cells | miRNAs that are low in elderly and diabetic patients inhibited S protein replication. The expression of these miRNAs increased after long periods of exercise. | Wang et al. [79] |
| miR-23a-3p, miR-26a-5p, miR-92a-3p, miR-103a-3p, and miR-181a-5p | Neuronal stem cells; human lung fibroblasts cell line LL24; human bronchial epithelial cell line Beas-2B; mouse microglial cell line BV2; human neuroblastoma cell line SK-N-BE(2)C; Vero cells | miRNAs of MSC-EVs inhibit viral replication in 3′UTR of SARS-CoV-2 genome (conserved region). | Park et al. [80] |
| miR-338-5p, miR-197-3p, miR-542-3p, miR-3168, let-7e-5p, miR-146a-5p, miR-221-3p, miR-15a-5p, and let-7c-5p | 100 blood samples: 20 patients with COVID-19 pneumonia, 20 patients with COVID-19 ARDS, 20 healthy donors, 28 patients with sepsis associated with ARDS, and 12 patients with bacterial community-acquired pneumonia | miR-338-5p targeted IL6 and OR52N2. Upregulated miR-542-3p increased furin activity. Downregulation of miR-3168 and let-7e-5p increased CXCL8 levels in severe COVID-19 infection. Inhibition of TLR4 by miR-146-5p in pneumonia and by let-7e-5p in COVID-19 ARDS. Downregulation of cytokines by miR-221-3p and miR-15a-5p in immunosuppressive state of COVID-19. | Meidert et al. [81] |
| miR-342-3p, miR-674-5p, miR-24-3p, miR-21a-5p, miR-23b-3p, miR-146-5p, miR-378a-5p, and let-7b-5p | Primary mouse peritoneal macrophages M2 (mMφ) isolated from peritoneal dialysis (PD) | Peritoneal M2-EVs promote reduction of pro-inflammatory cytokines levels. | Wang et al. [82] |
| miR-145 and miR-885 | 26 serum samples and umbilical vein endothelial cells (HUVECs) | HUVECs treated with serum of COVID-19 patients induce a reduction in miR-145 and miR-885 and an overexpression of tissue factor and Willebrand factor in endothelial cells. | Gambardella et al. [83] |
| miR-32-5p, miR-150-5p, miR-126-3p, miR-1-3p, miR-1228-5p, and miR-4433b-5p | 60 blood samples of COVID-19 patients | Increase in glycocalyx components and decrease in ADAMTS13 by downregulation of microRNAs. | Borrmann et al. [84] |
| let-7g-5p, miR-4454+miR-7975, hsa-miR-208a-3p, and hsa-miR-323-3p | Plasma samples of COVID-19 patients and cell culture (HEK-293 and HeLa cells) [72]; serum samples of 6 COVID-19 patients [85] | ExoACE2 with upregulation of let-7g-5p and hsa-miR-4454+miR-7975 and downregulation of hsa-miR-208a-3p and hsa-miR-323-3p, compared to non-ACE2-expressing exosomes. | El-Shennawy et al. [72], Mimmi et al. [85] * |
| hsa-let7b- 5p | HeLa cell line; 60 nasopharyngeal swabs (NPS) of COVID-19 patients | Low expression of hsa-let7b-5p in COVID-19 patients leads to a lack of regulation of genes (ACE2 and DPP4) exploited by SARS-CoV-2. | Latini et al. [86] * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, B.; Dias, T.R.; Teixeira, A.L.; Dias, F.; Medeiros, R. MicroRNAs Derived from Extracellular Vesicles: Keys to Understanding SARS-CoV-2 Vaccination Response in Cancer Patients? Cancers 2023, 15, 4017. https://doi.org/10.3390/cancers15164017
Almeida B, Dias TR, Teixeira AL, Dias F, Medeiros R. MicroRNAs Derived from Extracellular Vesicles: Keys to Understanding SARS-CoV-2 Vaccination Response in Cancer Patients? Cancers. 2023; 15(16):4017. https://doi.org/10.3390/cancers15164017
Chicago/Turabian StyleAlmeida, Beatriz, Tânia R. Dias, Ana Luísa Teixeira, Francisca Dias, and Rui Medeiros. 2023. "MicroRNAs Derived from Extracellular Vesicles: Keys to Understanding SARS-CoV-2 Vaccination Response in Cancer Patients?" Cancers 15, no. 16: 4017. https://doi.org/10.3390/cancers15164017





