Understanding DNA Damage Response and DNA Repair in Multiple Myeloma
Abstract
Simple Summary
Abstract
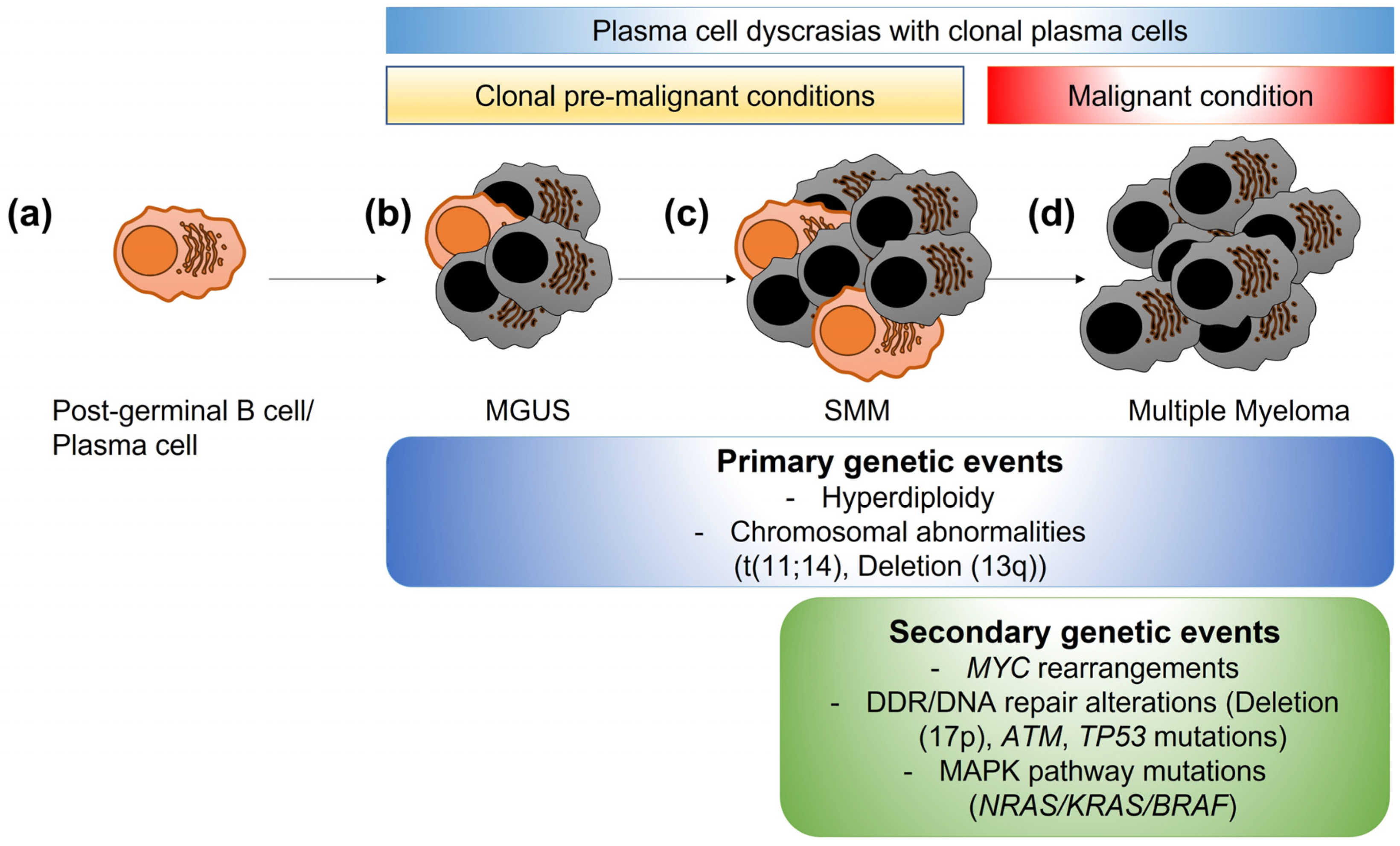
1. Introduction
2. DDR and DNA Repair in Normal Tissues and in MM
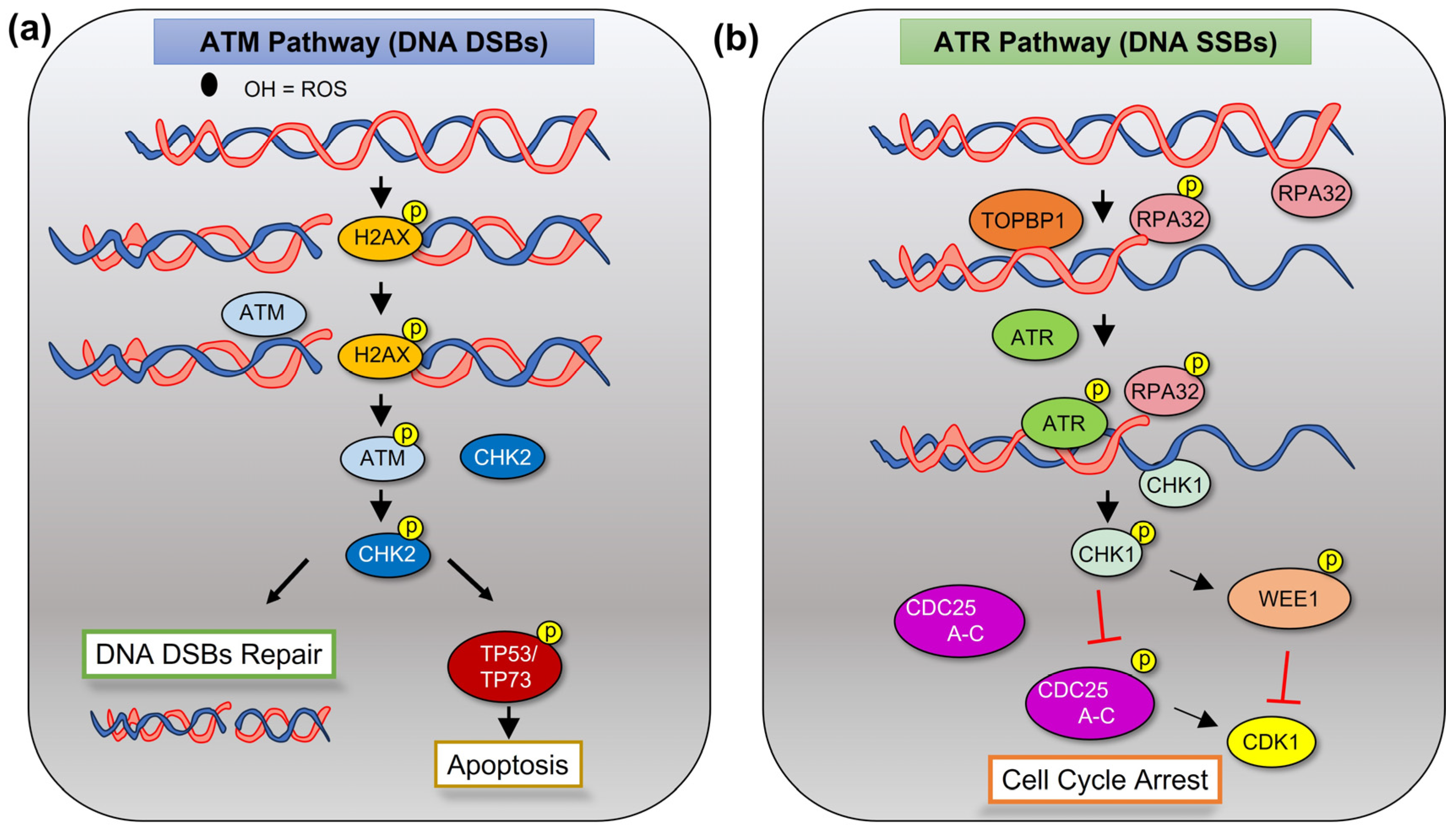
2.1. DDR and DNA Repair in Normal Tissues
2.2. Preventing Apoptotic DNA Damage Response (DDR) in MM
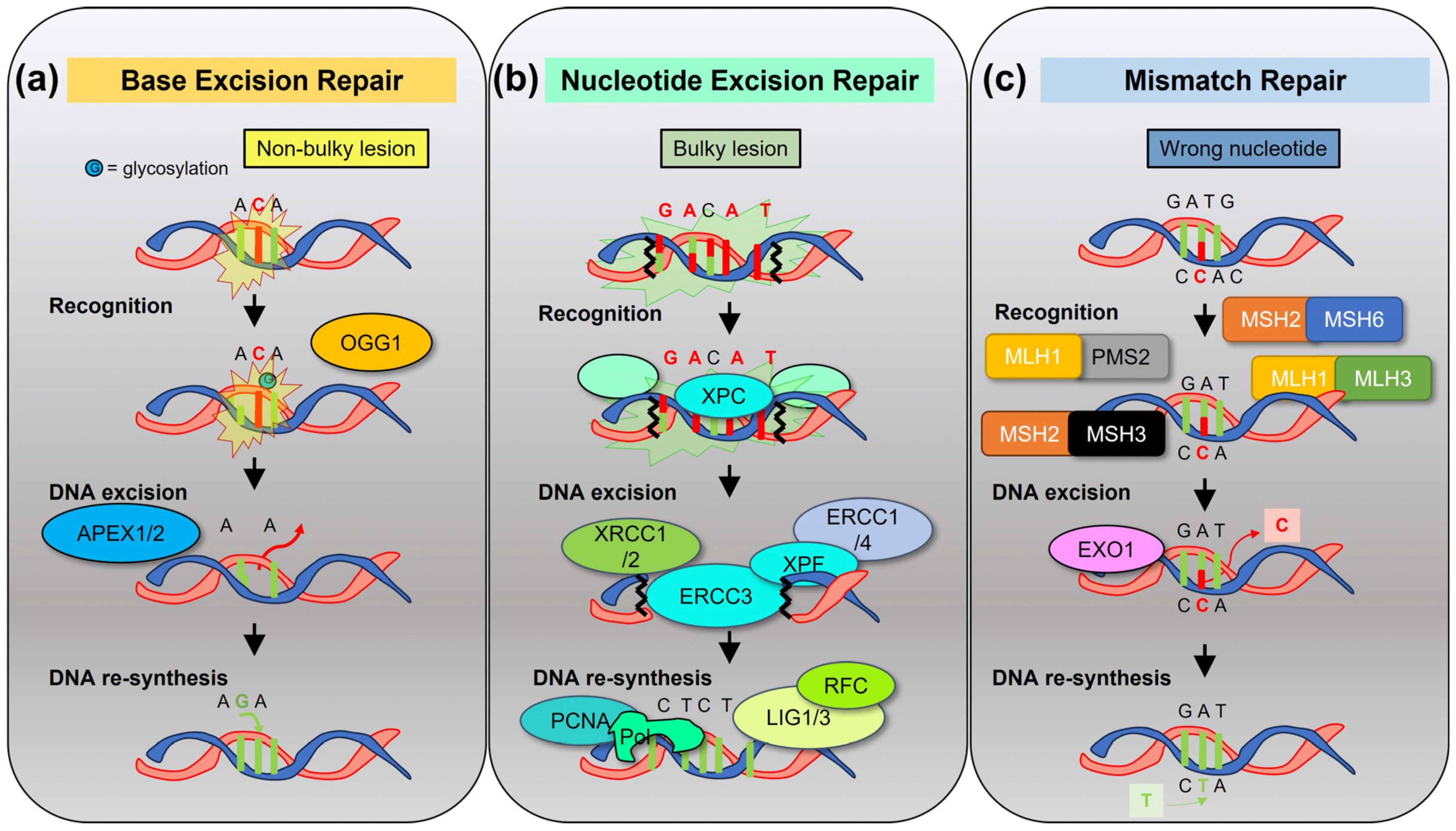
2.3. DNA Repair Pathways in MM
3. Reactive Oxygen Species in DDR and Immunogenic Cell Death
4. Mutations and Biomarkers of DNA Damage
4.1. Genomic Alterations Related to DDR and DNA Repair in MM
4.2. Biomarkers
4.3. Relationship with Myeloid Neoplasm Conditions
5. Therapeutic Interventions
6. Final Considerations on DNA Repair Inhibitors and Their Clinical Use in MM
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Rajkumar, S.V. Monoclonal gammopathy of undetermined significance. Br. J. Haematol. 2006, 134, 573–589. [Google Scholar] [CrossRef]
- Robiou du Pont, S.; Cleynen, A.; Fontan, C.; Attal, M.; Munshi, N.; Corre, J.; Avet-Loiseau, H. Genomics of Multiple Myeloma. J. Clin. Oncol. 2017, 35, 963–967. [Google Scholar] [CrossRef]
- Bustoros, M.; Sklavenitis-Pistofidis, R.; Park, J.; Redd, R.; Zhitomirsky, B.; Dunford, A.J.; Salem, K.; Tai, Y.T.; Anand, S.; Mouhieddine, T.H.; et al. Genomic Profiling of Smoldering Multiple Myeloma Identifies Patients at a High Risk of Disease Progression. J. Clin. Oncol. 2020, 38, 2380–2389. [Google Scholar] [CrossRef]
- Neben, K.; Jauch, A.; Hielscher, T.; Hillengass, J.; Lehners, N.; Seckinger, A.; Granzow, M.; Raab, M.S.; Ho, A.D.; Goldschmidt, H.; et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J. Clin. Oncol. 2013, 31, 4325–4332. [Google Scholar] [CrossRef]
- Abdallah, N.; Rajkumar, S.V.; Greipp, P.; Kapoor, P.; Gertz, M.A.; Dispenzieri, A.; Baughn, L.B.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; et al. Cytogenetic abnormalities in multiple myeloma: Association with disease characteristics and treatment response. Blood Cancer J. 2020, 10, 82. [Google Scholar] [CrossRef]
- Attal, M.; Lauwers-Cances, V.; Hulin, C.; Leleu, X.; Caillot, D.; Escoffre, M.; Arnulf, B.; Macro, M.; Belhadj, K.; Garderet, L.; et al. Lenalidomide, Bortezomib, and Dexamethasone with Transplantation for Myeloma. N. Engl. J. Med. 2017, 376, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.G.; Jacobus, S.J.; Weller, E.A.; Hassoun, H.; Lonial, S.; Raje, N.S.; Medvedova, E.; McCarthy, P.L.; Libby, E.N.; Voorhees, P.M.; et al. Triplet Therapy, Transplantation, and Maintenance until Progression in Myeloma. N. Engl. J. Med. 2022, 387, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Samur, M.K.; Roncador, M.; Aktas Samur, A.; Fulciniti, M.; Bazarbachi, A.H.; Szalat, R.E.; Shammas, M.A.; Sperling, A.S.; Richardson, P.G.; Magrangeas, F.; et al. High-Dose Melphalan Treatment Significantly Increases Mutational Burden at Relapse in Multiple Myeloma. Blood 2023, 141, 1724–1736. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef]
- Watanabe, N.; Broome, M.; Hunter, T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995, 14, 1878–1891. [Google Scholar] [CrossRef]
- Caldecott, K.W. DNA single-strand break repair and human genetic disease. Trends Cell Biol. 2022, 32, 733–745. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Horton, J.K.; Prasad, R.; Wilson, S.H. Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism. Annu. Rev. Biochem. 2019, 88, 137–162. [Google Scholar] [CrossRef]
- De Silva, I.U.; McHugh, P.J.; Clingen, P.H.; Hartley, J.A. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell Biol. 2000, 20, 7980–7990. [Google Scholar] [CrossRef] [PubMed]
- Evrard, C.; Tachon, G.; Randrian, V.; Karayan-Tapon, L.; Tougeron, D. Microsatellite Instability: Diagnosis, Heterogeneity, Discordance, and Clinical Impact in Colorectal Cancer. Cancers 2019, 11, 1567. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.D.; Grompe, M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 2003, 3, 23–34. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef]
- Ford, D.; Easton, D.F.; Peto, J. Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am. J. Hum. Genet. 1995, 57, 1457–1462. [Google Scholar]
- Gonzalez-Angulo, A.M.; Timms, K.M.; Liu, S.; Chen, H.; Litton, J.K.; Potter, J.; Lanchbury, J.S.; Stemke-Hale, K.; Hennessy, B.T.; Arun, B.K.; et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin. Cancer Res. 2011, 17, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Casadei, S.; Lee, M.K.; Pennil, C.C.; Nord, A.S.; Thornton, A.M.; Roeb, W.; Agnew, K.J.; Stray, S.M.; Wickramanayake, A.; et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 18032–18037. [Google Scholar] [CrossRef]
- Datta, A.; Biswas, K.; Sommers, J.A.; Thompson, H.; Awate, S.; Nicolae, C.M.; Thakar, T.; Moldovan, G.L.; Shoemaker, R.H.; Sharan, S.K.; et al. WRN helicase safeguards deprotected replication forks in BRCA2-mutated cancer cells. Nat. Commun. 2021, 12, 6561. [Google Scholar] [CrossRef]
- D’Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Kornepati, A.V.R.; Boyd, J.T.; Murray, C.E.; Saifetiarova, J.; de la Pena Avalos, B.; Rogers, C.M.; Bai, H.; Padron, A.S.; Liao, Y.; Ontiveros, C.; et al. Tumor Intrinsic PD-L1 Promotes DNA Repair in Distinct Cancers and Suppresses PARP Inhibitor-Induced Synthetic Lethality. Cancer Res. 2022, 82, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, Z.; Hu, R.; Dong, M.; Zhou, X.; Ren, S.; Zhang, Y.; Chen, C.; Huang, R.; Zhu, M.; et al. Metabolic Intervention Liposome Boosted Lung Cancer Radio-Immunotherapy via Hypoxia Amelioration and PD-L1 Restraint. Adv. Sci. 2023, 10, e2207608. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, Y.; Jiang, X.; Zheng, C.; Luo, W.; Xiang, X.; Qi, X.; Shen, J. Metformin modified chitosan as a multi-functional adjuvant to enhance cisplatin-based tumor chemotherapy efficacy. Int. J. Biol. Macromol. 2023, 224, 797–809. [Google Scholar] [CrossRef]
- Sato, H.; Niimi, A.; Yasuhara, T.; Permata, T.B.M.; Hagiwara, Y.; Isono, M.; Nuryadi, E.; Sekine, R.; Oike, T.; Kakoti, S.; et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017, 8, 1751. [Google Scholar] [CrossRef]
- Walters, D.K.; Wu, X.; Tschumper, R.C.; Arendt, B.K.; Huddleston, P.M.; Henderson, K.J.; Dispenzieri, A.; Jelinek, D.F. Evidence for ongoing DNA damage in multiple myeloma cells as revealed by constitutive phosphorylation of H2AX. Leukemia 2011, 25, 1344–1353. [Google Scholar] [CrossRef]
- Cottini, F.; Hideshima, T.; Xu, C.; Sattler, M.; Dori, M.; Agnelli, L.; ten Hacken, E.; Bertilaccio, M.T.; Antonini, E.; Neri, A.; et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat. Med. 2014, 20, 599–606. [Google Scholar] [CrossRef]
- Maruyama, J.; Inami, K.; Michishita, F.; Jiang, X.; Iwasa, H.; Nakagawa, K.; Ishigami-Yuasa, M.; Kagechika, H.; Miyamura, N.; Hirayama, J.; et al. Novel YAP1 Activator, Identified by Transcription-Based Functional Screen, Limits Multiple Myeloma Growth. Mol. Cancer Res. 2018, 16, 197–211. [Google Scholar] [CrossRef]
- Cottini, F.; Hideshima, T.; Suzuki, R.; Tai, Y.T.; Bianchini, G.; Richardson, P.G.; Anderson, K.C.; Tonon, G. Synthetic Lethal Approaches Exploiting DNA Damage in Aggressive Myeloma. Cancer Discov. 2015, 5, 972–987. [Google Scholar] [CrossRef]
- Abdallah, N.; Baughn, L.B.; Rajkumar, S.V.; Kapoor, P.; Gertz, M.A.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Dingli, D.; et al. Implications of MYC Rearrangements in Newly Diagnosed Multiple Myeloma. Clin. Cancer Res. 2020, 26, 6581–6588. [Google Scholar] [CrossRef]
- Affer, M.; Chesi, M.; Chen, W.G.; Keats, J.J.; Demchenko, Y.N.; Roschke, A.V.; Van Wier, S.; Fonseca, R.; Bergsagel, P.L.; Kuehl, W.M. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014, 28, 1725–1735. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.E.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Botrugno, O.A.; Bianchessi, S.; Zambroni, D.; Frenquelli, M.; Belloni, D.; Bongiovanni, L.; Girlanda, S.; Di Terlizzi, S.; Ferrarini, M.; Ferrero, E.; et al. ATR addiction in multiple myeloma: Synthetic lethal approaches exploiting established therapies. Haematologica 2020, 105, 2440–2447. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Boyle, E.M.; Wardell, C.P.; Murison, A.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Johnson, D.C.; Kaiser, M.F.; Melchor, L.; et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J. Clin. Oncol. 2015, 33, 3911–3920. [Google Scholar] [CrossRef]
- Galm, O.; Wilop, S.; Reichelt, J.; Jost, E.; Gehbauer, G.; Herman, J.G.; Osieka, R. DNA methylation changes in multiple myeloma. Leukemia 2004, 18, 1687–1692. [Google Scholar] [CrossRef]
- Sweiss, K.; Barwick, B.G.; Calip, G.S.; Rondelli, D.; Hofmeister, C.C.; Patel, P. Increased DNA Repair Gene Expression Correlates with MYC Expression and Inferior Progression-Free Survival in Multiple Myeloma Patients. Blood 2020, 136, 48–49. [Google Scholar] [CrossRef]
- Kumar, S.; Talluri, S.; Pal, J.; Yuan, X.; Lu, R.; Nanjappa, P.; Samur, M.K.; Munshi, N.C.; Shammas, M.A. Role of apurinic/apyrimidinic nucleases in the regulation of homologous recombination in myeloma: Mechanisms and translational significance. Blood Cancer J. 2018, 8, 92. [Google Scholar] [CrossRef]
- Gkotzamanidou, M.; Terpos, E.; Bamia, C.; Munshi, N.C.; Dimopoulos, M.A.; Souliotis, V.L. DNA repair of myeloma plasma cells correlates with clinical outcome: The effect of the nonhomologous end-joining inhibitor SCR7. Blood 2016, 128, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Szalat, R.; Samur, M.K.; Fulciniti, M.; Lopez, M.; Nanjappa, P.; Cleynen, A.; Wen, K.; Kumar, S.; Perini, T.; Calkins, A.S.; et al. Nucleotide excision repair is a potential therapeutic target in multiple myeloma. Leukemia 2018, 32, 111–119. [Google Scholar] [CrossRef]
- Velangi, M.R.; Matheson, E.C.; Morgan, G.J.; Jackson, G.H.; Taylor, P.R.; Hall, A.G.; Irving, J.A. DNA mismatch repair pathway defects in the pathogenesis and evolution of myeloma. Carcinogenesis 2004, 25, 1795–1803. [Google Scholar] [CrossRef][Green Version]
- Chim, C.S.; Liang, R.; Leung, M.H.; Kwong, Y.L. Aberrant gene methylation implicated in the progression of monoclonal gammopathy of undetermined significance to multiple myeloma. J. Clin. Pathol. 2007, 60, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Santon, A.; Garcia-Cosio, M.; Bellas, C. hMLH1 and MGMT inactivation as a mechanism of tumorigenesis in monoclonal gammopathies. Mod. Pathol. 2006, 19, 914–921. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Q.; van der Sluis, P.C.; Boulware, D.; Hazlehurst, L.A.; Dalton, W.S. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood 2005, 106, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, D.; Di Martino, M.T.; Amodio, N.; Morelli, E.; Montesano, M.; Botta, C.; Scionti, F.; Talarico, D.; Altomare, E.; Gallo Cantafio, M.E.; et al. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia 2019, 33, 487–498. [Google Scholar] [CrossRef]
- Caracciolo, D.; Scionti, F.; Juli, G.; Altomare, E.; Golino, G.; Todoerti, K.; Grillone, K.; Riillo, C.; Arbitrio, M.; Iannone, M.; et al. Exploiting MYC-induced PARPness to target genomic instability in multiple myeloma. Haematologica 2021, 106, 185–195. [Google Scholar] [CrossRef]
- Neri, P.; Ren, L.; Gratton, K.; Stebner, E.; Johnson, J.; Klimowicz, A.; Duggan, P.; Tassone, P.; Mansoor, A.; Stewart, D.A.; et al. Bortezomib-induced “BRCAness” sensitizes multiple myeloma cells to PARP inhibitors. Blood 2011, 118, 6368–6379. [Google Scholar] [CrossRef]
- Caracciolo, D.; Juli, G.; Riillo, C.; Coricello, A.; Vasile, F.; Pollastri, S.; Rocca, R.; Scionti, F.; Polera, N.; Grillone, K.; et al. Exploiting DNA Ligase III addiction of multiple myeloma by flavonoid Rhamnetin. J. Transl. Med. 2022, 20, 482. [Google Scholar] [CrossRef]
- Driscoll, J.E.A. Eosinophils Upregulate PD-L1 and PD-L2 Expression to Enhance the Immunosuppressive Microenvironment in Multiple Myeloma. Blood 2017, 130, 4417. [Google Scholar]
- Iwasa, M.; Harada, T.; Oda, A.; Bat-Erdene, A.; Teramachi, J.; Tenshin, H.; Ashtar, M.; Oura, M.; Sogabe, K.; Udaka, K.; et al. PD-L1 upregulation in myeloma cells by panobinostat in combination with interferon-gamma. Oncotarget 2019, 10, 1903–1917. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Sun, Y.; Torphy, R.J.; He, J.; Yanaga, K.; Edil, B.H.; Schulick, R.D.; Zhu, Y. Pomalidomide Inhibits PD-L1 Induction to Promote Antitumor Immunity. Cancer Res. 2018, 78, 6655–6665. [Google Scholar] [CrossRef]
- Gorgun, G.; Samur, M.K.; Cowens, K.B.; Paula, S.; Bianchi, G.; Anderson, J.E.; White, R.E.; Singh, A.; Ohguchi, H.; Suzuki, R.; et al. Lenalidomide Enhances Immune Checkpoint Blockade-Induced Immune Response in Multiple Myeloma. Clin. Cancer Res. 2015, 21, 4607–4618. [Google Scholar] [CrossRef] [PubMed]
- Yarde, D.N.; Oliveira, V.; Mathews, L.; Wang, X.; Villagra, A.; Boulware, D.; Shain, K.H.; Hazlehurst, L.A.; Alsina, M.; Chen, D.T.; et al. Targeting the Fanconi anemia/BRCA pathway circumvents drug resistance in multiple myeloma. Cancer Res. 2009, 69, 9367–9375. [Google Scholar] [CrossRef]
- Akcora-Yildiz, D.; Ozkan, T.; Ozen, M.; Gunduz, M.; Sunguroglu, A.; Beksac, M. Werner helicase is required for proliferation and DNA damage repair in multiple myeloma. Mol. Biol. Rep. 2023, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, M.O.; Perry, N.J.S.; Poulogiannis, G. DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 2018, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Barlow, C.; Dennery, P.A.; Shigenaga, M.K.; Smith, M.A.; Morrow, J.D.; Roberts, L.J.; Wynshaw-Boris, A.; Levine, R.L. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc. Natl. Acad. Sci. USA 1999, 96, 9915–9919. [Google Scholar] [CrossRef] [PubMed]
- Agathanggelou, A.; Weston, V.J.; Perry, T.; Davies, N.J.; Skowronska, A.; Payne, D.T.; Fossey, J.S.; Oldreive, C.E.; Wei, W.; Pratt, G.; et al. Targeting the Ataxia Telangiectasia Mutated-null phenotype in chronic lymphocytic leukemia with pro-oxidants. Haematologica 2015, 100, 1076–1085. [Google Scholar] [CrossRef]
- Alexander, A.; Cai, S.L.; Kim, J.; Nanez, A.; Sahin, M.; MacLean, K.H.; Inoki, K.; Guan, K.L.; Shen, J.; Person, M.D.; et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA 2010, 107, 4153–4158. [Google Scholar] [CrossRef]
- Lee, J.H.; Paull, T.T. Mitochondria at the crossroads of ATM-mediated stress signaling and regulation of reactive oxygen species. Redox Biol. 2020, 32, 101511. [Google Scholar] [CrossRef]
- Santos, C.X.; Tanaka, L.Y.; Wosniak, J.; Laurindo, F.R. Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: Roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid. Redox Signal. 2009, 11, 2409–2427. [Google Scholar] [CrossRef]
- Caillot, M.; Dakik, H.; Mazurier, F.; Sola, B. Targeting Reactive Oxygen Species Metabolism to Induce Myeloma Cell Death. Cancers 2021, 13, 2411. [Google Scholar] [CrossRef]
- Xiong, S.; Chng, W.J.; Zhou, J. Crosstalk between endoplasmic reticulum stress and oxidative stress: A dynamic duo in multiple myeloma. Cell. Mol. Life Sci. 2021, 78, 3883–3906. [Google Scholar] [CrossRef] [PubMed]
- Obeng, E.A.; Carlson, L.M.; Gutman, D.M.; Harrington, W.J., Jr.; Lee, K.P.; Boise, L.H. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 2006, 107, 4907–4916. [Google Scholar] [CrossRef] [PubMed]
- Raninga, P.V.; Di Trapani, G.; Vuckovic, S.; Bhatia, M.; Tonissen, K.F. Inhibition of thioredoxin 1 leads to apoptosis in drug-resistant multiple myeloma. Oncotarget 2015, 6, 15410–15424. [Google Scholar] [CrossRef] [PubMed]
- Gulla, A.; Morelli, E.; Samur, M.K.; Botta, C.; Hideshima, T.; Bianchi, G.; Fulciniti, M.; Malvestiti, S.; Prabhala, R.H.; Talluri, S.; et al. Bortezomib induces anti-multiple myeloma immune response mediated by cGAS/STING pathway activation. Blood Cancer Discov. 2021, 2, 468–483. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.V.; Garg, A.D.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Austen, B.; Barone, G.; Reiman, A.; Byrd, P.J.; Baker, C.; Starczynski, J.; Nobbs, M.C.; Murphy, R.P.; Enright, H.; Chaila, E.; et al. Pathogenic ATM mutations occur rarely in a subset of multiple myeloma patients. Br. J. Haematol. 2008, 142, 925–933. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia 2019, 33, 159–170. [Google Scholar] [CrossRef]
- Maura, F.; Petljak, M.; Lionetti, M.; Cifola, I.; Liang, W.; Pinatel, E.; Alexandrov, L.B.; Fullam, A.; Martincorena, I.; Dawson, K.J.; et al. Biological and prognostic impact of APOBEC-induced mutations in the spectrum of plasma cell dyscrasias and multiple myeloma cell lines. Leukemia 2018, 32, 1044–1048. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Murison, A.; Boyle, E.M.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Melchor, L.; Pawlyn, C.; Kaiser, M.F.; et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 2015, 6, 6997. [Google Scholar] [CrossRef]
- Talluri, S.; Samur, M.K.; Buon, L.; Kumar, S.; Potluri, L.B.; Shi, J.; Prabhala, R.H.; Shammas, M.A.; Munshi, N.C. Dysregulated APOBEC3G causes DNA damage and promotes genomic instability in multiple myeloma. Blood Cancer J. 2021, 11, 166. [Google Scholar] [CrossRef] [PubMed]
- Dumontet, C.; Landi, S.; Reiman, T.; Perry, T.; Plesa, A.; Bellini, I.; Barale, R.; Pilarski, L.M.; Troncy, J.; Tavtigian, S.; et al. Genetic polymorphisms associated with outcome in multiple myeloma patients receiving high-dose melphalan. Bone Marrow Transpl. 2010, 45, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Ushie, C.; Saitoh, T.; Iwasaki, A.; Moriyama, N.; Hattori, H.; Matsumoto, M.; Sawamura, M.; Isoda, J.; Handa, H.; Yokohama, A.; et al. The Polymorphisms of Base Excision Repair Genes Influence the Prognosis of Multiple Myeloma. Blood 2012, 120, 3981. [Google Scholar] [CrossRef]
- Struewing, J.P.; Hartge, P.; Wacholder, S.; Baker, S.M.; Berlin, M.; McAdams, M.; Timmerman, M.M.; Brody, L.C.; Tucker, M.A. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N. Engl. J. Med. 1997, 336, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Ferrara, K.; Barlogie, B.; Coleman, E.A.; Lynch, J.F.; Weisenburger, D.; Sanger, W.; Watson, P.; Nipper, H.; Witt, V.; et al. Familial myeloma. N. Engl. J. Med. 2008, 359, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.A.; Souliotis, V.L.; Anagnostopoulos, A.; Bamia, C.; Pouli, A.; Baltadakis, I.; Terpos, E.; Kyrtopoulos, S.A.; Sfikakis, P.P. Melphalan-induced DNA damage in vitro as a predictor for clinical outcome in multiple myeloma. Haematologica 2007, 92, 1505–1512. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maura, F.; Degasperi, A.; Nadeu, F.; Leongamornlert, D.; Davies, H.; Moore, L.; Royo, R.; Ziccheddu, B.; Puente, X.S.; Avet-Loiseau, H.; et al. A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun. 2019, 10, 2969. [Google Scholar] [CrossRef]
- Mailankody, S.; Pfeiffer, R.M.; Kristinsson, S.Y.; Korde, N.; Bjorkholm, M.; Goldin, L.R.; Turesson, I.; Landgren, O. Risk of acute myeloid leukemia and myelodysplastic syndromes after multiple myeloma and its precursor disease (MGUS). Blood 2011, 118, 4086–4092. [Google Scholar] [CrossRef]
- Palumbo, A.; Bringhen, S.; Kumar, S.K.; Lupparelli, G.; Usmani, S.; Waage, A.; Larocca, A.; van der Holt, B.; Musto, P.; Offidani, M.; et al. Second primary malignancies with lenalidomide therapy for newly diagnosed myeloma: A meta-analysis of individual patient data. Lancet Oncol. 2014, 15, 333–342. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Chitre, S.; Stolzel, F.; Cuthill, K.; Streetly, M.; Graham, C.; Dill, C.; Mohamedali, A.; Smith, A.; Schetelig, J.; Altmann, H.; et al. Clonal hematopoiesis in patients with multiple myeloma undergoing autologous stem cell transplantation. Leukemia 2018, 32, 2020–2024. [Google Scholar] [CrossRef] [PubMed]
- Wudhikarn, K.; Padrnos, L.; Lasho, T.; LaPlant, B.; Kumar, S.; Dispenzieri, A.; Lacy, M.; Rajkumar, S.V.; Gertz, M.; Mangaonkar, A.A.; et al. Clinical correlates and prognostic impact of clonal hematopoiesis in multiple myeloma patients receiving post-autologous stem cell transplantation lenalidomide maintenance therapy. Am. J. Hematol. 2021, 96, E157–E162. [Google Scholar] [CrossRef] [PubMed]
- Landau, H.J.; McNeely, S.C.; Nair, J.S.; Comenzo, R.L.; Asai, T.; Friedman, H.; Jhanwar, S.C.; Nimer, S.D.; Schwartz, G.K. The checkpoint kinase inhibitor AZD7762 potentiates chemotherapy-induced apoptosis of p53-mutated multiple myeloma cells. Mol. Cancer Ther. 2012, 11, 1781–1788. [Google Scholar] [CrossRef]
- Teoh, P.J.; An, O.; Chung, T.H.; Vaiyapuri, T.; Raju, A.; Hoppe, M.M.; Toh, S.H.M.; Wang, W.; Chan, M.C.; Fullwood, M.J.; et al. p53-NEIL1 co-abnormalities induce genomic instability and promote synthetic lethality with Chk1 inhibition in multiple myeloma having concomitant 17p13(del) and 1q21(gain). Oncogene 2022, 41, 2106–2121. [Google Scholar] [CrossRef]
- Mueller, S.; Haas-Kogan, D.A. WEE1 Kinase As a Target for Cancer Therapy. J. Clin. Oncol. 2015, 33, 3485–3487. [Google Scholar] [CrossRef] [PubMed]
- Hauge, S.; Naucke, C.; Hasvold, G.; Joel, M.; Rodland, G.E.; Juzenas, P.; Stokke, T.; Syljuasen, R.G. Combined inhibition of Wee1 and Chk1 gives synergistic DNA damage in S-phase due to distinct regulation of CDK activity and CDC45 loading. Oncotarget 2017, 8, 10966–10979. [Google Scholar] [CrossRef]
- Barbosa, R.S.S.; Dantonio, P.M.; Guimaraes, T.; de Oliveira, M.B.; Fook Alves, V.L.; Sandes, A.F.; Fernando, R.C.; Colleoni, G.W.B. Sequential combination of bortezomib and WEE1 inhibitor, MK-1775, induced apoptosis in multiple myeloma cell lines. Biochem. Biophys. Res. Commun. 2019, 519, 597–604. [Google Scholar] [CrossRef]
- Liang, L.; He, Y.; Wang, H.; Zhou, H.; Xiao, L.; Ye, M.; Kuang, Y.; Luo, S.; Zuo, Y.; Feng, P.; et al. The Wee1 kinase inhibitor MK1775 suppresses cell growth, attenuates stemness and synergises with bortezomib in multiple myeloma. Br. J. Haematol. 2020, 191, 62–76. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrilla, C.; Galloway, J.; Kudalkar, R.; Ismael, A.; Cottini, F. Understanding DNA Damage Response and DNA Repair in Multiple Myeloma. Cancers 2023, 15, 4155. https://doi.org/10.3390/cancers15164155
Petrilla C, Galloway J, Kudalkar R, Ismael A, Cottini F. Understanding DNA Damage Response and DNA Repair in Multiple Myeloma. Cancers. 2023; 15(16):4155. https://doi.org/10.3390/cancers15164155
Chicago/Turabian StylePetrilla, Cole, Joshua Galloway, Ruchi Kudalkar, Aya Ismael, and Francesca Cottini. 2023. "Understanding DNA Damage Response and DNA Repair in Multiple Myeloma" Cancers 15, no. 16: 4155. https://doi.org/10.3390/cancers15164155
APA StylePetrilla, C., Galloway, J., Kudalkar, R., Ismael, A., & Cottini, F. (2023). Understanding DNA Damage Response and DNA Repair in Multiple Myeloma. Cancers, 15(16), 4155. https://doi.org/10.3390/cancers15164155





