Simple Summary
The human genome produces various types of RNA molecules. A significant portion of these are long non-coding RNAs (lncRNAs) exceeding 200 nts without an obvious open reading frame. There are more than 200,000 lncRNAs, but we have limited understanding of their functions. LncRNAs can influence gene activity by interacting with proteins or nucleic acids. Some lncRNAs regulate genes involved in cancer, either promoting or suppressing tumor growth. One lncRNA of interest is ANRIL (Antisense Noncoding RNA in the INK4 Locus), which can affect gene expression through different ways. However, ANRIL’s exact role in cancer is complex and varies in different situations, making it a challenging area to study. This review strives to offer a thorough comprehension of ANRIL’s role in regulating genes and its impact on the development of cancer.
Abstract
ANRIL (Antisense Noncoding RNA in the INK4 Locus), a long non-coding RNA encoded in the human chromosome 9p21 region, is a critical factor for regulating gene expression by interacting with multiple proteins and miRNAs. It has been found to play important roles in various cellular processes, including cell cycle control and proliferation. Dysregulation of ANRIL has been associated with several diseases like cancers and cardiovascular diseases, for instance. Understanding the oncogenic role of ANRIL and its potential as a diagnostic and prognostic biomarker in cancer is crucial. This review provides insights into the regulatory mechanisms and oncogenic significance of the 9p21 locus and ANRIL in cancer.
Keywords:
LncRNA; ANRIL; 9p21 locus; cancer; gene regulation; epigenetics; competitive endogenous RNA 1. LncRNAs and Cancers
The human genome is extensively transcribed into thousands of RNAs. A significant part of the resulting transcriptome corresponds to the long non-coding RNAs (lncRNAs) defined as RNAs longer than 200 nts and lacking obvious open reading frames. The number of lncRNAs exceeds 200,000 and the field lacks evidence supporting the functionality of most of them [1,2,3,4]. Compared to mRNAs, lncRNAs exhibit stronger tissue-specific expression and often function in a tissue-specific manner [5]. The broad definition of lncRNAs encompasses a large and highly heterogeneous collection of transcripts that differ at least in their subcellular localizations and functions. Cytoplasmic lncRNAs mainly modulate gene expression by affecting the different stages of mRNA life, including stability and translation, while nuclear lncRNAs mainly associate with the genome or with premature RNAs in synthesis to regulate gene expression and alternative splicing at the chromatin level, respectively [1]. For instance, by recruiting epigenetic writers, chromatin remodelers, transcription and splicing factors, several lncRNAs control the expression of genes nearby or/and distant from their hosting locus (-cis and -trans activity, respectively) [6,7]. The diversity of activities of the lncRNAs places them at a crossroad between the genomic and the epigenetic regulations qualified as supragenomic regulations. This refers to the genomic information beyond the level of individual genes, such as the non-coding regions of the genome, involved in the modulation of the organization and regulation of the genomic elements [8].
Cellular experiments have shown that lncRNAs are involved in a wide spectrum of biological processes ranging from cell proliferation, apoptosis and nutrient sensing to cell differentiation [9,10]. Consequently, the deregulation of their expression can affect cell homeostasis and may favor the occurrence and/or development of pathologies. This is consistent with the fact that lncRNAs have been associated with key aspects of the cancer biology such as uncontrollable proliferation, evasion of cell death, metastasis and drug resistance [11]. According to “the lncRNADisease 2.0 database”, lncRNAs are associated with 529 pathologies divided into several categories, including three major ones corresponding to cancers (44.2%), cardiovascular pathologies (11.6%) and neurodegenerative diseases (7.3%) [4]. For these reasons, several lncRNAs are already used as biomarkers as their expression rate correlates with the diagnostic nature of certain pathologies and more importantly they definitively may prove to be valuable therapeutic cancer targets [12,13].
Cancer is one of the leading causes of death worldwide and refers to a group of diseases characterized by the uncontrolled growth and spread of abnormal cells capable or not of invading nearby tissues and organs. Aberrant expressions and splicing profiles of lncRNAs have been found in various types of cancer in agreement with the fact that some lncRNAs act as oncogenes, promoting cancer development and progression, while others act as tumor suppressors, inhibiting cancer growth and metastasis [14,15]. For example, the lncRNA HOTAIR (HOX Transcript Antisense Intergenic RNA) has been found to be overexpressed in several types of cancer and promotes metastasis by interacting with chromatin-modifying enzymes and promoting epigenetic changes [16]. In contrast, the lncRNA GAS5 (Growth Arrest-Specific 5) has been shown to be downregulated in cancer and acts as a tumor suppressor by inhibiting cell proliferation and promoting apoptosis [17].
In this review, we focused on the implication of the lncRNA ANRIL (Antisense Noncoding RNA in the INK4 Locus, CDKN2B-AS1) in cancers. ANRIL is transcribed from the 9p21 locus in several linear and circular spliced variants mainly made of repetitive elements derived from LINE, SINE and LTR sequences [18,19,20]. The overexpression of some of them is correlated with pathologies such as cardiovascular disease (CVD), type 2 diabetes (T2D) and cancers [21,22].
ANRIL can modulate gene expression at the post-transcriptional level by acting as a competing element RNA (ceRNA) of miRNAs and proteins [23,24]. Additionally, ANRIL affects gene expression at the chromatin level negatively and positively by guiding the recruitment of chromatin modifiers or transcriptional activators at specific loci [25,26]. Recently, converging data showed that ANRIL is also able to affect the patterns of alternative splicing in HEK293 and HUVEC cells [20,27]. These regulatory activities are often prone to enhancing cell proliferation, migration, invasion, and metastasis, and to suppressing apoptosis and senescence mainly attributed to the modulation of the expression of key cancer-related genes involved, for instance, in the p53 axis. To our knowledge, ANRIL is the lncRNA with the highest frequency of alterations in the context of cancer development and progression.
The proposed roles of ANRIL in cancer biology are still unclear. This is mainly due to differences in the types of cancer studied and/or the methods used to evaluate ANRIL expression likely to exclude the functional contribution of the individual ANRIL isoforms for instance. It is also possible that ANRIL has different activities in different stages of cancer progression or in different cell types. This drastically complicates the understanding of the mechanisms through which ANRIL promotes cancer development. In this review, we addressed these points and made efforts to formalize a clearer vision of the intertwined domain related to ANRIL and cancers.
2. ANRIL and the 9p21 Locus
2.1. Discovery of the 9p21 Locus as a Region of High Interest
The 9p21 locus is located on the short arm (p) of chromosome 9 at region 21 and spans over 170 kilobases (kb). This locus has drawn particular attention due to the frequent observation of homozygous deletions or epigenetic modifications, such as DNA methylation-induced transcriptional silencing resulting in the inactivation of this genomic region in multiple cancer types. Additionally, single-nucleotide polymorphisms (SNPs) within this region are linked to several age-related disorders, CVD and cancers. These SNPs are predominantly found within the genes themselves and the adjacent gene desert region spanning approximately 0.3 megabases [28]. Note that the precise mechanisms through which these SNPs exert their effects remain largely unknown.
The 9p21 locus was initially described as containing the CDKN2A/ARF and CDKN2B genes which encode three proteins involved in cell cycle regulation: the p14/ARF, p16INK4a/CDKN2A and p15INK4b/CDKN2B proteins, respectively [29]. Initially, the CDKN2A/ARF gene was found to be frequently inactivated or mutated in melanoma and other cancers, at least in part due to its ability to modulate the p53 axis [30]. Further investigations allowed researchers to link the 9p21 locus to various other abnormal situations, including atherosclerosis, T2D, Alzheimer’s disease, lupus erythematosus, epilepsy, glaucoma, obsessive compulsive disorder and sepsis [31,32,33,34,35,36,37,38]. The specific genetic variants and mechanisms underlying these associations are complex and continue to be the subject of ongoing research.
Pasmant and colleagues made a significant finding in 2007 by identifying the long non-coding RNA ANRIL encoded within the 9p21 locus. It was promptly recognized as a critical factor in controlling the expression of this locus [31]. As previously stated, this review is directed towards the association between ANRIL and cancers. For comprehensive insights into ANRIL and its involvement in other pathologies, we suggest referring to the reviews authored by Hannou et al., 2015 and Kong et al., 2018 for instance [22,39].
2.2. The lncRNA ANRIL and the Genes of the 9p21 Locus
As mentioned above, the 9p21 locus includes the gene that encodes the long non-coding RNA ANRIL. This gene is orientated in the opposite direction (antisense) to the genes within the locus and extends over a region of 126 kb, covering the CDKN2B gene located within its first intron. The intergenic region separating the ANRIL gene from the CDKN2A/ARF gene is a bidirectional promoter long to approximately 300 bp [31,40,41].
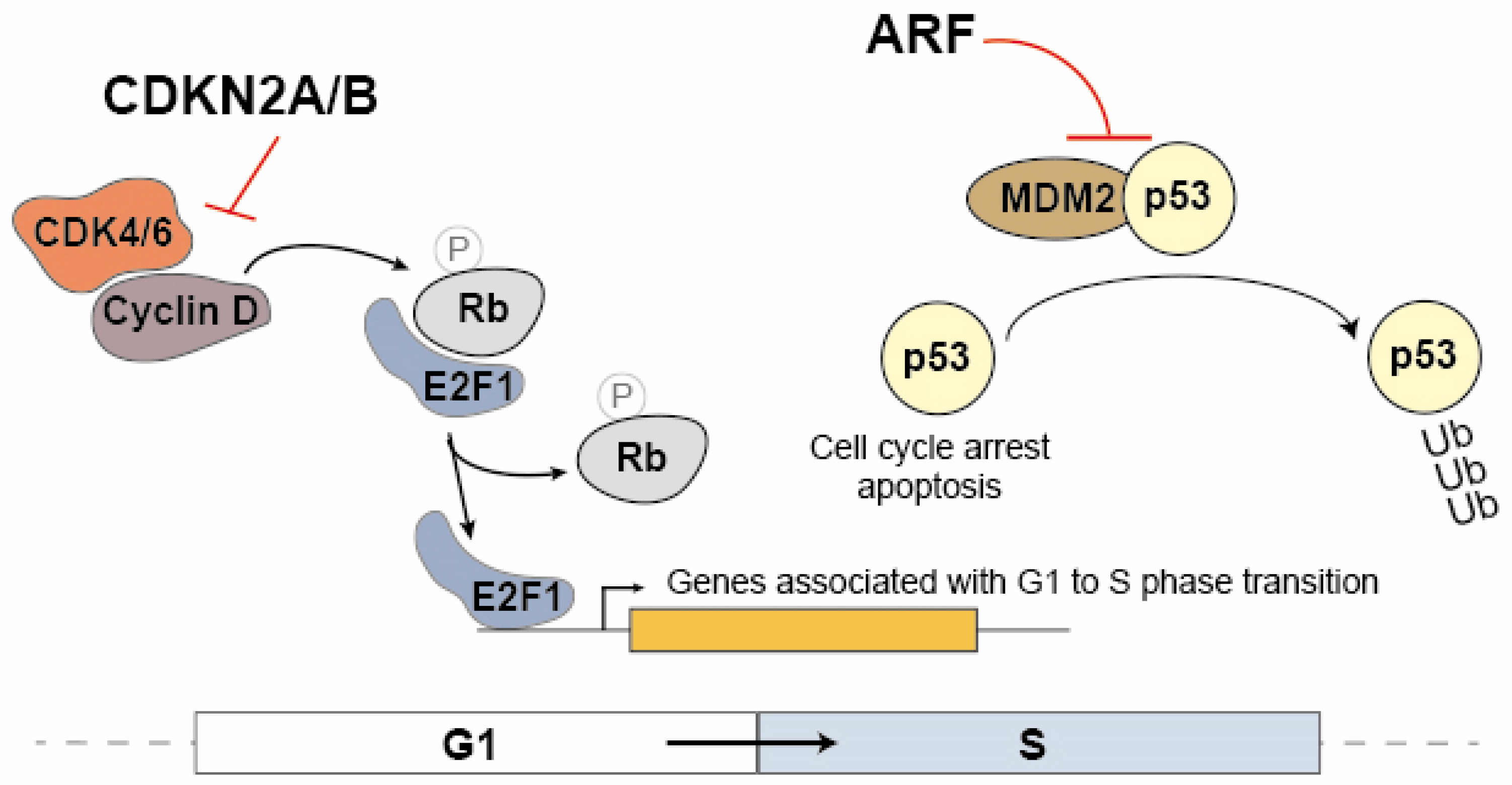
The CDKN2A and CDKN2B proteins are cyclin inhibitors that regulate cell cycle progression in the G1/S phase by inhibiting the association between CDK4/6 and cyclin D (Figure 1). In the G1 phase, the Rb protein (Retinoblastoma Protein) sequesters the E2F1 transcription factor. During the transition to the S phase, the Rb protein is phosphorylated by the CDK4/6 and cyclin D complex, leading to the dissociation of E2F1. As a result, E2F1 becomes capable of activating the transcription of genes associated with the transition to the S phase [29,42]. ARF also contributes to cell cycle modulation by promoting the dissociation of the MDM2 ubiquitin ligase from p53, leading to its stabilization and consequently to the activation of the cell cycle arrest at the G1/S (Figure 1) [43]. These proteins play a crucial role in regulating the cell cycle, and any dysfunctions in their expression may have significant implications for cancer. Given that ANRIL can modulate the expression of multiple genes, including CDKN2B, it is compelling to consider ANRIL a critical contributor to many of the pathological processes dependent on the 9p21 locus.
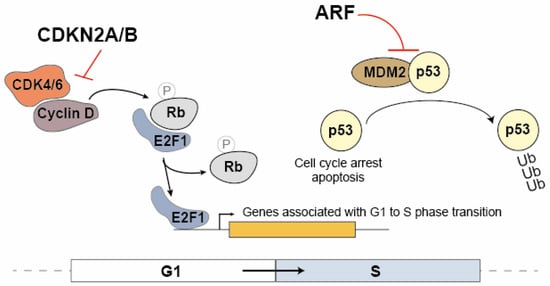
Figure 1.
The regulatory mechanisms involving CDKN2A, CDKN2B and ARF on the cell cycle progression. In the G1 phase, the Rb protein (Retinoblastoma Protein) binds to the E2F1 transcription factor, sequestering it. Upon transitioning to the S phase, the CDK4/6 and cyclin D complex phosphorylates the Rb protein, resulting in the release of E2F1. Consequently, E2F1 becomes active and promotes the transcription of genes involved in the transition to the S phase. CDKN2A/2B inhibits the association between CDK4/6 and cyclin D, therefore acting as cyclin inhibitors to control cell cycle progression during the G1/S phase. Additionally, ARF plays a role in cell cycle modulation by promoting the dissociation of the MDM2 ubiquitin ligase from p53, leading to p53 stabilization. This stabilization activates cell cycle arrest at the G1/S barrier.
3. ANRIL Expression and Abundance
As the vast majority of the lncRNAs, ANRIL expression is modest (less than 1000 copies per HEK293 cells) and tissue-specific [44,45]. ANRIL is also transcribed by the RNA polymerase II and processed through the canonical splicing, capping and polyadenylation pathways. The transcriptomic analysis of publicly available data by the GTEx consortium revealed that among 53 normal human tissues, ANRIL is more expressed in transverse colon, pituitary glands, small intestine and testis compared to the other analyzed tissues [46]. Beside tissue-specific expression, the ANRIL rate also appears to be dependent on the development stage. For instance, ANRIL is expressed from the toddler stage in testis (2–10 years) [47].
Since ANRIL is crucial for maintaining cellular homeostasis, the regulation of its expression is a very sensitive issue [48,49]. First, it has been shown that the 9p21 locus is contained within a single topologically associated domain (TAD) [50,51]. This specific arrangement depending at least on the CDKN2A promoter and 3 CTCF binding sequences within CDKN2B exon-1, ARF exon-1b, and CDKN2A exon-3 is likely to play a critical role in regulating the expression of the CDKN2A/ARF, CDKN2B, ANRIL genes in HUVEC cells and in the colon-cancer-related cell lines GES1, BGC823 and H1299 [50,52,53]. A recent study has demonstrated that the formation of this TAD relies on RNAs and CTCF, with both actors being essential for strengthening TAD insulation, facilitating interactions between enhancers and promoters, and promoting gene expression within the 9p21 locus [54].
ANRIL expression is regulated by multiple factors and stimuli-dependent mechanisms such as genotoxic stress and inflammatory response [26,41,52,55]. For instance, DNA damage activates the E2F1 transcription factor, leading to increased ANRIL expression and subsequently cell proliferation upon DNA repair [41]. HUVEC cells treated with TNF-α or IFNγ (interferon γ) show induction of ANRIL expression by NF-kB and STAT1 activation, respectively, both capable of associating response elements located within its promoter [26,52]. In retinoblastoma, hypoxia induces the direct HIF-1α binding to the ANRIL promoter region to transcriptionally activate its expression [56].
As mentioned before, the bidirectional promoter of the CDKN2A/ARF-ANRIL genes contains DNase I hypersensitivity regions and CpG islands. Interestingly, the methylation of these elements has been shown to inhibit the association of CTCF, resulting in a decrease in ANRIL expression associated with H3K4me3 reduction [57]. Note that validating a causal link between DNA methylation of the CDKN2A/ARF promoter and ANRIL transcription requires further clarification and could benefit from methods including CpG mutagenesis, reporter assays, and/or epigenetic editing techniques [58].
Conversely, the interaction between ERα (Estrogen Receptor Alpha) and its putative binding site located in this promoter region is facilitated by CpG methylation, resulting in increased ANRIL expression upon β-estradiol treatment of SW872 cells [59]. The methylation status of the ANRIL promoter region also influences the association of the transcription factors SMAD3/4, thereby affecting ANRIL expression levels in SaOS-2 osteosarcoma cells [60]. Additionally, TET2 (Tet Methylcytosine Dioxygenase 2), a tumor suppressor factor responsible for CpG demethylation, decreases ANRIL expression, leading to the inhibition of gastric cancer cell growth [61].
ANRIL expression is further modulated by the oncogenic transcription factors c-MYC, SOX2, and SP1 in lung, pharynx, and liver cancers, respectively [62,63,64]. Moreover, ANRIL expression is likely to be repressed by different factors, including Androgen Receptor (AR) and Phospholipase D (PLD) in prostate and lung cancer, respectively [65,66]. Finally, in the context of colon cancer, when HCT-8 and HCT116 cells were treated with Qingjie Fuzheng Granule (QFG), it was found that ANRIL expression decreased together with TGF-β1, phosphorylated (p)-SMAD2/3, SMAD4, and N-cadherin reduction [67].
Additionally, post-transcriptional mechanisms govern ANRIL abundance. In the context of colon cancer, it has been proposed that the association between AUF1 and ANRIL may lead to a detrimental effect on ANRIL stability and that the presence of P14AS lncRNA reduces the interaction between AUF1 and ANRIL. This competitive binding of P14AS lncRNA to AUF1 leads to an increase in ANRIL expression level [68]. In the same line, ANRIL is stabilized by IGF2BP3, favoring proliferation and metastasis in renal cancer tissues and related cell lines [69].
4. ANRIL Phylogeny, Evolution and Transposable Elements
LncRNAs exhibit lower levels of conservation compared to protein coding genes, and ANRIL is no exception to this trend. Through a phylogenetic analysis involving 27 species (from zebrafish to human), it was determined that ANRIL originated in placental mammals with a limited number of exons. As a result of specific evolutionary processes, an increased number of exons became evident within the haplorhines cladus, while exons’ erosion was observed during rodent evolution, limiting the use of murine models to study the association between ANRIL and cancers. The primate-specific expansion was attributed to the insertion of transposable elements (TE) within existing exons or introns, resulting in the modification of its functional capabilities [70]. Indeed, the evolution of the lncRNAs is a complex process that is influenced by a variety of factors, including TEs, which are DNA sequences that have the ability to move through a genome [71]. They make up a significant portion of many genomes, including those of humans in the range of more than 60% [72]. Interestingly, the proportion of TEs positively correlates with the number of lncRNA genes, suggesting that the generation and the evolution of the lncRNA genes highly depend on the presence of the TEs [72]. The latter have the ability to insert themselves into the genome, which, via exaptation, may lead to the creation of new transcription units by providing transcription factor binding sites or transcriptional start sites [73]. The resulting “junk RNA” produced via pervasive transcription may serve as a source for the evolution of lncRNA through non-adaptive or neutral processes. Over time, the lncRNA gene repertoire may undergo genome modifications that promote the acquisition of protein/DNA/RNA binding sites, thus conferring relevant functional capabilities to the resulting lncRNAs. These newly evolved lncRNAs can be recruited as new components of existing biological systems or may gain genetic elements controlling its expression in a tissue-specific manner [74]. This is consistent with the Repeat Insertion Domains of LncRNAs (RIDLs) hypothesis, which suggests that these repeat insertion domains act as functional RNA domains through two distinct mechanisms: a specific secondary structure that mediates (1) interaction with proteins, and (2) hybridization to nucleic acids [75]. Interestingly, TEs cover 35% of the ANRIL sequence, and the Exon8, which is 70% covered by the subcategory of LTR named ERVL-MaLR, has been described to be involved in ANRIL genomic occupancy [19,20]. Consequently, it appears that ANRIL adheres to the RIDL hypothesis.
5. ANRIL Exons and Isoforms
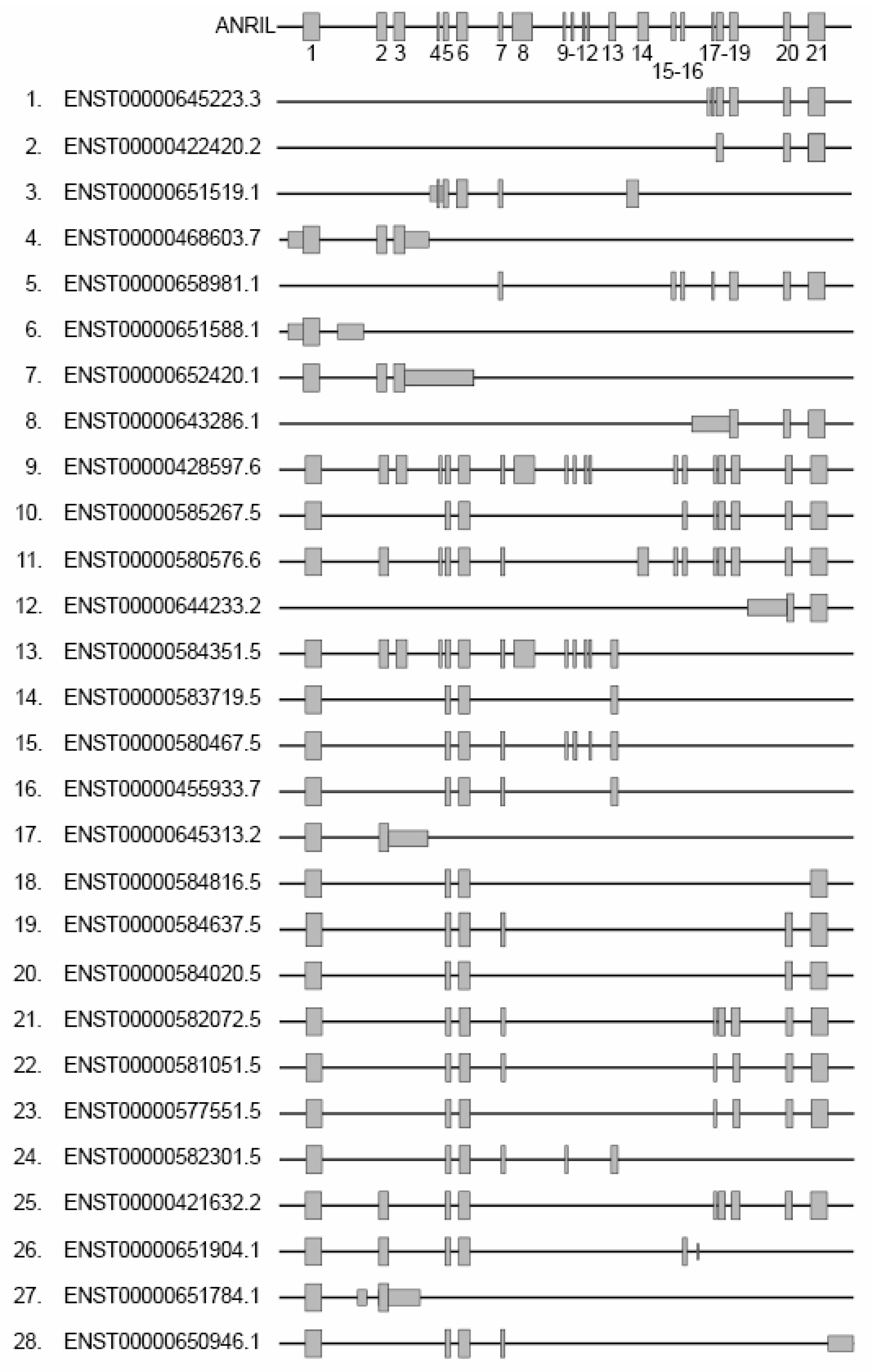
The ANRIL gene consists of 21 exons, with lengths ranging from 74 to 696 nucleotides (mean length of 202 nucleotides) (Figure 2). These exons undergo alternative splicing (AS), resulting in the generation of at least 28 different linear isoforms with varying lengths [76] (from 602 to 7713 nts). To date, the specific mechanism that regulates the alternative splicing of ANRIL is largely unknown. Only one recently published study proposes that m6A post-transcriptional modifications, which involve the addition of a methyl group to RNA molecules, may play a role in the regulation of ANRIL AS. These modifications could facilitate the recruitment of splicing regulators such as SRSF3, which contribute to the splicing process in pancreatic cancer [77].
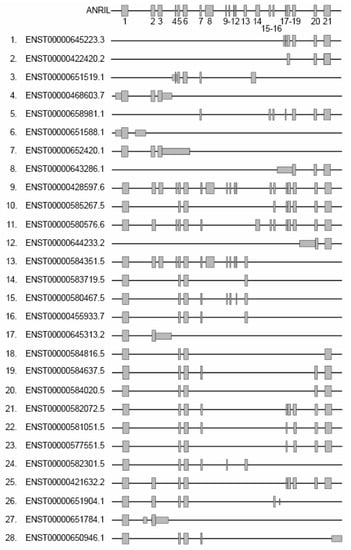
Figure 2.
Representation of the different linear isoforms of ANRIL. Grey rectangles represent exons, numbered from 1 to 21. The left side displays isoform accession numbers from the Ensembl database along with alternative nomenclature ranging from 1 to 28.
Figure 2 shows each linear isoform that we numbered from 1 to 28 in addition to the Ensembl accession numbers. Out of the 21 exons, exons 1, 5, and 6 are present in the majority of linear isoforms. These isoforms can be categorized into two groups based on their 3′ extremities and exon composition: the isoforms that include both exons 1 and 21 (isoforms 9–11, 18–23 and 25) or exons 1 and 13 (isoforms 13–16 and 24). When studying a melanoma cell line, it was found that the inclusion of exons in the isoforms varied, indicating a heterogeneous expression pattern. This suggests that multiple isoforms, possibly specific to different tissues, coexist within the cell with potential variable activities [44,45,77,78]. This is consistent with the fact that lncRNAs often consist of multiple exons arranged for creating distinct modules. As an example, the well-known lncRNA HOTAIR implicated in gene regulation and chromatin remodeling acts as a scaffold to recruit two distinct chromatin-modifying complexes and modulate proliferation in cell glioblastoma [79]. In this line, the ANRIL isoforms 9 (ENTS0000428597.6 or NR_003529) and 13 (ENTS00000584351.5 or DQ485454) have been recognized as regulators of gene expression with antagonistic roles. This duality in their functions has been observed in human endothelial cell lines in the context of CVD. When the isoform 9 is overexpressed, it leads to the down-regulation of the EZR and CXCL11 genes, as well as the up-regulation of TMEM106B. Conversely, overexpression of the isoform 13 has the opposite effect on these genes. These antagonistic activities are also evident at the cellular level of physiological processes. The expression of the isoform 9 appears to promote transmigration and cellular adhesion, while the isoform 13 elicits contrasting effects [80].
In addition, at least 30 circular isoforms have been identified, which include exons 4 to 16. Circular RNAs (circRNAs) are a type of lncRNA that are abundant, evolutionarily conserved, and often exhibit tissue-specific expression patterns. They are generated through a process called back splicing, and possess unique properties such as resistance to exonuclease degradation due to their lack of 5′ and 3′ ends [81]. CircRNAs can act as competitive endogenous RNA (ceRNA) by sequestering one or multiple RNA-binding proteins (RBP) or miRNAs [82]. Analysis of the ratio between circular and linear ANRIL isoforms within a cohort of patients with CVD and melanoma revealed a significant difference compared to the control groups [78,83]. In addition, Holdt and colleagues discovered a circANRIL, which exhibits anti-atherosclerotic properties by sequestrating PES1, a factor involved in the assembly of the 60S ribosomal subunit. The retention of PES1 promotes apoptosis and halts cell proliferation, both of which are functions associated with the development of atherosclerosis [24]. These findings suggest that the linear/circular ratio is inversely correlated with the progression of the disease, indicating an antagonistic relationship between these two isoform categories. Hence, maintaining an accurate equilibrium in the expression of ANRIL isoforms is believed to be deemed vital for appropriately modulating gene regulation, whereas an imbalance in their expression may profoundly impact cell physiology. Future investigations focusing on the comprehensive characterization of ANRIL isoforms in different cancer subtypes hold the promise of providing crucial insights into the contribution of ANRIL in cancer development.
6. ANRIL and Cancer
6.1. ANRIL Expression in Cancer
ANRIL has undergone extensive analysis, with researchers employing various methodologies such as RTqPCR, RNA sequencing, or microarrays across numerous cancer cell lines and tissues. In an effort to gain clarity, a review of the literature has been performed and 76 articles have been sorted based on several criteria, such as the accessibility of primer sequences employed in RTqPCR, to compile a comprehensive inventory of the ANRIL isoforms identified in each investigation. The results are indicated in Table 1 and demonstrate that the vast majority of the linear ANRIL isoforms (1, 2, 4, 6, 7, 9–11, 13–28) are upregulated in a wide range of cancer types, including lung (LC), gastric (GC), breast (BC), ovarian (OC), cervical (CC), colorectal (CRC), bladder (BladC), thyroid (TC), brain (BrC), osteosarcoma (OS), myeloma (MM), prostate (PC), leukemia (ATL/AML), melanoma, endometrial (EC), renal (RC), retinoblastoma (RB), head/neck (HNSCC/LSCC), intrahepatic cholangiocarcinoma (iCCA) and hepatocellular (HCC) cancers. The overexpression of ANRIL is associated with poor prognosis and a lower overall survival in GC, LC, HCC, HNSCC/LSCC, RC, iCCA, EC, AML, OS, CC and OC (Table 1). One meta-analysis published in 2022, based on a sample of 1708 cancer patients extracted from 23 studies across three databases, could also established a clear correlation between high ANRIL expression, adverse overall survival rates, larger tumor size, advanced TNM stage, and lymph node metastasis [84].
As mentioned before, it is possible that ANRIL has different activities in different stages of cancer progression. This is consistent with the fact that ANRIL expression is significantly correlated with a higher TNM stage of cancers, including LC, GC, HCC, LSCC, OS, CRC, CC, OC, HCC, BladC and OS (Table 1). In addition, while five articles demonstrate upregulation of ANRIL in non-small cell lung cancer and adenocarcinoma (NSCLC/LUAD) [48,66,85,86,87], one study reports the opposite effect in idiopathic pulmonary fibrosis (IPF), an independent risk factor of lung cancer with NSCLC being the main pathological type [88,89,90]. These differences may arise from variations in the analyzed tissues (NSCLC/LUAD vs. IPF) but possibly from the detection of distinct subsets of ANRIL isoforms too. Indeed, in NSCLC and LUAD, isoforms 1, 2, 4, 5, 7–13, 17, 19–23, and 25–27 were identified, whereas in IPF, only the detection of isoforms 3, 9, 11, 13, 15, 16, 19, 21, 22, 24, and 28 was performed. In consequence, an abundance of the isoforms 3, 15, 16, 24, and 28 was exclusively evaluated in IPF, suggesting that these isoforms may be downregulated in this condition. This observation is in favor of the existence of separate functional units within ANRIL isoforms, which may contribute to different activities involved in the physiological and pathological outcomes of lung-related conditions.
Altogether, these data strongly suggest that the majority of the linear ANRIL isoforms likely possess tumor-promoting capabilities. Since linear isoforms are mainly nuclear, one reasonably hypothesizes that ANRIL over-expression generates excess ANRIL molecules capable of reinforcing the regulatory activity it exerts on its direct targets or even on additional genomic regions. The molecular aberrations thus generated, including, for instance, inappropriate gene modulation, could be responsible for the appearance and/or reinforcement of pathological processes. In agreement, cell line experiments have shown that a reduction in ANRIL leads to increased apoptosis and senescence while decreasing cell proliferation, invasion, and migration.
To conclude, note that the relative levels of circular isoforms of ANRIL have not been systematically explored in most studies. Only two articles have reported an upregulation of circular ANRIL isoforms in cancer, specifically in CC and melanoma [78,91]. Although their association with cancer remains somewhat poorly assessed, it is conceivable to hypothesize that these circular forms may exert a tumor-promoting influence akin to linear isoforms. This may contrast with findings in CVD and T2D, where linear and circular forms of ANRIL have been described to exhibit opposing activities [24,92].

Table 1.
Summary of the ANRIL expression associated with cancers. It includes information on cancer types, samples used in the studies, the methodologies used for ANRIL detection, the ANRIL isoforms detected, and the corresponding references.
Table 1.
Summary of the ANRIL expression associated with cancers. It includes information on cancer types, samples used in the studies, the methodologies used for ANRIL detection, the ANRIL isoforms detected, and the corresponding references.
| Cancers | ANRIL Expression | Tissues | Cell Lines | ANRIL Detection RTqPCR (Fw_Rv), RNAseq, Microarrays | Detected Isoforms | References |
|---|---|---|---|---|---|---|
| LC | Up | 1/87 NSCLC tissues, 2/TNM I stage LUAD | A549, H460, H1299, H1975, SPC-A1, H1650 | Ex17_Ex18, Ex1_Ex2, Ex20_Ex20, Ex12_Ex16 | 1, 2, 4, 5, 7, 8, 9, 10, 11, 12, 13, 17, 19, 20, 21, 22, 23, 25, 26, 27 | [48,66,85,86,87] |
| Down | 24 IPF | Ex6_Ex7 | 3, 9, 11, 13, 15, 16, 19, 21, 22, 24, 28 | [88] | ||
| GC | Up | 1/20 paired GC, 2/19,317 GC patients and Lymph nodes, 3/83GC, 4/120GC | AGS, BGC823, MGC80–3, MKN-45, SGC-7901, HGC-27, HSC-39, FU97 | Ex1/2_Ex2, Ex1_Ex2, Ex11/12_Ex13, Ex1_Ex1, Ex14_Ex15, RNAseq | 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [93,94,95,96,97,98] |
| BC | Up | 1/787 early BC patients, 2/37 TNBC | MCF10A, MCF7, T47D, MDA-MB-231, BT549, HS578T, SKBR3, BT474, BT20 | Ex12-Ex15, Ex12_16, Ex5_Ex6/7, Ex3_Ex4, Ex1_Ex1, Ex17/18_Ex18, Hs01390879_m1 (Ex1_Ex2), RNAseq | 1, 3, 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [31,99,100,101,102,103,104] |
| OC | Up | 1/18 OC, 2/86 OC, 3/102 EOC tissues, including 68 SOC tissues | SKOV3, OVCAR3, HO-8910, SKOV3, A2780, Hey, OVCA429, OVCA433 | Ex21_Ex21, Ex1_Ex2, Ex4_Ex5 | 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 12, 13, 17, 18, 19, 20, 21, 22, 23, 25, 26, 27 | [105,106,107,108] |
| CC | Up | 41 high-grade squamous intraepithelial lesions (HSILs), and 75 cervical cancer tissues | CaSki, SiHa | Divergent Ex2_Ex4 | CircRNA | [91] |
| Up | 53CC | HeLa, CaSki, SiHa, ME-180, H1299 | Ex1_Ex2 | 4, 7, 9, 11, 13, 17, 25, 26, 27 | [86,109] | |
| CRC | Up | 20 from CRC, 20 from adenomatous polyp patient, 172CC | Ex1_Ex1, Ex3_Ex4 | 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [67,110] | |
| Down | Meta-Analysis from 10 sets of RNAseq, 40 patients with CC (with 10 patients each in stages I, II, III and IV) | Caco2 | Ex1_Ex1, RNASeq | 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [111,112] | |
| BladC | Up | 1/30 BC, 2/51 BC | EJ | Ex1_Ex1, Ex1_Ex2 | 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [113,114] |
| No misregulation | 85 NMIBC | 97-1, 97-7, MGH-U3, MGH-U4, RT112, RT 4, UMU-UC5, UMU-UC7, VM-CUB1, 5637 | Ex17/18_Ex18 | 1, 9, 10, 11, 21, 25 | [115] | |
| TC | Up | 510TC, 502TC | TPC-1, HTH83, FTC-133 | Ex2_Ex3, RNAseq | 4, 7, 9, 13 (+RNAseq) | [116,117] |
| BrC | Up | 1/15G, 2/142G, 3/10 all stages each, 4/19G | A172, LN18, T98G, U251, LN229, U87 | Ex9_Ex12, Ex1_Ex2, Ex13_Ex13, RNAseq/uArray | 4, 7, 9, 11, 13, 14, 15, 16, 17, 24, 25, 26, 27 (+uArray and RNAseq) | [118,119,120,121,122] |
| OS | Up | 1/56OS, 2/19OS (IIB, III), 3/30OS, 4/57OS, 5/53OS | SW1353, MG-63, SAOS2, HOS, U2OS | Ex2_Ex3, Ex1_Ex1/5, Ex12_Ex13, Ex5_Ex6, Ex1_Ex2 | 3, 4, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [123,124,125,126,127,128,129] |
| MM | Up | 1/80MM, 2/70MM | U266, MM.1S, NCI-H929 | Ex3/4_Ex4, Ex12_Ex15 | 9, 13 | [130,131] |
| PC | Up | 10PC | LNCap, PC3, DU145 | Ex12_Ex15, Ex17/18_Ex18, Ex6_Ex6/7 | 1, 3, 9, 10, 11, 13, 15, 16, 19, 21, 22, 24, 25, 28 | [132] |
| ATL/AML | Up | 1/178AML, 2/100AML, 3/109AML, 4/6ATL, 5/27T-ALL | MOLM-13, HL-60, MT-2, MT-4, C8166, MT-1, HPB-ATL-2, HPB-ATL-T, ED, TL-Om1, MOLT4s, CCRF-CEM, KOPT-K1 | Ex1_Ex2, Ex1_Ex1, Ex17/18_Ex18 | 1, 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [133,134,135,136,137] |
| Melanoma | Up | NZM, OM431, A375 | Ex1_Ex1, Ex5_Ex6 | 1, 3, 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [138] | |
| Up | NZM | Outward facing primers targeted against exons 2, 4, 6, 8, 14 and 16 | CirRNA: More than 30 isoforms differentially expressed in melanoma cells | [78] | ||
| Fusion MTAP | 174 cell lines included in this study were derived from metastasized tumors of 134 melanoma patients | [139,140] | ||||
| EC | Up | 1/87EC, 2/20EC, 3/Transcriptome data from 552UC, 575UC | HEC-1A, RL95-2 | Ex1_Ex2, Ex1_Ex1, Ex6_Ex6 | 3, 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [141,142,143,144] |
| iCCA | Up | 39iCCA | Ex19_Ex20 | 1, 5, 8, 9, 10, 11, 21, 22, 23, 25 | [145] | |
| RC | Up | 1/42KIRC, 2/108ccRCC | 769-P, ACHN, 786-O, Caki-1, Caki-2, ACHN | Ex1_Ex1, Ex5_Ex6, Hs03300540_m1, HCR and RNaseq | 3, 4, 6, 7, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [69,146] |
| RB | Up | 28RB | HXO-RB44, Y79 | Ex1_Ex2, Ex11_Ex15 | 4, 7, 9, 11, 13, 17, 25, 26, 27 | [56,147] |
| HNSCC/LSCC | Up | 1/60LSCC, 2/54LSCC, 3/28LSCC, 4/35NPC, 5/522HNSCC | Tu177, HN4, AMC-HN-8, NP69, FaDu, CAL27, CNE1, CNE2, S18, HONE1, 5–8F, AMC-HN-8, SNU-899, HNEC | Ex2_Ex3, Ex21_Ex21, Ex1_Ex1, Ex1_Ex2, Ex1/5_Ex6, uArray, RNAseq | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [148,149,150,151,152,153,154] |
| HCC | Up | 1/30HCC, 2/34LC, 3/100HCC, 4/85HCC, 5/50Cirrhosis, 6/130HCC, 7/77HCC, 8/317HCC | Huh7, Hep3B, Sk-Hep1, MHCC97H, SMMC-7721, HepG2 | Ex21_Ex21, Ex1_Ex1, Ex14_Ex15, Ex15_Ex16, Ex1_Ex2, Ex12_Ex15, RNAseq | 1, 2, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 | [64,155,156,157,158,159,160,161,162] |
(LC: lung cancer, GC: gastric cancer, BC: breast cancer, OC: ovarian cancer, CC: cervical cancer, CRC: colorectal cancer, BladC: bladder cancer, TC: thyroid cancer, BrC: brain cancer, OS: osteosarcoma, MM: myeloma, PC: prostate cancer, ATL/AML: leukemia, EC: endometrial cancer, RC: renal cancer, RB: retinoblastoma, HNSCC/LSCC: head/neck cancer, iCCA: intrahepatic cholangiocarcinoma and HCC: hepatocellular cancer).
6.2. MTAP-ANRIL Fusion in Cancer
The contribution of the MTAP-ANRIL fusion is also critical when assessing the involvement of ANRIL in cancer biology. This fusion gene, documented to exhibit a frequency exceeding 7% in melanoma, results from a chromosomal rearrangement between ANRIL and MTAP, a tumor suppressor gene involved in purine metabolism (methylthioadenosine phosphorylase) [139]. In this context, recent research has demonstrated that the MTAP-ANRIL gene fusion leads to the suppression of the wild-type MTAP expression and facilitates an epithelial–mesenchymal transition-like process through the activation of JNK and p38 MAPK pathways, both in vitro and in vivo [140].
6.3. Polymorphisms of ANRIL in Cancer
Since its first association with pathologies in 2007 [31], the 9p21 locus have been the topic of several genome-wide association studies (GWAS), unveiling ANRIL sequences as a hotspot for single nucleotides polymorphisms (SNPs) that are susceptibility factors for several pathologies such as CVD, T2D and cancers [163,164]. Focusing on the cancer-related pathologies, 34 SNPs were identified to be associated with higher risk of developing different types of cancer including LC, GC, BC, CC, TC, BrC, MM, PC, ATL/AML/B-ALL, melanoma/BCC, EC, ADC, HNSCC/LSCC/ESCC, PancC, OS and overall cancer risk (Table 2). In addition to this trait, SNPs are also correlated, for instance, with reduced overall survival (rs1333049) in ESCC [165], with larger tumor size (rs11333048) in TC [166], with higher TMN stage (rs3217992) in OS [167], and linked to the presence of metastases (rs1063192) in TC [168] (Table 2). Note that despite this clear association, most of the studies do not allow researchers to discern whether the observed associations reflect causality or if they indicate other underlying complexities within the biological system. This observation underscores the importance of conducting comprehensive investigations, including functional studies and mechanistic analyses similar to the exhaustive investigations conducted for instances such as rs10811656/rs10757278 in the context of CVD [52].

Table 2.
Summary of the SNPs found within the ANRIL gene and their connection to various types of cancers. The summary provides details about the specific cancer types involved and includes the relevant references.
A total of 76% of the SNPs are localized in introns, 17% in downstream, and 6% in upstream sequences. Note that among all these SNPs, the expression of ANRIL has not been systematically evaluated, which makes the correlation between risky genotype and ANRIL expression difficult. However, it is possible to consider two levels that are likely to be affected by these variations. Firstly, at the DNA level, these variations may have an impact on the expression of the 9p21 locus through the disruption of binding sites for transcriptional regulators and/or enhancer sequences (rs10757278, rs10811656, rs4977757, rs1333045, rs1537373) as it is described for CVD [18,52,217,218]. In cancer, studies have predicted that the intronic risk allele rs17694493 disrupts both the transcription factors (STAT1 and RUNX1) and androgen receptor-binding motifs in PC. This may actively participate in the cell cycle regulation by modulating the expression of the CDKN2B-CDKN2A gene cluster, thereby playing a causal role in predisposing cancer risk [65,219]. As mentioned previously, the expression of the 9p21 locus also depends on the methylation state of the CpG island which modulates the association of CTCF to the ANRIL promoter [57]. One reasonably hypothesizes that the presence of SNPs may interfere with CTCF binding. In this case, this would be reminiscent of the SNP rs12936231 that has been found to disrupt the CTCF binding site within the ZPBP2 gene in the context of asthma [220].
At the RNA level, sequence variations have the potential to impact protein binding sites and structural elements. This concept has been investigated in the context of a risky genotype associated with myocardial infarction, specifically with regard to rs10965215 and rs10738605. It is hypothesized that these variations reduce the free energy of ANRIL secondary structures which may interfere with the binding of proteins and consequently with the ANRIL activities [221]. SNPs may also disrupt the interaction between splicing factors (such as SR or hnRNPs) and regulatory elements (Exonic Splicing Enhancer ESE, Exonic Splicing Silencer ESS, Intronic Splicing Silencer ISS and Intronic Splicing Enhancer ISE) within ANRIL. This may lead to a similar effect observed for PSMD13, where the presence of the SNP rs7128029 affects its splicing pattern, leading to the exclusion of an exon in EC [222]. If so, the heterogeneity of ANRIL isoforms observed in pathological situations may be attributed to the presence of peculiar SNPs.
7. ANRIL Activities
7.1. Cytoplasmic Activities of ANRIL
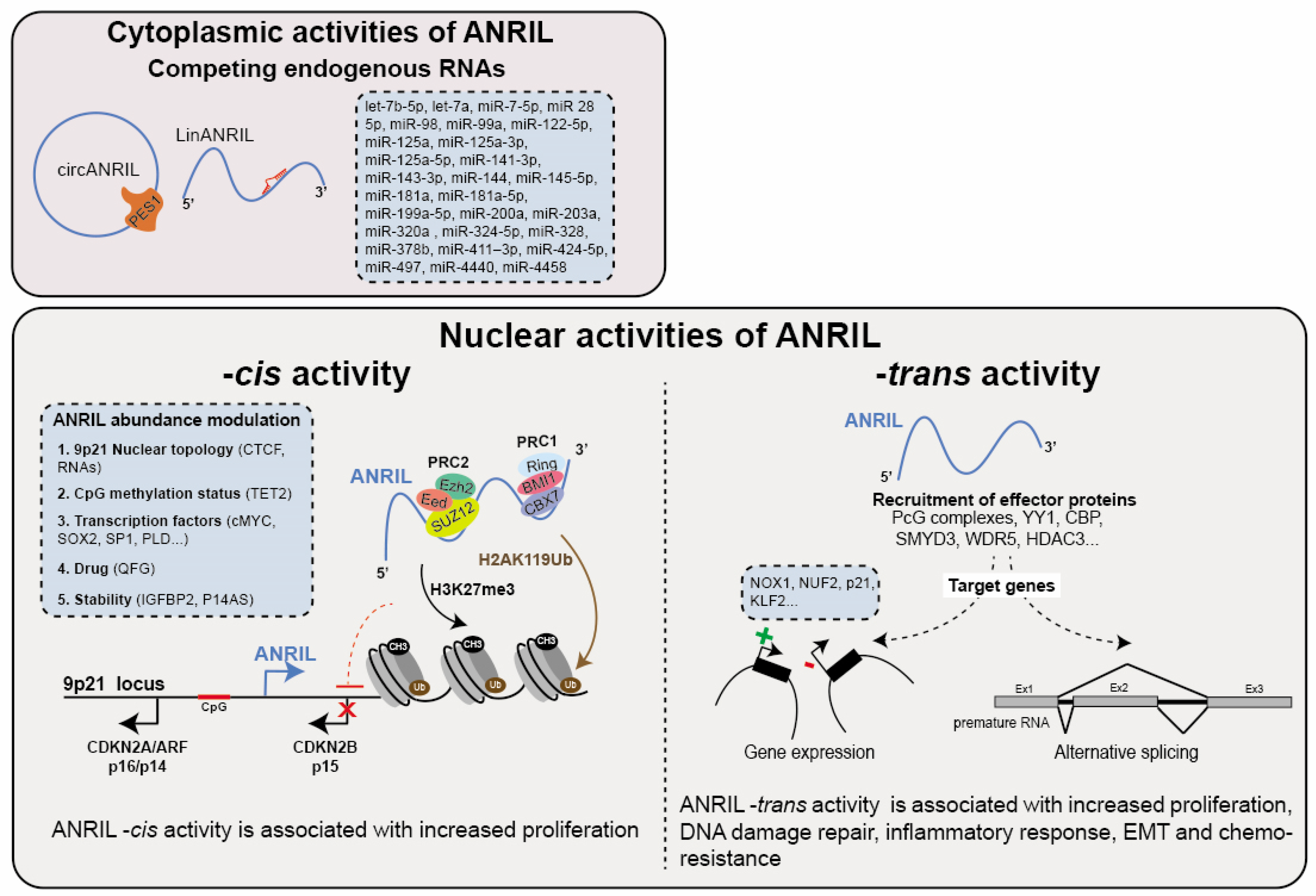
The association between cancer biology and the cytoplasmic isoforms of ANRIL primarily stems from their capacities to function as competing endogenous RNAs (ceRNA). This functionality arises from ANRIL’s ability to form hybrid complexes with miRNAs when it is overexpressed in cancer cells. Consequently, this interaction results in the decreased activity of tumor-suppressor microRNAs, ultimately leading to the upregulation of genes that are normally repressed by these miRNAs (Table 3). To date, the proposed ceRNA role of ANRIL involves a single interaction with a total of 28 miRNAs which hybridize with 10 ANRIL exons predominantly occurring on exons 1, 11, and 20 (let-7b-5p, let-7a, miR-7-5p, miR-28-5p, miR-98, miR-99a, miR-122-5p, miR-125a, miR-125a-3p, miR-125a-5p, miR-141-3p, miR-143-3p, miR-144, miR-145-5p, miR-181a, miR-181a-5p, miR-199a-5p, miR-200a, miR-203a, miR-320a, miR-324-5p, miR-328, miR-378b, miR-411–3p, miR-424-5p, miR-497, miR-4440, miR-4458) (Figure 3). The potential ANRIL/miRNA associations have been correlated with multiple cancers including GC, BC, OC, CC, CRC, TC, BrC, OS, PC, MM, PC, ATL/AML, EC, RC, RB, HNSCC/LSCC, HCC, LC and some are even associated with higher TMN stage (Table 3). These observations strongly support the role of ANRIL as an oncogene by virtue of its ceRNA activity. For instance, this activity has been implicated in modulating the miR-320a/HMGB1 axis, a pathway associated with inflammation in TC [223]. Moreover, the interactions between ANRIL and let-7 miRNA (7a, 7b-5p), miR-122-5p and miR-181a have been shown to increase cancer stem cell proliferation and epithelial–mesenchymal transition (EMT) by affecting multiple pathways including Wnt, NOTCH, STAT3/NF, MAPK/ERK, PI3K/AKT, and glycolysis in BrC, OC, TC, CR and HNSCC [106,116,120,151,152,224]. In the context of the let-7 miRNAs, ANRIL competitively interacts with pivotal components within the aforementioned pathways, such as the TCF-4, CCND1, HMGA2, NUMB, and c-Myc mRNAs. This counteracts their role as tumor suppressors, thus hampering their inherent tumor-suppressive functionality [151,224]. Additionally, ANRIL positively influences cell proliferation through the miR-181a-5p/CCNG1, miR-203a/CDK2, miR-141-3p/CCND1/2, and miR-497/CDK6 axes in GC, HCC, RC, and HNSCC, respectively [96,146,225,226].

Table 3.
Summary detailing the connection between miRNAs and ANRIL. It covers various aspects such as the types of cancer involved, the influence of ANRIL/miRNA interactions on different cancer-related processes, the target genes, the specific location of hybridization on ANRIL, and the relevant references.
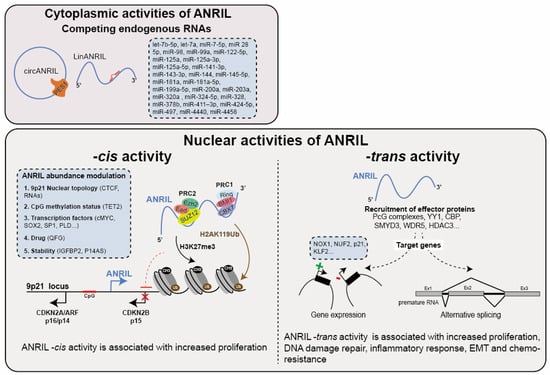
Figure 3.
Functions of ANRIL. ANRIL exerts various regulatory roles both in the cytoplasm and nucleus. In the cytoplasm, ANRIL (circANRIL: circular ANRIL and linANRIL: linear ANRIL) acts as a competing endogenous RNA (ceRNA) for miRNAs and proteins, thereby modulating gene expression at the post-transcriptional level. Within the nucleus, ANRIL functions as a cis-regulator, facilitating the recruitment of Polycomb group proteins (PcG) to the 9p21 locus. This leads to transcriptional repression of genes including CDKN2B, achieved through the deposition of H2AK119ub (PRC1) and H3K27me3 (PRC2) histone marks at the 9p21 locus. Furthermore, ANRIL modulates gene expression at the chromatin level in -trans by guiding the recruitment of chromatin modifiers or transcriptional activators (such as PcG, YY1…) to specific loci (e.g., NOX1, KLF2…). Emerging evidence indicates also that ANRIL impacts alternative splicing patterns. Overall, these regulatory activities predominantly enhance cell proliferation, migration, invasion, and metastasis while suppressing apoptosis and senescence, primarily attributed to the modulation of key cancer-related gene expression.
Interestingly, the ceRNA activity of ANRIL is associated with chemoresistance. It has been shown that ANRIL acts as ceRNA for the miR-125a in the development of TNBC and its chemoresistance of doxorubicin by enhancing glycolytic activity, specifically targeting enolase 1 (ENO1) [99]. ANRIL is also likely to act on the multidrug transporters miR-125a-5p/MRP4 axis, which are important players in EC paclitaxel resistance [141]. In the context of CRC, ceRNA activity of ANRIL has been proposed to promote resistance to 5-Fluorouracil by inhibiting the ABCC1/let-7a pathway [247]. Finally, ANRIL may favor cisplatin resistance in LC and OS by modulating the miR-328/ABCG2/MDR1 and miR-125a-5p/STAT3 signaling pathways, respectively [125,228].
Radiotherapy is an important treatment for cancer, mainly by triggering DNA double-strand breaks to induce cell death. Promoting DNA damage repair can decrease the radiosensitivity of tumor cells. Since ANRIL has been described to be a key regulator in DNA damage repair (HDR), it is well suited to play a role in this process. In agreement, ANRIL positively acts on HDR by modulating the miR-7-5p/PARP1/RAD51 and miR-145-5p/MMP1 pathways in LC and TNBC respectively [100,228].
Although the majority of studies do not distinguish between circular and linear isoforms of ANRIL, and only a limited number of them assess the functional interactions between ANRIL and miRNAs through techniques such as dual-luciferase assays, these findings emphasize the complex regulatory interplay between ANRIL, miRNAs and cancer.
7.2. Nuclear Activities of ANRIL
7.2.1. -cis Activity
ANRIL functions as a cis-regulator by facilitating the recruitment of the Polycomb group proteins (PcG) to the 9p21 locus. This results in the transcriptional repression of, at least, the CDKN2B gene through the deposition of H2AK119ub (PRC1) and H3K27me3 (PRC2) histone marks at the promoter region of the CDKN2A/ARF-ANRIL genes in PC and normal lung fibroblast (Figure 3) [25,248]. While the exact molecular mechanism and sequential elements involved in this association remain incompletely understood, the recruitment of CBX7 (PRC1) by ANRIL appears to depend on a stem-loop structure [25]. Note that this activity has been attributed to the linear isoforms of ANRIL. Conversely, a particular circular isoform has been identified to interact with EZH2 (a component of PRC2), leading to the displacement of EZH2 from the CDKN2A/ARF-ANRIL promoter. This alteration in EZH2 localization influences the deposition of H3K27me3 on the latter, resulting in the increased expression of ANRIL in human primary fibroblasts undergoing oncogene-induced senescence [249].
An intriguing aspect that remains to be fully understood is the mechanism through which ANRIL interacts with protein binders to carry out its functions. Notably, it has been demonstrated that the association between ANRIL and PRC1 relies on chaperone proteins such as MOV10, whose helicase activity is essential for recruiting the ANRIL/PRC1 complex to the CDKN2A/ARF promoter [250]. One hypothesis suggests that MOV10 might influence the structural conformation of ANRIL, promoting its interaction with the PRC1. If so, it would be reminiscent of the lncRNA roXes (RNA on the X) involved in dosage compensation in Drosophila melanogaster. In this context, the RNA helicase MLE known to be crucial for unwinding conserved stem-loop structures facilitates the association between the roXes and MSL2, a critical protein involved in the assembly and regulation of the MSL complex [251].
Another point which also remains to be elucidated is how ANRIL associates the genome to exert its functions at specific loci. It has been proposed that the -cis activity depends on the formation of a triple helix via a triplex-forming oligonucleotide (TFO) located in ANRIL Exon1 and the CDKN2B promoter [252]. Triplexes are formed when a single-stranded RNA fragment accommodates the major groove of the double stranded DNA. Since this association relies on base-pairing interactions, such as Hoogsteen or reverse Hoogsteen hydrogen bonds occurring between ANRIL and the CDKN2B promoter, such an anchoring mechanism may explain, at least in part, how ANRIL recognizes specifically the 9p21 locus [253].
7.2.2. -trans Activity of ANRIL
ANRIL Regulation of Genes Located Outside the 9p21 Locus
Several studies have provided evidence suggesting that ANRIL exerts direct regulation on gene expression beyond the 9p21 locus (Figure 3). Indeed, RNA-FISH experiments performed in PC and EC have demonstrated that ANRIL localization is not restricted to the 9p21 locus [25,254]. The overexpression of ANRIL fragments in HEK293 cells has shown to upregulate 219 genes and downregulate 708 genes involved in processes such as development, adhesion, proliferation, and apoptosis [19]. In a more recent transcriptomic analysis of vascular smooth muscle cells (VSMCs), Lo Sardo et al. identified altered expression profiles in approximately 3000 genes located outside the 9p21 locus upon depletion of the 3′ end of the ANRIL gene compared to wild-type cells again involved in proliferation and apoptosis [255]. To distinguish between direct and indirect genomic targets of ANRIL, Alfeghaly and colleagues combined ChIRP-seq genomic occupancy data with transcriptomic analysis in ANRIL-knockdown HEK293 cells. This analysis could identify 123 downregulated and 65 upregulated genes as direct targets of ANRIL [20]. Similar to its -cis activity, ANRIL is thought to exert its repressive -trans activity by interacting with Polycomb group proteins (PcG). Consistent with this notion, ANRIL is capable of epigenetically suppressing the transcription of the p21 and KLF2 (Kruppel-like factor 2) genes in HCC and LC. This repression occurs through ANRIL binding to the PRC2 complex and subsequently to its recruitment to the promoters of these genes [64,256]. It is important to note that among the direct target genes found to be silenced in the study conducted by Alfeghaly (2021), only 17% of them are likely to be regulated through a PcG-dependent mechanism [20]. This finding suggests that ANRIL may employ PcG-independent mechanisms to silence gene expression. Further in-depth investigation is needed to fully understand these mechanisms.
As previously mentioned, ANRIL has the potential to activate gene expression. According to Zhou et al., they elucidated the mechanism through which the ANRIL/YY1 complex contributes to the activation of IL6 and 8 (interleukins 6 and 8) genes in HUVEC cells in response to a TNF-α pro-inflammatory signal [26]. Furthermore, ANRIL likely enhances the growth, migration, and invasion abilities of HNSCC and TC cancer cells by positively regulating the TGF-β1/Smad signaling pathway [257,258]. Regarding its role in RC, ANRIL recruits the CBP (CREB-binding protein) and SMYD3 (SET and MYND domain-containing 3) epigenetic-modifying complex to activate the transcription of NUF2 at the chromatin level. This activation occurs through the deposition of local modifications of H3K27ac and H3K4me3 histone marks [69]. Moreover, ANRIL was found to associate with WDR5 and HDAC3 proteins in human aortic smooth muscle cells (HASMC) [259]. This complex is necessary for the deposition of H3K9Ac and H3K4me3 histone marks on the promoter region of the NOX1 gene, resulting in its transcriptional activation.
Note that approximately 24% of the miRNAs identified as ANRIL targets in the context of its ceRNA activity do not exhibit apparent hybridization sites on ANRIL (miR-99a, miR-125a-3p, miR-144, miR-145-5p, miR-200a, miR-203a, miR-384) [94,100,121,225,231,237,239,243,245]. This observation suggests the possibility of indirect effects of ANRIL on the miRNA abundance or may highlight the involvement of ANRIL -trans activity in epigenetically down-regulating the expression of these miRNAs. Such mechanisms have been already reported in various contexts, including HCC, where ANRIL enhances cancer migration and proliferation by modulating the expression of the miR-191 and by subsequently affecting the vimentin expression [260]. Additionally, ANRIL has been found to epigenetically silence miR-99a and miR-449a via PcG complexes, promoting the CDK6/E2F1 pathway and establishing a positive feedback loop for its own expression, continuing, therefore, to promote gastric cancer cell proliferation [98]. The mechanism engaged by ANRIL for specifically associating the genomic loci in the context of its -trans activity is not fully understood yet. However, the ChIRP-seq data generated by Alfeghaly and colleagues allowed for the identification of 3227 binding sites across the genome in HEK293 cells. Interestingly, among the sequences of these binding sites, 98% are enriched with G/A repeats. Moreover, ANRIL exon 8, which is 70% covered by the subcategory of LTR named ERVL-MaLR, is involved in its genomic occupancy in HEK293 cells by forming triple helices for instance [20]. However, this genome recognition mode cannot solely explain ANRIL binding to the chromatin, and alternative modes engaged by ANRIL to associate with the genome have to be considered. LncRNA–chromatin recognition can take place through the interaction with specific protein partners that serve as bridge between the DNA and the lncRNA as the heterogeneous nuclear RiboNucleoProtein U (hnRNP U) matrix protein [261,262]. Another mechanism relies on the direct interaction of the lncRNA with the DNA molecule via RNA–DNA hybrid duplexes formed via canonical Watson–Crick base-pairing. The resulting hybrid is named R-loop [263,264]. Further investigations are needed to identify potential R-loops or ANRIL protein-binders involved in its genomic association.
Role of ANRIL in Modulating the Alternative Splicing
Genomic occupancy assays performed in HEK293 cells revealed that the majority of the binding sites for ANRIL are located in non-coding regions, including introns and intergenic regions. Remarkably, 40.3% of the ANRIL sites are intronic, and, more importantly, 24% of the genes contacted by ANRIL in HEK293 are affected in terms of alternative splicing (AS) upon ANRIL knockdown [20]. In addition, the overexpression of ANRIL in HUVEC cells, coupled with RNA-seq and AS analysis, revealed significant impacts on the AS patterns of numerous genes. These affected genes play essential roles in translation, DNA repair, RNA processing, and participate in the NFκB signaling pathway [27]. These findings highlight the significant influence of ANRIL on the AS landscape (Figure 3). It is noteworthy that this activity is reminiscent of the lncRNA asFGFR2 described to act on the AS of the FGFR2 transcript [265]. In this case, asFGFR2 modulates the chromatin structure at the FGFR2 locus, resulting in the limitation of the binding of the MRG15-PTB chromatin adapter complex to exon IIIb. As a consequence, this modulation promotes the inclusion of exon IIIb in the FGFR2 mRNA. Interestingly, this isoform plays an antitumoral role in HCC, which definitively links lncRNA, AS modulation and cancer fields in a concrete manner [266].
Cancer-specific splicing events have been detected in diverse cancer types such as BC, LC, PC, and AML/ALL [15,267,268]. The combinatorial nature of the AS relies on the selective utilization of splicing sites within the pre-mRNA, which is mediated by factors including RNA Binding Proteins (RBPs) such as the SR and hnRNP proteins, which function as -trans acting regulatory factors [269]. In addition, a close association between chromatin characteristics and AS has been identified. Nucleosomes, through their specific patterns of histone modifications, have the ability to influence AS [270]. For instance, exons exhibit a higher occurrence of certain histone modifications such as H3K27me2, H3K36me3, and H4K20me3. The presence of these modifications can impact AS by interacting with protein factors that facilitate the recruitment of trans-acting regulatory factors, or even directly recruiting components of the spliceosome [271,272,273,274,275,276,277]. Even though the specific molecular mechanism employed by ANRIL to regulate AS remains unknown, its ability to associate with proteins and modulate chromatin landscape highly suggests that ANRIL plays a role in the AS process in a direct manner. If so, one reasonably hypothesizes that this capacity might also contribute to ANRIL involvement as a susceptibility factor in cancer development.
8. Conclusions
In conclusion, ANRIL is a critical factor in maintaining the balance of cellular functions despite its relatively simple nature. It plays a crucial role in regulating the progression of the cell cycle by interacting with PcG and guiding them to specific genomic loci, resulting in the silencing of genes like CDKN2B. Additionally, ANRIL acts as a ceRNA, impacting the activity of over 20 miRNAs involved in various cellular processes such as cell proliferation, invasion, inflammation, and EMT. Consequently, the dysregulation of ANRIL disrupts normal cell cycle control, leading to uncontrolled cell growth and the development of cancer. This explains why ANRIL is found to be upregulated in more than 20 types of cancer, as well as in conditions like CVD and T2D. To the best of our knowledge, ANRIL is the lncRNA most strongly associated with these pathological conditions.
While substantial progress has been made in unraveling the functional roles of ANRIL in cancer, there is still much to discover. The precise molecular mechanisms underlying ANRIL activities, particularly its PcG-independent functions and its role on the splicing modulation, remain incompletely understood. In addition, the complexity of ANRIL isoforms allows for versatility in its interactions with different proteins, RNA molecules, and genomic regions, thereby influencing various biological processes. The investigation of the contribution at the molecular level of each isoform individually represents a major challenge in the field. Thus, ANRIL stands as a fascinating and multifaceted molecule with diverse implications in cancer biology. Its dysregulation makes it a promising area for continued investigation and the development of innovative therapeutic approaches.
Since ANRIL is predominantly overexpressed, a potential therapeutic approach could involve reducing its intracellular abundance within cancerous lesions. This could be achieved through methods such as RNA interference (RNAi), antisense oligonucleotides (ASOs), the RNA-targeting CRISPR-Cas system, or epigenetic modulators employed to alter the epigenetic profile of the ANRIL gene [278]. Another strategy could involve utilizing small molecules designed to hinder ANRIL functions, achieved through modifications of lncRNA–protein interactions or the induction of structural changes [279]. It is worth noting that the primary challenge lies in crafting a molecule that guarantees effectiveness while minimizing unintended effects. The creation of such a molecule relies on a comprehensive understanding of the lncRNA activities at the molecular level, which still need to be thoroughly elucidated for ANRIL. Also, given the complexity of ANRIL interactions and effects, combination therapies that target multiple aspects of its activity could be more effective than single-target approaches. In the end, this could lead to the development of personalized treatments, enabling the adaptation of strategies for reducing or inhibiting the expression patterns or activities, respectively, of specific ANRIL isoforms in cancers.
The key role of lncRNAs in cancer development and progression has made these classes of RNA a major point of interest for researchers. As ANRIL, many are found overexpressed in cancer tissues or cancer lines in culture. Hence, the advancements in ANRIL biology should provide knowledge about its multiple activities but may also lead to the discovery of more generic activities attributed to lncRNAs.
Author Contributions
A.S., J.L., G.G., L.A. and S.M.: manuscript writing; literature search, manuscript writing and final approval; A.S, J.L., G.G., L.A. and S.M.: critical reading and final approval; A.S., J.L., G.G., L.A. and S.M.: literature search. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CNRS-UL and SATT SAYENS, grant “Projet incitatif”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
ABCC1: ATP-binding cassette sub-family C member 1, ANRIL: Antisense Non-coding RNA in the INK4 Locus, ACHN: Adenocarcinoma Human Kidney, ARF: Alternative Reading Frame, AUF1: AU-rich Element RNA-binding Protein 1, CBX7: Chromobox Homolog 7, CC: Cervical Cancer, CCA: Cholangiocarcinoma, CCND1/2: Cyclin D1/D2, CDK6: Cyclin-dependent Kinase 6, CDKN2A-2B: Cyclin-dependent Kinase Inhibitor 2A-2B, c-MYC: Cellular Myelocytomatosis Oncogene, CTCF: CCCTC-binding Factor, CVD: Cardiovascular Disease, CXCL11: C-X-C Motif Chemokine Ligand 11, E2F1: E2F Transcription Factor 1, EMT: Epithelial–mesenchymal Transition, ERK: Extracellular Signal-Regulated Kinase, ERVL-MaLR: Endogenous Retrovirus-Like MaLR, ERα: Estrogen Receptor Alpha, ESE: Exonic Splicing Enhancer, ESS: Exonic Splicing Silencer, EZH2: Enhancer of Zeste Homolog 2, EZR: Ezrin, FGFR2: Fibroblast Growth Factor Receptor 2, FISH: Fluorescence In Situ Hybridization, GAS5: Growth Arrest-Specific 5, GC: Gastric Cancer, GTEx: Genotype-Tissue Expression, GWAS: Genome-Wide Association Study, H3K27ac: Histone H3 Lysine 27 Acetylation, H3K27me2: Histone H3 Lysine 27 Dimethylation, H3K36me3: Histone H3 Lysine 36 Trimethylation, H3K37me3: Histone H3 Lysine 37 Trimethylation, H3K4me3: Histone H3 Lysine 4 Trimethylation, H3K9ac: Histone H3 Lysine 9 Acetylation, H4K20me3: Histone H4 Lysine 20 Trimethylation, HCC: Hepatocellular Carcinoma, HIF-1α: Hypoxia-Inducible Factor 1 Alpha, HMGB1: High Mobility Group Box 1, hnRNP U: Heterogeneous Nuclear Ribonucleoprotein U, HNSCC/LSCC: Head and Neck Squamous Cell Carcinoma/Lung Squamous Cell Carcinoma, HOTAIR: HOX Transcript Antisense Intergenic RNA, IFNA21: Interferon Alpha 21, IFN-γ: Interferon Gamma, IGF2BP3: Insulin-like Growth Factor 2 mRNA-Binding Protein 3, INK4: Inhibitor of Cyclin-Dependent Kinase 4, IPF: Idiopathic Pulmonary Fibrosis, ISE: Intronic Splicing Enhancer, ISS: Intronic Splicing Silencer, JNK: c-Jun N-terminal Kinase, KLF2: Kruppel-Like Factor 2, LC: Lung Cancer, LINE: Long Interspersed Nuclear Element, lncRNAs: Long Non-Coding RNAs, LTR: Long Terminal Repeat, LUAD: Lung Adenocarcinoma, MAPK: Mitogen-Activated Protein Kinase, MDR1: Multidrug Resistance Protein 1, miRNAs: MicroRNAs, MLE: Maleless, MM: Myeloma, MMP1: Matrix Metallopeptidase 1, MOV10: Moloney Leukemia Virus 10, MRG15: Mortality Factor on Chromosome 4 Gene 15, MRP4: Multidrug Resistance-Associated Protein 4, MSL2: Male-Specific Lethal 2, MTAP: Methylthioadenosine Phosphorylase, NF-kB: Nuclear Factor-kappa B, NOTCH: Neurogenic Locus Notch Homolog Protein, NOX1: NADPH Oxidase 1, NSCLC: Non-Small Cell Lung Cancer, NUF2: NDC80 Kinetochore Complex Component Nuf2, OC: Ovarian Cancer, OS: Osteosarcoma, PARP1: Poly(ADP-ribose) Polymerase 1, PC: Prostate Cancer, PancC: Pancreatic Cancer, PcG: Polycomb Group, PES1: Pescadillo Ribosomal Biogenesis Factor 1, PLD: Phospholipase D, PRC1: Polycomb Repressive Complex 1, PRC2: Polycomb Repressive Complex 2, PSMD13: 26S Proteasome Non-ATPase Regulatory Subunit 13, PTB: Polypyrimidine Tract-Binding Protein, QFG: Qingjie Fuzheng Granule, Rb: Retinoblastoma Protein, RB: Retinoblastoma, RBP: RNA-Binding Protein, RC: Renal Cancer, RIDL: Repeat Insertion Domains of LncRNAs, RNP: Ribonucleoprotein, roXes: RNA on X, RTqPCR: Reverse Transcription Quantitative Polymerase Chain Reaction, RUNX1: Runt-Related Transcription Factor 1, SINE: Short Interspersed Nuclear Element, SMAD3/4: SMAD Family Member 3/4, SMYD3: SET and MYND Domain-Containing Protein 3, SNPs: Single-Nucleotide Polymorphisms, SOX2: SRY-Box Transcription Factor 2, SP1: Specificity Protein 1, SR: Splicing Regulator, SRSF3: Serine/Arginine-Rich Splicing Factor 3, STAT1: Signal Transducer and Activator of Transcription 1, T2D: Type 2 Diabetes, TAD: Topologically Associated Domain, TC: Thyroid Cancer, TE: Transposable Element, TET2: Tet Methylcytosine Dioxygenase 2, TFO: Triplex Forming Oligonucleotide, TGF-β1: Transforming Growth Factor Beta 1, TMEM106B: Transmembrane Protein 106B, TNBC: Triple-Negative Breast Cancer, TNF-α: Tumor Necrosis Factor Alpha, VSMC: Vascular Smooth Muscle Cell, Wnt: Wingless/Integrated, YY1: Yin Yang 1, ZPBP2: Zona Pellucida-Binding Protein.
References
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell. Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a Complete Map of the Human Long Non-Coding RNA Transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A Comprehensive Annotation Database for Long Non-Coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An Updated Database of Long Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Achour, C.; Aguilo, F. Long Non-Coding RNA and Polycomb: An Intricate Partnership in Cancer Biology. Front. Biosci. 2018, 23, 2106–2132. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated LncRNA Function upon Genomic and Epigenomic Regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef]
- Clark, M.B.; Mattick, J.S. Long Noncoding RNAs in Cell Biology. Semin. Cell. Dev. Biol. 2011, 22, 366–376. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Lemos, A.E.G.; da Matos, A.R.; Ferreira, L.B.; Gimba, E.R.P. The Long Non-Coding RNA PCA3: An Update of Its Functions and Clinical Applications as a Biomarker in Prostate Cancer. Oncotarget 2019, 10, 6589–6603. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef] [PubMed]
- McCabe, E.M.; Rasmussen, T.P. LncRNA Involvement in Cancer Stem Cell Function and Epithelial-Mesenchymal Transitions. Semin. Cancer Biol. 2021, 75, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Climente-González, H.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell. Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef]
- Chen, L.; Qian, X.; Wang, Z.; Zhou, X. The HOTAIR LncRNA: A Remarkable Oncogenic Promoter in Human Cancer Metastasis. Oncol. Lett. 2021, 21, 302. [Google Scholar] [CrossRef]
- Lin, G.; Wu, T.; Gao, X.; He, Z.; Nong, W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front. Oncol. 2022, 12, 846497. [Google Scholar] [CrossRef]
- Jarinova, O.; Stewart, A.F.R.; Roberts, R.; Wells, G.; Lau, P.; Naing, T.; Buerki, C.; McLean, B.W.; Cook, R.C.; Parker, J.S.; et al. Functional Analysis of the Chromosome 9p21.3 Coronary Artery Disease Risk Locus. Arter. Thromb. Vasc. Biol. 2009, 29, 1671–1677. [Google Scholar] [CrossRef]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef]
- Alfeghaly, C.; Sanchez, A.; Rouget, R.; Thuillier, Q.; Igel-Bourguignon, V.; Marchand, V.; Branlant, C.; Motorin, Y.; Behm-Ansmant, I.; Maenner, S. Implication of Repeat Insertion Domains in the Trans-Activity of the Long Non-Coding RNA ANRIL. Nucleic Acids Res. 2021, 49, 4954–4970. [Google Scholar] [CrossRef]
- Razeghian-Jahromi, I.; Karimi Akhormeh, A.; Zibaeenezhad, M.J. The Role of ANRIL in Atherosclerosis. Dis. Markers 2022, 2022, 8859677. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A LncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-M.; Ju, H.-Y.; Wu, Y.-T.; Guo, W.; Mao, L.; Ma, H.-L.; Xia, W.-Y.; Hu, J.-Z.; Ren, G.-X. Long Non-Coding RNA ANRIL Promotes Tumorgenesis through Regulation of FGFR1 Expression by Sponging MiR-125a-3p in Head and Neck Squamous Cell Carcinoma. Am. J. Cancer Res. 2018, 8, 2296–2310. [Google Scholar] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-Coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cell. 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Wittfeldt, A.; Sun, J.; Liu, C.; Wang, X.; Gan, L.-M.; Cao, H.; Liang, Z. Long Non-Coding RNA ANRIL Regulates Inflammatory Responses as a Novel Component of NF-ΚB Pathway. RNA Biol. 2016, 13, 98–108. [Google Scholar] [CrossRef]
- Wufuer, A.; Luohemanjiang, X.; Du, L.; Lei, J.; Shabier, M.; Han, D.F.; Ma, J. ANRIL Overexpression Globally Induces Expression and Alternative Splicing of Genes Involved in Inflammation in HUVECs. Mol. Med. Rep. 2023, 27, 27. [Google Scholar] [CrossRef]
- Farooq, U.; Notani, D. Transcriptional Regulation of INK4/ARF Locus by Cis and Trans Mechanisms. Front. Cell. Dev. Biol. 2022, 10, 948351. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. CDK Inhibitors: Positive and Negative Regulators of G1-Phase Progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef]
- Pacifico, A.; Leone, G. Role of P53 and CDKN2A Inactivation in Human Squamous Cell Carcinomas. J. Biomed. Biotechnol. 2007, 2007, 43418. [Google Scholar] [CrossRef]
- Pasmant, E.; Laurendeau, I.; Héron, D.; Vidaud, M.; Vidaud, D.; Bièche, I. Characterization of a Germ-Line Deletion, Including the Entire INK4/ARF Locus, in a Melanoma-Neural System Tumor Family: Identification of ANRIL, an Antisense Noncoding RNA Whose Expression Coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef] [PubMed]
- Akbari, M.; Hussen, B.M.; Eslami, S.; Neishabouri, S.M.; Ghafouri-Fard, S. Association between ANRIL Polymorphisms and Risk of Obsessive-Compulsive Disorder. Heliyon 2023, 9, e14081. [Google Scholar] [CrossRef] [PubMed]
- AbdAllah, N.B.; Al Ageeli, E.; Shbeer, A.; Abdulhakim, J.A.; Toraih, E.A.; Salman, D.O.; Fawzy, M.S.; Nassar, S.S. Long Non-Coding RNAs ANRIL and HOTAIR Upregulation Is Associated with Survival in Neonates with Sepsis in a Neonatal Intensive Care Unit. Int. J. Gen. Med. 2022, 15, 6237–6247. [Google Scholar] [CrossRef]
- Huang, G.; Liang, D.; Luo, L.; Lan, C.; Luo, C.; Xu, H.; Lai, J. Significance of the LncRNAs MALAT1 and ANRIL in Occurrence and Development of Glaucoma. J. Clin. Lab. Anal. 2022, 36, e24215. [Google Scholar] [CrossRef] [PubMed]
- Mirzajani, S.; Ghafouri-Fard, S.; Habibabadi, J.M.; Glassy, M.C.; Sayad, A.; Taheri, M. Altered ANRIL Methylation in Epileptic Patients. J. Mol. Neurosci. 2021, 71, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Safari, M.; Taheri, M.; Samadian, M. Expression of Linear and Circular LncRNAs in Alzheimer’s Disease. J. Mol. Neurosci. 2022, 72, 187–200. [Google Scholar] [CrossRef]
- Abd-Elmawla, M.A.; Fawzy, M.W.; Rizk, S.M.; Shaheen, A.A. Role of Long Non-Coding RNAs Expression (ANRIL, NOS3-AS, and APOA1-AS) in Development of Atherosclerosis in Egyptian Systemic Lupus Erythematosus Patients. Clin. Rheumatol. 2018, 37, 3319–3328. [Google Scholar] [CrossRef]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef]
- Hannou, S.A.; Wouters, K.; Paumelle, R.; Staels, B. Functional Genomics of the CDKN2A/B Locus in Cardiovascular and Metabolic Disease: What Have We Learned from GWASs? Trends Endocrinol. Metab. 2015, 26, 176–184. [Google Scholar] [CrossRef]
- Sato, K.; Nakagawa, H.; Tajima, A.; Yoshida, K.; Inoue, I. ANRIL Is Implicated in the Regulation of Nucleus and Potential Transcriptional Target of E2F1. Oncol. Rep. 2010, 24, 701–707. [Google Scholar] [CrossRef][Green Version]
- Wan, G.; Mathur, R.; Hu, X.; Liu, Y.; Zhang, X.; Peng, G.; Lu, X. Long Non-Coding RNA ANRIL (CDKN2B-AS) Is Induced by the ATM-E2F1 Signaling Pathway. Cell. Signal. 2013, 25, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Ghafouri-Fard, S. Antisense Non-Coding RNA in the INK4 Locus (ANRIL) in Human Cancers. Int. J. Cancer Manag. 2018, 11, e67864. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, Y.; Yarbrough, W.G. ARF Promotes MDM2 Degradation and Stabilizes P53: ARF-INK4a Locus Deletion Impairs Both the Rb and P53 Tumor Suppression Pathways. Cell 1998, 92, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Kyriakou, T.; Goel, A.; Peden, J.; Mälarstig, A.; Paulsson-Berne, G.; Hamsten, A.; Watkins, H.; Franco-Cereceda, A.; Gabrielsen, A.; et al. Relationship between CAD Risk Genotype in the Chromosome 9p21 Locus and Gene Expression. Identification of Eight New ANRIL Splice Variants. PLoS ONE 2009, 4, e7677. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of Linear and Novel Circular Forms of an INK4/ARF-Associated Non-Coding RNA Correlates with Atherosclerosis Risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef]
- GTEx Consortium Human Genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascenção, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene Expression across Mammalian Organ Development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef]
- Lin, L.; Gu, Z.-T.; Chen, W.-H.; Cao, K.-J. Increased Expression of the Long Non-Coding RNA ANRIL Promotes Lung Cancer Cell Metastasis and Correlates with Poor Prognosis. Diagn. Pathol. 2015, 10, 14. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, Z.-P.; Li, H.; Zhang, H.-Q.; Ma, F.-Q. ANRIL Is Associated with the Survival Rate of Patients with Colorectal Cancer, and Affects Cell Migration and Invasion in Vitro. Mol. Med. Rep. 2016, 14, 1714–1720. [Google Scholar] [CrossRef]
- Hirosue, A.; Ishihara, K.; Tokunaga, K.; Watanabe, T.; Saitoh, N.; Nakamoto, M.; Chandra, T.; Narita, M.; Shinohara, M.; Nakao, M. Quantitative Assessment of Higher-Order Chromatin Structure of the INK4/ARF Locus in Human Senescent Cells. Aging Cell. 2012, 11, 553–556. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Harismendy, O.; Notani, D.; Song, X.; Rahim, N.G.; Tanasa, B.; Heintzman, N.; Ren, B.; Fu, X.-D.; Topol, E.J.; Rosenfeld, M.G.; et al. 9p21 DNA Variants Associated with Coronary Artery Disease Impair Interferon-γ Signalling Response. Nature 2011, 470, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Ma, W.; Wang, X.; Qiao, J.; Zhang, B.; Cui, C.; Liu, Z.; Deng, D. Coordinated Transcription of ANRIL and P16 Genes Is Silenced by P16 DNA Methylation. Chin. J. Cancer Res. 2018, 30, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Saravanan, B.; Walavalkar, K.; Farooq, U.; Singh, A.K.; Radhakrishnan, S.; Thakur, J.; Pandit, A.; Henikoff, S.; Notani, D. Active Enhancers Strengthen Insulation by RNA-Mediated CTCF Binding at Chromatin Domain Boundaries. Genome Res. 2023, 33, 1–17. [Google Scholar] [CrossRef]
- Özgür, E.; Mert, U.; Isin, M.; Okutan, M.; Dalay, N.; Gezer, U. Differential Expression of Long Non-Coding RNAs during Genotoxic Stress-Induced Apoptosis in HeLa and MCF-7 Cells. Clin. Exp. Med. 2013, 13, 119–126. [Google Scholar] [CrossRef]
- Yin, X.; Liao, Y.; Xiong, W.; Zhang, Y.; Zhou, Y.; Yang, Y. Hypoxia-Induced LncRNA ANRIL Promotes Cisplatin Resistance in Retinoblastoma Cells through Regulating ABCG2 Expression. Clin. Exp. Pharmacol. Physiol. 2020, 47, 1049–1057. [Google Scholar] [CrossRef]
- Rodriguez, C.; Borgel, J.; Court, F.; Cathala, G.; Forné, T.; Piette, J. CTCF Is a DNA Methylation-Sensitive Positive Regulator of the INK/ARF Locus. Biochem. Biophys. Res. Commun. 2010, 392, 129–134. [Google Scholar] [CrossRef]
- Rahme, G.J.; Javed, N.M.; Puorro, K.L.; Xin, S.; Hovestadt, V.; Johnstone, S.E.; Bernstein, B.E. Modeling Epigenetic Lesions That Cause Gliomas. Cell 2023, 186, 3674–3685.e14. [Google Scholar] [CrossRef]
- Lillycrop, K.; Murray, R.; Cheong, C.; Teh, A.L.; Clarke-Harris, R.; Barton, S.; Costello, P.; Garratt, E.; Cook, E.; Titcombe, P.; et al. ANRIL Promoter DNA Methylation: A Perinatal Marker for Later Adiposity. EBioMedicine 2017, 19, 60–72. [Google Scholar] [CrossRef]
- Curtis, E.M.; Murray, R.; Titcombe, P.; Cook, E.; Clarke-Harris, R.; Costello, P.; Garratt, E.; Holbrook, J.D.; Barton, S.; Inskip, H.; et al. Perinatal DNA Methylation at CDKN2A Is Associated with Offspring Bone Mass: Findings From the Southampton Women’s Survey. J. Bone Min. Res. 2017, 32, 2030–2040. [Google Scholar] [CrossRef]
- Deng, W.; Wang, J.; Zhang, J.; Cai, J.; Bai, Z.; Zhang, Z. TET2 Regulates LncRNA-ANRIL Expression and Inhibits the Growth of Human Gastric Cancer Cells. IUBMB Life 2016, 68, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, X.; Xu, L.; Rong, C.; Shen, C.; Bian, W. Long Noncoding RNA ANRIL Could Be Transactivated by C-Myc and Promote Tumor Progression of Non-Small-Cell Lung Cancer. Onco. Targets Ther. 2016, 9, 3077–3084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, J.-H.; Tang, J.-M.; Li, J.; Li, X.-W. Upregulation of SOX2-Activated LncRNA ANRIL Promotes Nasopharyngeal Carcinoma Cell Growth. Sci. Rep. 2018, 8, 3333. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chen, W.; Qi, F.; Xia, R.; Sun, M.; Xu, T.; Yin, L.; Zhang, E.; De, W.; Shu, Y. Long Non-Coding RNA ANRIL Is Upregulated in Hepatocellular Carcinoma and Regulates Cell Apoptosis by Epigenetic Silencing of KLF2. J. Hematol. Oncol. 2015, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, J.; Qu, M.; Gao, X.; Zhang, W.; Wang, Z.; Zhao, L.; Wang, Y.; Li, B.; Li, J.; et al. Integrative Epigenome Map of the Normal Human Prostate Provides Insights into Prostate Cancer Predisposition. Front. Cell. Dev. Biol. 2021, 9, 723676. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-H.; Kim, D.; Jin, E.-J. Down-Regulation of Phospholipase D Stimulates Death of Lung Cancer Cells Involving Up-Regulation of the Long NcRNA ANRIL. Anticancer Res. 2015, 35, 2795–2803. [Google Scholar]
- Zhang, L.; Liu, J.; Lin, S.; Tan, J.; Huang, B.; Lin, J. Qingjie Fuzheng Granule Inhibited the Migration and Invasion of Colorectal Cancer Cells by Regulating the LncRNA ANRIL/Let-7a/TGF-Β1/Smad Axis. Evid. Based Complement. Altern. Med. 2020, 2020, 5264651. [Google Scholar] [CrossRef]
- Ma, W.; Qiao, J.; Zhou, J.; Gu, L.; Deng, D. Characterization of Novel LncRNA P14AS as a Protector of ANRIL through AUF1 Binding in Human Cells. Mol. Cancer 2020, 19, 42. [Google Scholar] [CrossRef]
- Xie, X.; Lin, J.; Fan, X.; Zhong, Y.; Chen, Y.; Liu, K.; Ren, Y.; Chen, X.; Lai, D.; Li, X.; et al. LncRNA CDKN2B-AS1 Stabilized by IGF2BP3 Drives the Malignancy of Renal Clear Cell Carcinoma through Epigenetically Activating NUF2 Transcription. Cell Death Dis. 2021, 12, 201. [Google Scholar] [CrossRef]
- He, S.; Gu, W.; Li, Y.; Zhu, H. ANRIL/CDKN2B-AS Shows Two-Stage Clade-Specific Evolution and Becomes Conserved after Transposon Insertions in Simians. BMC Evol. Biol. 2013, 13, 247. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten Things You Should Know about Transposable Elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Feschotte, C. Volatile Evolution of Long Noncoding RNA Repertoires: Mechanisms and Biological Implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-Coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Guigó, R. The RIDL Hypothesis: Transposable Elements as Functional Domains of Long Noncoding RNAs. RNA 2014, 20, 959–976. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-W.; Pan, J.-J.; Hu, J.-F.; Zhang, J.-Q.; Huang, L.; Huang, Y.; Liao, C.-Y.; Yang, C.; Chen, Z.-W.; Wang, Y.-D.; et al. SRSF3-Mediated Regulation of N6-Methyladenosine Modification-Related LncRNA ANRIL Splicing Promotes Resistance of Pancreatic Cancer to Gemcitabine. Cell. Rep. 2022, 39, 110813. [Google Scholar] [CrossRef]
- Sarkar, D.; Oghabian, A.; Bodiyabadu, P.K.; Joseph, W.R.; Leung, E.Y.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggests Variations in Processing. Int. J. Mol. Sci. 2017, 18, 1378. [Google Scholar] [CrossRef]
- Zhao, J.; Jin, W.; Yi, K.; Wang, Q.; Zhou, J.; Tan, Y.; Xu, C.; Xiao, M.; Hong, B.; Xu, F.; et al. Combination LSD1 and HOTAIR-EZH2 Inhibition Disrupts Cell Cycle Processes and Induces Apoptosis in Glioblastoma Cells. Pharmacol. Res. 2021, 171, 105764. [Google Scholar] [CrossRef]
- Cho, H.; Li, Y.; Archacki, S.; Wang, F.; Yu, G.; Chakrabarti, S.; Guo, Y.; Chen, Q.; Wang, Q.K. Splice Variants of LncRNA RNA ANRIL Exert Opposing Effects on Endothelial Cell Activities Associated with Coronary Artery Disease. RNA Biol. 2020, 17, 1391–1401. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of CircRNA Biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Lu, D.; Xu, A. The Interaction of CircRNAs and RNA Binding Proteins: An Important Part of CircRNA Maintenance and Function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Razeghian-Jahromi, I.; Zibaeenezhad, M.J.; Karimi Akhormeh, A.; Dara, M. Expression Ratio of Circular to Linear ANRIL in Hypertensive Patients with Coronary Artery Disease. Sci. Rep. 2022, 12, 1802. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, L.; Zhao, W.; Zhou, Y.; Shao, S. High Expression of ANRIL Correlated with the Poor Prognosis in Patients with Cancer: A Meta-Analysis. Medicine 2022, 101, e30531. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, Y.; Huang, Y.; Cao, K.; Liu, T.; Shen, H.; Cui, J.; Li, B.; Cai, J.; Gao, F.; et al. Long Non-Coding RNA ANRIL Promotes Homologous Recombination-Mediated DNA Repair by Maintaining ATR Protein Stability to Enhance Cancer Resistance. Mol. Cancer 2021, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Naemura, M.; Murasaki, C.; Inoue, Y.; Okamoto, H.; Kotake, Y. Long Noncoding RNA ANRIL Regulates Proliferation of Non-Small Cell Lung Cancer and Cervical Cancer Cells. Anticancer Res. 2015, 35, 5377–5382. [Google Scholar]
- Tian, Z.; Wen, S.; Zhang, Y.; Shi, X.; Zhu, Y.; Xu, Y.; Lv, H.; Wang, G. Identification of Dysregulated Long Non-Coding RNAs/MicroRNAs/MRNAs in TNM I Stage Lung Adenocarcinoma. Oncotarget 2017, 8, 51703–51718. [Google Scholar] [CrossRef]
- Du, Y.; Hao, X.; Liu, X. Low Expression of Long Noncoding RNA CDKN2B-AS1 in Patients with Idiopathic Pulmonary Fibrosis Predicts Lung Cancer by Regulating the P53-Signaling Pathway. Oncol. Lett. 2018, 15, 4912–4918. [Google Scholar] [CrossRef]
- Kato, E.; Takayanagi, N.; Takaku, Y.; Kagiyama, N.; Kanauchi, T.; Ishiguro, T.; Sugita, Y. Incidence and Predictive Factors of Lung Cancer in Patients with Idiopathic Pulmonary Fibrosis. ERJ Open Res. 2018, 4, 00111–02016. [Google Scholar] [CrossRef]
- Artinian, V.; Kvale, P.A. Cancer and Interstitial Lung Disease. Curr. Opin. Pulm. Med. 2004, 10, 425–434. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Yang, S.; Cen, Y.; Zhu, T.; Wang, L.; Xia, L.; Liu, Y.; Zou, J.; Xu, J.; et al. CircCDKN2B-AS1 Interacts with IMP3 to Stabilize Hexokinase 2 MRNA and Facilitate Cervical Squamous Cell Carcinoma Aerobic Glycolysis Progression. J. Exp. Clin. Cancer Res. 2020, 39, 281. [Google Scholar] [CrossRef]
- MacMillan, H.J.; Kong, Y.; Calvo-Roitberg, E.; Alonso, L.C.; Pai, A.A. High-Throughput Analysis of ANRIL CircRNA Isoforms in Human Pancreatic Islets. Sci. Rep. 2022, 12, 7745. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Guan, B.; Shang, H.; Peng, J.; Yang, H.; Lin, J. Babao Dan Inhibits Lymphangiogenesis of Gastric Cancer in Vitro and in Vivo via LncRNA-ANRIL/VEGF-C/VEGFR-3 Signaling Axis. Biomed. Pharmacother. 2022, 154, 113630. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, M.; Niu, Q.; Zhang, F.; Yang, Y.; Jiang, X. Knockdown of Long Non-Coding RNA ANRIL Inhibits Tumorigenesis in Human Gastric Cancer Cells via MicroRNA-99a-Mediated down-Regulation of BMI1. Braz. J. Med. Biol. Res. 2018, 51, e6839. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, D.; Yu, Z.; Li, H.; Zhao, S.; Hainisayimu, T.; Liu, L.; Wang, K. A Nomogram Model Based on the Number of Examined Lymph Nodes-Related Signature to Predict Prognosis and Guide Clinical Therapy in Gastric Cancer. Front. Immunol. 2022, 13, 947802. [Google Scholar] [CrossRef]
- Hu, X.; Lou, T.; Yuan, C.; Wang, Y.; Tu, X.; Wang, Y.; Zhang, T. Effects of LncRNA ANRIL-Knockdown on the Proliferation, Apoptosis and Cell Cycle of Gastric Cancer Cells. Oncol. Lett. 2021, 22, 621. [Google Scholar] [CrossRef]
- Lan, W.-G.; Xu, D.-H.; Xu, C.; Ding, C.-L.; Ning, F.-L.; Zhou, Y.-L.; Ma, L.-B.; Liu, C.-M.; Han, X. Silencing of Long Non-Coding RNA ANRIL Inhibits the Development of Multidrug Resistance in Gastric Cancer Cells. Oncol. Rep. 2016, 36, 263–270. [Google Scholar] [CrossRef]
- Zhang, E.; Kong, R.; Yin, D.; You, L.; Sun, M.; Han, L.; Xu, T.; Xia, R.; Yang, J.; De, W.; et al. Long Noncoding RNA ANRIL Indicates a Poor Prognosis of Gastric Cancer and Promotes Tumor Growth by Epigenetically Silencing of MiR-99a/MiR-449a. Oncotarget 2014, 5, 2276–2292. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, W.; Zhang, H.; Chu, Z.; Liu, H.; Fang, X.; Tang, D. Long Non-Coding RNA ANRIL Promotes Chemoresistance in Triple-Negative Breast Cancer via Enhancing Aerobic Glycolysis. Life Sci. 2022, 306, 120810. [Google Scholar] [CrossRef]
- Luo, Z.-B.; Lai, G.-E.; Jiang, T.; Cao, C.-L.; Peng, T.; Liu, F.-E. A Competing Endogenous RNA Network Reveals Novel LncRNA, MiRNA and MRNA Biomarkers With Diagnostic and Prognostic Value for Early Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820983293. [Google Scholar] [CrossRef]
- Xu, S.-T.; Xu, J.-H.; Zheng, Z.-R.; Zhao, Q.-Q.; Zeng, X.-S.; Cheng, S.-X.; Liang, Y.-H.; Hu, Q.-F. Long Non-Coding RNA ANRIL Promotes Carcinogenesis via Sponging MiR-199a in Triple-Negative Breast Cancer. Biomed. Pharmacother. 2017, 96, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, J.H.; Chae, Y.S.; Park, H.Y.; Kim, W.W.; Lee, S.J.; Jeong, J.-H.; Kang, S.H. Long Noncoding RNA SnaR Regulates Proliferation, Migration and Invasion of Triple-Negative Breast Cancer Cells. Anticancer Res. 2016, 36, 6289–6295. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Chen, K.; Fan, H.; Jin, F. The Synergistic Effect of CDKN2B-AS1 and SPC25 on Triple-Negative Breast Cancer. Ann. Transl. Med. 2022, 10, 783. [Google Scholar] [CrossRef]
- Mehta-Mujoo, P.M.; Cunliffe, H.E.; Hung, N.A.; Slatter, T.L. Long Non-Coding RNA ANRIL in the Nucleus Associates with Periostin Expression in Breast Cancer. Front. Oncol. 2019, 9, 885. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhai, J.; Fu, Y. LncRNA CDKN2B-AS1 Promotes the Progression of Ovarian Cancer by MiR-143-3p/SMAD3 Axis and Predicts a Poor Prognosis. Neoplasma 2020, 67, 782–793. [Google Scholar] [CrossRef]
- Miao, J.-T.; Gao, J.-H.; Chen, Y.-Q.; Chen, H.; Meng, H.-Y.; Lou, G. LncRNA ANRIL Affects the Sensitivity of Ovarian Cancer to Cisplatin via Regulation of Let-7a/HMGA2 Axis. Biosci. Rep. 2019, 39, BSR20182101. [Google Scholar] [CrossRef]
- Qiu, J.-J.; Wang, Y.; Liu, Y.-L.; Zhang, Y.; Ding, J.-X.; Hua, K.-Q. The Long Non-Coding RNA ANRIL Promotes Proliferation and Cell Cycle Progression and Inhibits Apoptosis and Senescence in Epithelial Ovarian Cancer. Oncotarget 2016, 7, 32478–32492. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Liu, H.; Su, D.; Luo, F.; Zhou, F. Long Noncoding RNA CDKN2B-AS1 Interacts with MiR-411-3p to Regulate Ovarian Cancer in Vitro and in Vivo through HIF-1a/VEGF/P38 Pathway. Biochem. Biophys. Res. Commun. 2019, 514, 44–50. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, G.; Zhang, H.; Tian, J.; Li, Y. Long Non-Coding RNA ANRIL Indicates a Poor Prognosis of Cervical Cancer and Promotes Carcinogenesis via PI3K/Akt Pathways. Biomed. Pharmacother. 2017, 85, 511–516. [Google Scholar] [CrossRef]
- Sadri, S.; Rejali, L.; Hadizadeh, M.; Aghdaei, H.A.; Young, C.; Nazemalhosseini-Mojarad, E.; Zali, M.R.; Bonab, M.A. ANRIL as a Prognostic Biomarker in Colon Pre-Cancerous Lesion Detection via Non-Invasive Sampling. Genes Genet. Syst. 2022, 96, 285–292. [Google Scholar] [CrossRef]
- Akbari, F.; Peymani, M.; Salehzadeh, A.; Ghaedi, K. Integrative in Silico and in Vitro Transcriptomics Analysis Revealed New LncRNAs Related to Intrinsic Apoptotic Genes in Colorectal Cancer. Cancer Cell Int. 2020, 20, 546. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, W.; Huang, F.; Sun, J.; Li, K.P.; Shi, J.; Yang, J.; Li, J.; Li, Y.; Hu, N.; et al. Comprehensive Analysis of the Expression Profiles of Long Non-Coding RNAs with Associated CeRNA Network Involved in the Colon Cancer Staging and Progression. Sci. Rep. 2019, 9, 16910. [Google Scholar] [CrossRef] [PubMed]
- Abbastabar, M.; Sarfi, M.; Golestani, A.; Karimi, A.; Pourmand, G.; Khalili, E. Tumor-Derived Urinary Exosomal Long Non-Coding RNAs as Diagnostic Biomarkers for Bladder Cancer. EXCLI J. 2020, 19, 301–310. [Google Scholar] [CrossRef]
- Zhu, H.; Li, X.; Song, Y.; Zhang, P.; Xiao, Y.; Xing, Y. Long Non-Coding RNA ANRIL Is up-Regulated in Bladder Cancer and Regulates Bladder Cancer Cell Proliferation and Apoptosis through the Intrinsic Pathway. Biochem. Biophys. Res. Commun. 2015, 467, 223–228. [Google Scholar] [CrossRef]
- Martínez-Fernández, M.; Feber, A.; Dueñas, M.; Segovia, C.; Rubio, C.; Fernandez, M.; Villacampa, F.; Duarte, J.; López-Calderón, F.F.; Gómez-Rodriguez, M.J.; et al. Analysis of the Polycomb-Related LncRNAs HOTAIR and ANRIL in Bladder Cancer. Clin. Epigenet. 2015, 7, 109. [Google Scholar] [CrossRef]
- Wu, Q.; He, Y.; Liu, X.; Luo, F.; Jiang, Y.; Xiang, M.; Zhao, R. Cancer Stem Cell-like Cells-Derived Exosomal LncRNA CDKN2B-AS1 Promotes Biological Characteristics in Thyroid Cancer via MiR-122-5p/P4HA1 Axis. Regen. Ther. 2023, 22, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Yi, C.; Lin, H.; Zhao, J.; Yuan, J.; Chen, Y.; Chen, H.; Lin, L.; Zhao, Y. LncRNA CDKN2B-AS1 Could Be an Indicator to Identify Prognosis and Status of Immune Microenvironment in Thyroid Cancer. Dis. Markers 2022, 2022, 4317480. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, Y.; Wen, L.; Zhou, Q.; Yan, S. LncRNA CDKN2B-AS1 Contributes to Glioma Development by Regulating the MiR-199a-5p/DDR1 Axis. J. Gene Med. 2022, 24, e3389. [Google Scholar] [CrossRef]
- Sun, Y.; Jing, Y.; Zhang, Y. Serum LncRNA-ANRIL and SOX9 Expression Levels in Glioma Patients and Their Relationship with Poor Prognosis. World J. Surg. Oncol. 2021, 19, 287. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, H.; Hou, D. Sevoflurane Represses Proliferation and Migration of Glioma Cells by Regulating the ANRIL/Let-7b-5p Axis. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Dai, W.; Tian, C.; Jin, S. Effect of LncRNA ANRIL Silencing on Anoikis and Cell Cycle in Human Glioma via MicroRNA-203a. Onco. Targets Ther. 2018, 11, 5103–5109. [Google Scholar] [CrossRef] [PubMed]
- Paul, Y.; Thomas, S.; Patil, V.; Kumar, N.; Mondal, B.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Mahalingam, K.; Somasundaram, K. Genetic Landscape of Long Noncoding RNA (LncRNAs) in Glioblastoma: Identification of Complex LncRNA Regulatory Networks and Clinically Relevant LncRNAs in Glioblastoma. Oncotarget 2018, 9, 29548–29564. [Google Scholar] [CrossRef]
- Lee, A.M.; Ferdjallah, A.; Moore, E.; Kim, D.C.; Nath, A.; Greengard, E.; Huang, R.S. Long Non-Coding RNA ANRIL as a Potential Biomarker of Chemosensitivity and Clinical Outcomes in Osteosarcoma. Int. J. Mol. Sci. 2021, 22, 11168. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Tao, H.; Jin, L.; Xiang, W.; Guo, W. CDKN2B-AS1 Exerts Oncogenic Role in Osteosarcoma by Promoting Cell Proliferation and Epithelial to Mesenchymal Transition. Cancer Biother. Radiopharm. 2020, 35, 58–65. [Google Scholar] [CrossRef]
- Li, G.; Zhu, Y. Effect of LncRNA ANRIL Knockdown on Proliferation and Cisplatin Chemoresistance of Osteosarcoma Cells in Vitro. Pathol. Res. Pract. 2019, 215, 931–938. [Google Scholar] [CrossRef]
- Cheng, S.; Huang, T.; Li, P.; Zhang, W.; Wang, Z.; Chen, Y. Long Non-Coding RNA ANRIL Promotes the Proliferation, Migration and Invasion of Human Osteosarcoma Cells. Exp. Ther. Med. 2017, 14, 5121–5125. [Google Scholar] [CrossRef]
- Guan, H.; Mei, Y.; Mi, Y.; Li, C.; Sun, X.; Zhao, X.; Liu, J.; Cao, W.; Li, Y.; Wang, Y. Downregulation of LncRNA ANRIL Suppresses Growth and Metastasis in Human Osteosarcoma Cells. Onco. Targets Ther. 2018, 11, 4893–4899. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Liu, G.; Yuan, D.; Dai, J.; Cui, Y.; Tang, X. Long Non-Coding RNA ANRIL Is Associated with a Poor Prognosis of Osteosarcoma and Promotes Tumorigenesis via PI3K/Akt Pathway. J. Bone Oncol. 2018, 11, 51–55. [Google Scholar] [CrossRef]
- Abula, A.; Saimaiti, G.; Maimaiti, X.; Wuqikun, W.; Abulaiti, A.; Ren, P.; Yusufu, A. The Stimulative Function of Long Noncoding RNA CDKN2B-AS1 in Osteosarcoma by Targeting the MicroRNA-122/CCNG1 Axis. J. Recept. Signal. Transduct. Res. 2022, 42, 71–79. [Google Scholar] [CrossRef]
- Yang, L.-H.; Du, P.; Liu, W.; An, L.-K.; Li, J.; Zhu, W.-Y.; Yuan, S.; Wang, L.; Zang, L. LncRNA ANRIL Promotes Multiple Myeloma Progression and Bortezomib Resistance by EZH2-Mediated Epigenetically Silencing of PTEN. Neoplasma 2021, 68, 788–797. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, H.-Y.; Zhang, J.-L.; Wan, D.-M.; Li, Y.-M.; Jiang, Z.-X. Dysregulation of LncRNA ANRIL Mediated by MiR-411-3p Inhibits the Malignant Proliferation and Tumor Stem Cell like Property of Multiple Myeloma via Hypoxia-Inducible Factor 1α. Exp. Cell. Res. 2020, 396, 112280. [Google Scholar] [CrossRef] [PubMed]
- Drak Alsibai, K.; Vacher, S.; Meseure, D.; Nicolas, A.; Lae, M.; Schnitzler, A.; Chemlali, W.; Cros, J.; Longchampt, E.; Cacheux, W.; et al. High Positive Correlations between ANRIL and P16-CDKN2A/P15-CDKN2B/P14-ARF Gene Cluster Overexpression in Multi-Tumor Types Suggest Deregulated Activation of an ANRIL-ARF Bidirectional Promoter. Noncoding RNA 2019, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zhu, K.; Yin, Y.; Luo, Z. Long Non-coding RNA ANRIL Is a Potential Indicator of Disease Progression and Poor Prognosis in Acute Myeloid Leukemia. Mol. Med. Rep. 2021, 23, 112. [Google Scholar] [CrossRef] [PubMed]
- Gamaleldin, M.A.; Ghallab, O.; Abo Elwafa, R.A. Prognostic Significance of Long Non Coding RNA ANRIL and SNHG14 in Acute Myeloid Leukemia. Asian Pac. J. Cancer Prev. 2021, 22, 3763–3771. [Google Scholar] [CrossRef]
- Sun, L.-Y.; Li, X.-J.; Sun, Y.-M.; Huang, W.; Fang, K.; Han, C.; Chen, Z.-H.; Luo, X.-Q.; Chen, Y.-Q.; Wang, W.-T. LncRNA ANRIL Regulates AML Development through Modulating the Glucose Metabolism Pathway of AdipoR1/AMPK/SIRT1. Mol. Cancer 2018, 17, 127. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wu, W.; Chen, M.; Cheng, W.; Yu, J.; Fang, J.; Xu, L.; Yasunaga, J.-I.; Matsuoka, M.; Zhao, T. Long Noncoding RNA ANRIL Supports Proliferation of Adult T-Cell Leukemia Cells through Cooperation with EZH2. J. Virol. 2018, 92, e00909–e00918. [Google Scholar] [CrossRef]
- Li, G.; Gao, L.; Zhao, J.; Liu, D.; Li, H.; Hu, M. LncRNA ANRIL/MiR-7-5p/TCF4 Axis Contributes to the Progression of T Cell Acute Lymphoblastic Leukemia. Cancer Cell. Int. 2020, 20, 335. [Google Scholar] [CrossRef]
- Xu, S.; Wang, H.; Pan, H.; Shi, Y.; Li, T.; Ge, S.; Jia, R.; Zhang, H.; Fan, X. ANRIL LncRNA Triggers Efficient Therapeutic Efficacy by Reprogramming the Aberrant INK4-Hub in Melanoma. Cancer Lett. 2016, 381, 41–48. [Google Scholar] [CrossRef]
- Xie, H.; Rachakonda, P.S.; Heidenreich, B.; Nagore, E.; Sucker, A.; Hemminki, K.; Schadendorf, D.; Kumar, R. Mapping of Deletion Breakpoints at the CDKN2A Locus in Melanoma: Detection of MTAP-ANRIL Fusion Transcripts. Oncotarget 2016, 7, 16490–16504. [Google Scholar] [CrossRef]
- Lin, Z.; Lei, Y.; Wen, M.; He, Q.; Tian, D.; Xie, H. MTAP-ANRIL Gene Fusion Promotes Melanoma Epithelial-Mesenchymal Transition-like Process by Activating the JNK and P38 Signaling Pathways. Sci. Rep. 2023, 13, 9073. [Google Scholar] [CrossRef]
- Shang, C.; Ao, C.N.; Cheong, C.C.; Meng, L. Long Non-Coding RNA CDKN2B Antisense RNA 1 Gene Contributes to Paclitaxel Resistance in Endometrial Carcinoma. Front. Oncol. 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Ma, J.; Ma, X.-X. CDKN2B-AS1 Promotes Malignancy as a Novel Prognosis-Related Molecular Marker in the Endometrial Cancer Immune Microenvironment. Front. Cell. Dev. Biol. 2021, 9, 721676. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, H.; Duan, Y.; Zhu, J.; Dai, H. M6A-Related Long Noncoding RNAs Predict Prognosis and Indicate Therapeutic Response in Endometrial Carcinoma. J. Clin. Lab. Anal. 2023, 37, e24813. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Fan, W.; Sui, X.; Wang, J.; Zhao, J. Necroptosis-Related LncRNA Signatures for Prognostic Prediction in Uterine Corpora Endometrial Cancer. Reprod. Sci. 2023, 30, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Angenard, G.; Merdrignac, A.; Louis, C.; Edeline, J.; Coulouarn, C. Expression of Long Non-Coding RNA ANRIL Predicts a Poor Prognosis in Intrahepatic Cholangiocarcinoma. Dig. Liver Dis. 2019, 51, 1337–1343. [Google Scholar] [CrossRef]
- Dasgupta, P.; Kulkarni, P.; Majid, S.; Hashimoto, Y.; Shiina, M.; Shahryari, V.; Bhat, N.S.; Tabatabai, L.; Yamamura, S.; Saini, S.; et al. LncRNA CDKN2B-AS1/MiR-141/Cyclin D Network Regulates Tumor Progression and Metastasis of Renal Cell Carcinoma. Cell Death Dis. 2020, 11, 660. [Google Scholar] [CrossRef]
- Ren, H.; Guo, X.; Li, F.; Xia, Q.; Chen, Z.; Xing, Y. Four Autophagy-Related Long Noncoding RNAs Provide Coexpression and CeRNA Mechanisms in Retinoblastoma through Bioinformatics and Experimental Evidence. ACS Omega 2021, 6, 33976–33984. [Google Scholar] [CrossRef]
- Liu, F.; Xiao, Y.; Ma, L.; Wang, J. Regulating of Cell Cycle Progression by the LncRNA CDKN2B-AS1/MiR-324-5p/ROCK1 Axis in Laryngeal Squamous Cell Cancer. Int. J. Biol. Markers 2020, 35, 47–56. [Google Scholar] [CrossRef]
- Chen, H.; Xin, Y.; Zhou, L.; Huang, J.; Tao, L.; Cheng, L.; Tian, J. Cisplatin and Paclitaxel Target Significant Long Noncoding RNAs in Laryngeal Squamous Cell Carcinoma. Med. Oncol. 2014, 31, 246. [Google Scholar] [CrossRef]
- Matsunaga, N.; Wakasaki, T.; Yasumatsu, R.; Kotake, Y. Long Noncoding RNA, ANRIL, Regulates the Proliferation of Head and Neck Squamous Cell Carcinoma. Anticancer Res. 2019, 39, 4073–4077. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, N.; Luo, J. Downregulation of LncRNA ANRIL Represses Tumorigenicity and Enhances Cisplatin-Induced Cytotoxicity via Regulating MicroRNA Let-7a in Nasopharyngeal Carcinoma. J. Biochem. Mol. Toxicol. 2017, 31, e21904. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.-R.; Zhang, D.-J.; Fu, Z.-M.; Guo, Y.-Y.; Guan, G.-F. Long Non-Coding RNA ANRIL Promotes Proliferation, Clonogenicity, Invasion and Migration of Laryngeal Squamous Cell Carcinoma by Regulating MiR-181a/Snai2 Axis. Regen. Ther. 2019, 11, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-W.; Hu, Z.-L.; Li, H.; Tan, Q.-F.; Tong, J.; Zhang, Y.-Q. Knockdown of LncRNA ANRIL Suppresses the Production of Inflammatory Cytokines and Mucin 5AC in Nasal Epithelial Cells via the MiR-15a-5p/JAK2 Axis. Mol. Med. Rep. 2021, 23, 145. [Google Scholar] [CrossRef] [PubMed]
- Kopczyńska, M.; Kolenda, T.; Guglas, K.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Mackiewicz, A.; Mackiewicz, J.; Lamperska, K. PRINS LncRNA Is a New Biomarker Candidate for HPV Infection and Prognosis of Head and Neck Squamous Cell Carcinomas. Diagnostics 2020, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Li, Y.; He, F.; Kong, J. LncRNA CDKN2B-AS1 Promotes Cell Viability, Migration, and Invasion of Hepatocellular Carcinoma via Sponging MiR-424-5p. Cancer Manag. Res. 2020, 12, 6807–6819. [Google Scholar] [CrossRef]
- Xu, L.; Wu, H.; Pan, J.; Chen, Z.; Du, L. Significance of LncRNA CDKN2B-AS1 in Interventional Therapy of Liver Cancer and the Mechanism under Its Participation in Tumour Cell Growth via MiR-199a-5p. J. Oncol. 2022, 2022, 2313416. [Google Scholar] [CrossRef]
- Huang, Y.; Xiang, B.; Liu, Y.; Wang, Y.; Kan, H. LncRNA CDKN2B-AS1 Promotes Tumor Growth and Metastasis of Human Hepatocellular Carcinoma by Targeting Let-7c-5p/NAP1L1 Axis. Cancer Lett. 2018, 437, 56–66. [Google Scholar] [CrossRef]
- Li, K.; Zhao, B.; Wei, D.; Cui, Y.; Qian, L.; Wang, W.; Liu, G. Long Non-Coding RNA ANRIL Enhances Mitochondrial Function of Hepatocellular Carcinoma by Regulating the MiR-199a-5p/ARL2 Axis. Environ. Toxicol. 2020, 35, 313–321. [Google Scholar] [CrossRef]
- Han, W.; Wang, Q.; Zheng, L.; Hong, H.; Yan, B.; Ma, Y.; Li, X.; Zhou, D. The Role of LncRNA ANRIL in the Progression of Hepatocellular Carcinoma. J. Pharm. Pharmacol. 2021, 73, 1033–1038. [Google Scholar] [CrossRef]
- Sabry, H.S.; Tayel, S.I.; Enar, M.E.; Elabd, N.S. Differential Expression of Long Noncoding RNA in Hepatocellular Carcinoma on Top of Chronic HCV and HBV Infections. Clin. Exp. Hepatol. 2021, 7, 337–350. [Google Scholar] [CrossRef]
- Hua, L.; Wang, C.-Y.; Yao, K.-H.; Chen, J.-T.; Zhang, J.-J.; Ma, W.-L. High Expression of Long Non-Coding RNA ANRIL Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3076–3082. [Google Scholar] [PubMed]
- Sui, J.; Miao, Y.; Han, J.; Nan, H.; Shen, B.; Zhang, X.; Zhang, Y.; Wu, Y.; Wu, W.; Liu, T.; et al. Systematic Analyses of a Novel LncRNA-Associated Signature as the Prognostic Biomarker for Hepatocellular Carcinoma. Cancer Med. 2018, 7, 3240–3256. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, M.S.; Santibanez Koref, M.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010, 6, e1000899. [Google Scholar] [CrossRef]
- Aguilo, F.; Di Cecilia, S.; Walsh, M.J. Long Non-Coding RNA ANRIL and Polycomb in Human Cancers and Cardiovascular Disease. Curr. Top. Microbiol. Immunol. 2016, 394, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Ghobadi, N.; Mehramiz, M.; ShahidSales, S.; Rezaei Brojerdi, A.; Anvari, K.; Khazaei, M.; Rezayi, M.; Sadegh Khorrami, M.; Joudi-Mashhad, M.; Ramshini, H.; et al. A Genetic Variant in CDKN2A/2B Locus Was Associated with Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. J. Cell. Physiol. 2019, 234, 5070–5076. [Google Scholar] [CrossRef]
- Maruei-Milan, R.; Heidari, Z.; Aryan, A.; Asadi-Tarani, M.; Salimi, S. Long Non-Coding RNA ANRIL Polymorphisms in Papillary Thyroid Cancer and Its Severity. Br. J. Biomed. Sci. 2021, 78, 58–62. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, J.-S.; Hu, W.-F. Functional Genetic Single-Nucleotide Polymorphisms (SNPs) in Cyclin-Dependent Kinase Inhibitor 2A/B (CDKN2A/B) Locus Are Associated with Risk and Prognosis of Osteosarcoma in Chinese Populations. Med. Sci. Monit. 2019, 25, 1307–1313. [Google Scholar] [CrossRef]
- Barbieri, R.B.; Bufalo, N.E.; Secolin, R.; Assumpção, L.V.M.; Maciel, R.M.B.; Cerutti, J.M.; Ward, L.S. Polymorphisms of Cell Cycle Control Genes Influence the Development of Sporadic Medullary Thyroid Carcinoma. Eur. J. Endocrinol. 2014, 171, 761–767. [Google Scholar] [CrossRef]
- Lv, X.; Cui, Z.; Li, H.; Li, J.; Yang, Z.; Bi, Y.; Gao, M.; Zhang, Z.; Wang, S.; Zhou, B.; et al. Association between Polymorphism in CDKN2B-AS1 Gene and Its Interaction with Smoking on the Risk of Lung Cancer in a Chinese Population. Hum. Genom. 2019, 13, 58. [Google Scholar] [CrossRef]
- Tritto, V.; Ferrari, L.; Esposito, S.; Zuccotti, P.; Bianchessi, D.; Natacci, F.; Saletti, V.; Eoli, M.; Riva, P. Non-Coding RNA and Tumor Development in Neurofibromatosis Type 1: ANRIL Rs2151280 Is Associated with Optic Glioma Development and a Mild Phenotype in Neurofibromatosis Type 1 Patients. Genes 2019, 10, 892. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, L.; Li, N.; Wang, M.; Yao, L.; Dong, S.; Zhang, M.; Yang, P.; Hao, Q.; Wu, Y.; et al. Impact of Four LncRNA Polymorphisms (Rs2151280, Rs7763881, Rs1136410, and Rs3787016) on Glioma Risk and Prognosis: A Case-Control Study. Mol. Carcinog. 2019, 58, 2218–2229. [Google Scholar] [CrossRef] [PubMed]
- Poi, M.J.; Li, J.; Sborov, D.W.; VanGundy, Z.; Cho, Y.K.; Lamprecht, M.; Pichiorri, F.; Phelps, M.A.; Hofmeister, C.C. Polymorphism in ANRIL Is Associated with Relapse in Patients with Multiple Myeloma after Autologous Stem Cell Transplant. Mol. Carcinog. 2017, 56, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Seligson, N.; Zhang, X.; Johnson, J.; Vangundy, Z.; Wang, D.; Phelps, M.; Hofmeister, C.; Sadee, W.; Poi, M.J. Association of ANRIL Polymorphism with Overall Survival in Adult Patients with Hematologic Malignancies After Allogeneic Hematopoietic Stem Cell Transplantation. Anticancer Res. 2020, 40, 5707–5713. [Google Scholar] [CrossRef] [PubMed]
- Stacey, S.N.; Sulem, P.; Masson, G.; Gudjonsson, S.A.; Thorleifsson, G.; Jakobsdottir, M.; Sigurdsson, A.; Gudbjartsson, D.F.; Sigurgeirsson, B.; Benediktsdottir, K.R.; et al. New Common Variants Affecting Susceptibility to Basal Cell Carcinoma. Nat. Genet. 2009, 41, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Petkevicius, V.; Streleckiene, G.; Balciute, K.; Link, A.; Leja, M.; Malfertheiner, P.; Skieceviciene, J.; Kupcinskas, J. Association of Long Non-Coding RNA Polymorphisms with Gastric Cancer and Atrophic Gastritis. Genes 2020, 11, 1505. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Pouresmaeili, F.; Omrani, M.D.; Habibi, M.; Sarrafzadeh, S.; Noroozi, R.; Rakhshan, A.; Sayad, A.; Ghafouri-Fard, S. Association of ANRIL Gene Polymorphisms with Prostate Cancer and Benign Prostatic Hyperplasia in an Iranian Population. Biomark. Med. 2017, 11, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, W.; Shao, Z. Association between Long Non-Coding RNA Polymorphisms and Cancer Risk: A Meta-Analysis. Biosci. Rep. 2018, 38, BSR20180365. [Google Scholar] [CrossRef]
- Sun, Q.; Chong, F.; Jiang, X.; Wang, Y.; Xu, K.; Zou, Y.; Song, C. Association Study of SNPs in LncRNA CDKN2B-AS1 with Breast Cancer Susceptibility in Chinese Han Population. Int. J. Biochem. Cell. Biol. 2022, 143, 106139. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Kim, J.; Kim, S.-W.; Park, S.K.; Ahn, S.H.; Lee, M.H.; Suh, Y.J.; Noh, D.-Y.; Son, B.H.; Cho, Y.U.; et al. BRCA1/2-Negative, High-Risk Breast Cancers (BRCAX) for Asian Women: Genetic Susceptibility Loci and Their Potential Impacts. Sci. Rep. 2018, 8, 15263. [Google Scholar] [CrossRef]
- Timofeeva, M.N.; Hung, R.J.; Rafnar, T.; Christiani, D.C.; Field, J.K.; Bickeböller, H.; Risch, A.; McKay, J.D.; Wang, Y.; Dai, J.; et al. Influence of Common Genetic Variation on Lung Cancer Risk: Meta-Analysis of 14,900 Cases and 29,485 Controls. Hum. Mol. Genet. 2012, 21, 4980–4995. [Google Scholar] [CrossRef]
- Omrani, M.D.; Mohammad-Rahimi, H.; Basiri, A.; Fallahian, M.; Noroozi, R.; Taheri, M.; Ghafouri-Fard, S. Discrimination of Patients with Prostate Cancer from Healthy Persons Using a Set of Single Nucleotide Polymorphisms. Urol. J. 2021, 18, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Fehringer, G.; Kraft, P.; Pharoah, P.D.; Eeles, R.A.; Chatterjee, N.; Schumacher, F.R.; Schildkraut, J.M.; Lindström, S.; Brennan, P.; Bickeböller, H.; et al. Cross-Cancer Genome-Wide Analysis of Lung, Ovary, Breast, Prostate, and Colorectal Cancer Reveals Novel Pleiotropic Associations. Cancer. Res. 2016, 76, 5103–5114. [Google Scholar] [CrossRef] [PubMed]
- Campa, D.; Barrdahl, M.; Gaudet, M.M.; Black, A.; Chanock, S.J.; Diver, W.R.; Gapstur, S.M.; Haiman, C.; Hankinson, S.; Hazra, A.; et al. Genetic Risk Variants Associated with in Situ Breast Cancer. Breast Cancer Res. 2015, 17, 82. [Google Scholar] [CrossRef]
- Long, J.; Zhang, B.; Signorello, L.B.; Cai, Q.; Deming-Halverson, S.; Shrubsole, M.J.; Sanderson, M.; Dennis, J.; Michailidou, K.; Easton, D.F.; et al. Evaluating Genome-Wide Association Study-Identified Breast Cancer Risk Variants in African-American Women. PLoS ONE 2013, 8, e58350. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, V.; Rizzolo, P.; Scarnò, M.; Chillemi, G.; Navazio, A.S.; Valentini, V.; Zelli, V.; Zanna, I.; Saieva, C.; Masala, G.; et al. Novel and Known Genetic Variants for Male Breast Cancer Risk at 8q24.21, 9p21.3, 11q13.3 and 14q24.1: Results from a Multicenter Study in Italy. Eur. J. Cancer 2015, 51, 2289–2295. [Google Scholar] [CrossRef]
- Antoniou, A.C.; Kuchenbaecker, K.B.; Soucy, P.; Beesley, J.; Chen, X.; McGuffog, L.; Lee, A.; Barrowdale, D.; Healey, S.; Sinilnikova, O.M.; et al. Common Variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 Are Associated with Breast Cancer Risk for BRCA1 and/or BRCA2 Mutation Carriers. Breast Cancer Res. 2012, 14, R33. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Lü, B.; Ruan, W.; Zhu, Y.; Sheng, H.; Lai, M. Genetic Polymorphisms and Breast Cancer Risk: Evidence from Meta-Analyses, Pooled Analyses, and Genome-Wide Association Studies. Breast Cancer Res. Treat. 2011, 127, 309–324. [Google Scholar] [CrossRef]
- Dahlin, A.M.; Wibom, C.; Andersson, U.; Hougaard, D.M.; Bybjerg-Grauholm, J.; Deltour, I.; Hultman, C.M.; Kähler, A.K.; Karlsson, R.; Hjalmars, U.; et al. Genetic Variants in the 9p21.3 Locus Associated with Glioma Risk in Children, Adolescents, and Young Adults: A Case-Control Study. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1252–1258. [Google Scholar] [CrossRef]
- Chen, Y.-D.; Zhang, N.; Qiu, X.-G.; Yuan, J.; Yang, M. LncRNA CDKN2BAS Rs2157719 Genetic Variant Contributes to Medulloblastoma Predisposition. J. Gene Med. 2018, 20, e3000. [Google Scholar] [CrossRef]
- Adel Fahmideh, M.; Lavebratt, C.; Schüz, J.; Röösli, M.; Tynes, T.; Grotzer, M.A.; Johansen, C.; Kuehni, C.E.; Lannering, B.; Prochazka, M.; et al. CCDC26, CDKN2BAS, RTEL1 and TERT Polymorphisms in Pediatric Brain Tumor Susceptibility. Carcinogenesis 2015, 36, 876–882. [Google Scholar] [CrossRef]
- Li, W.-Q.; Pfeiffer, R.M.; Hyland, P.L.; Shi, J.; Gu, F.; Wang, Z.; Bhattacharjee, S.; Luo, J.; Xiong, X.; Yeager, M.; et al. Genetic Polymorphisms in the 9p21 Region Associated with Risk of Multiple Cancers. Carcinogenesis 2014, 35, 2698–2705. [Google Scholar] [CrossRef] [PubMed]
- Park, S.L.; Fesinmeyer, M.D.; Timofeeva, M.; Caberto, C.P.; Kocarnik, J.M.; Han, Y.; Love, S.-A.; Young, A.; Dumitrescu, L.; Lin, Y.; et al. Pleiotropic Associations of Risk Variants Identified for Other Cancers with Lung Cancer Risk: The PAGE and TRICL Consortia. J. Natl. Cancer Inst. 2014, 106, dju061. [Google Scholar] [CrossRef] [PubMed]
- Wibom, C.; Späth, F.; Dahlin, A.M.; Langseth, H.; Hovig, E.; Rajaraman, P.; Johannesen, T.B.; Andersson, U.; Melin, B. Investigation of Established Genetic Risk Variants for Glioma in Prediagnostic Samples from a Population-Based Nested Case-Control Study. Cancer Epidemiol. Biomark. Prev. 2015, 24, 810–816. [Google Scholar] [CrossRef]
- Qi, X.; Wan, Y.; Zhan, Q.; Yang, S.; Wang, Y.; Cai, X. Effect of CDKN2A/B Rs4977756 Polymorphism on Glioma Risk: A Meta-Analysis of 16 Studies Including 24077 Participants. Mamm. Genome 2016, 27, 1–7. [Google Scholar] [CrossRef]
- Lu, H.; Yang, Y.; Wang, J.; Liu, Y.; Huang, M.; Sun, X.; Ke, Y. The CDKN2A-CDKN2B Rs4977756 Polymorphism and Glioma Risk: A Meta-Analysis. Int. J. Clin. Exp. Med. 2015, 8, 17480–17488. [Google Scholar]
- Viana-Pereira, M.; Moreno, D.A.; Linhares, P.; Amorim, J.; Nabiço, R.; Costa, S.; Vaz, R.; Reis, R.M. Replication of GWAS Identifies RTEL1, CDKN2A/B, and PHLDB1 SNPs as Risk Factors in Portuguese Gliomas Patients. Mol. Biol. Rep. 2020, 47, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Ghasimi, S.; Wibom, C.; Dahlin, A.M.; Brännström, T.; Golovleva, I.; Andersson, U.; Melin, B. Genetic Risk Variants in the CDKN2A/B, RTEL1 and EGFR Genes Are Associated with Somatic Biomarkers in Glioma. J. Neurooncol. 2016, 127, 483–492. [Google Scholar] [CrossRef]
- Shete, S.; Hosking, F.J.; Robertson, L.B.; Dobbins, S.E.; Sanson, M.; Malmer, B.; Simon, M.; Marie, Y.; Boisselier, B.; Delattre, J.-Y.; et al. Genome-Wide Association Study Identifies Five Susceptibility Loci for Glioma. Nat. Genet. 2009, 41, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Peng, Y.; Zhao, X. An Updated and Comprehensive Meta-Analysis of Association Between Seven Hot Loci Polymorphisms from Eight GWAS and Glioma Risk. Mol. Neurobiol. 2016, 53, 4397–4405. [Google Scholar] [CrossRef]
- Dahlin, A.M.; Wibom, C.; Ghasimi, S.; Brännström, T.; Andersson, U.; Melin, B. Relation between Established Glioma Risk Variants and DNA Methylation in the Tumor. PLoS ONE 2016, 11, e0163067. [Google Scholar] [CrossRef]
- Bei, J.-X.; Su, W.-H.; Ng, C.-C.; Yu, K.; Chin, Y.-M.; Lou, P.-J.; Hsu, W.-L.; McKay, J.D.; Chen, C.-J.; Chang, Y.-S.; et al. A GWAS Meta-Analysis and Replication Study Identifies a Novel Locus within CLPTM1L/TERT Associated with Nasopharyngeal Carcinoma in Individuals of Chinese Ancestry. Cancer Epidemiol. Biomark. Prev. 2016, 25, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.Y.-Y.; Johansson, G.; Wibom, C.; Brännström, T.; Malmström, A.; Henriksson, R.; Golovleva, I.; Bondy, M.L.; Andersson, U.; Dahlin, A.M.; et al. The Genetic Architecture of Gliomagenesis-Genetic Risk Variants Linked to Specific Molecular Subtypes. Cancers 2019, 11, 2001. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Camino, A.; Martin-Guerrero, I.; Garcia de Andoin, N.; Sastre, A.; Carbone Bañeres, A.; Astigarraga, I.; Navajas, A.; Garcia-Orad, A. Confirmation of Involvement of New Variants at CDKN2A/B in Pediatric Acute Lymphoblastic Leukemia Susceptibility in the Spanish Population. PLoS ONE 2017, 12, e0177421. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Sazzini, M.; Garagnani, P.; Ferrari, A.; Boattini, A.; Lonetti, A.; Papayannidis, C.; Mantovani, V.; Marasco, E.; Ottaviani, E.; et al. A Polymorphism in the Chromosome 9p21 ANRIL Locus Is Associated to Philadelphia Positive Acute Lymphoblastic Leukemia. Leuk. Res. 2011, 35, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Davari, D.R.; Orlow, I.; Kanetsky, P.A.; Luo, L.; Edmiston, S.N.; Conway, K.; Parrish, E.A.; Hao, H.; Busam, K.J.; Sharma, A.; et al. Disease-Associated Risk Variants in ANRIL Are Associated with Tumor-Infiltrating Lymphocyte Presence in Primary Melanomas in the Population-Based GEM Study. Cancer Epidemiol. Biomark. Prev. 2021, 30, 2309–2316. [Google Scholar] [CrossRef]
- Gu, F.; Pfeiffer, R.M.; Bhattacharjee, S.; Han, S.S.; Taylor, P.R.; Berndt, S.; Yang, H.; Sigurdson, A.J.; Toro, J.; Mirabello, L.; et al. Common Genetic Variants in the 9p21 Region and Their Associations with Multiple Tumours. Br. J. Cancer 2013, 108, 1378–1386. [Google Scholar] [CrossRef]
- Giaccherini, M.; Farinella, R.; Gentiluomo, M.; Mohelnikova-Duchonova, B.; Kauffmann, E.F.; Palmeri, M.; Uzunoglu, F.; Soucek, P.; Petrauskas, D.; Cavestro, G.M.; et al. Association between a Polymorphic Variant in the CDKN2B-AS1/ANRIL Gene and Pancreatic Cancer Risk. Int. J. Cancer 2023, 153, 373–379. [Google Scholar] [CrossRef]
- Gong, W.-J.; Yin, J.-Y.; Li, X.-P.; Fang, C.; Xiao, D.; Zhang, W.; Zhou, H.-H.; Li, X.; Liu, Z.-Q. Association of Well-Characterized Lung Cancer LncRNA Polymorphisms with Lung Cancer Susceptibility and Platinum-Based Chemotherapy Response. Tumour Biol. 2016, 37, 8349–8358. [Google Scholar] [CrossRef]
- Seifi, S.; Pouya, F.; Rahmani, M.; Mehramiz, M.; Rastgar-Moghadam, A.; Gharib, M.; Rahmani, F.; Shahidsales, S.; Hassanian, S.M.; Khazaei, M.; et al. Association of Cyclin-Dependent Kinase Inhibitor 2A/B with Increased Risk of Developing Breast Cancer. J. Cell. Physiol. 2020, 235, 5141–5145. [Google Scholar] [CrossRef]
- Lesseur, C.; Diergaarde, B.; Olshan, A.F.; Wünsch-Filho, V.; Ness, A.R.; Liu, G.; Lacko, M.; Eluf-Neto, J.; Franceschi, S.; Lagiou, P.; et al. Genome-Wide Association Analyses Identify New Susceptibility Loci for Oral Cavity and Pharyngeal Cancer. Nat. Genet. 2016, 48, 1544–1550. [Google Scholar] [CrossRef]
- Wrensch, M.; Jenkins, R.B.; Chang, J.S.; Yeh, R.-F.; Xiao, Y.; Decker, P.A.; Ballman, K.V.; Berger, M.; Buckner, J.C.; Chang, S.; et al. Variants in the CDKN2B and RTEL1 Regions Are Associated with High-Grade Glioma Susceptibility. Nat. Genet. 2009, 41, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Feng, F.-T.; Xu, M.; Liu, W.-S.; Yao, Y.-Y.; Xie, S.-H.; Li, X.-Z.; Ye, Z.-L.; Feng, Q.-S.; Chen, L.-Z.; et al. Nasopharyngeal Carcinoma Risk Prediction via Salivary Detection of Host and Epstein-Barr Virus Genetic Variants. Oncotarget 2017, 8, 95066–95074. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A Genome-Wide Association Study of Nasopharyngeal Carcinoma Identifies Three New Susceptibility Loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Driver, K.E.; Song, H.; Lesueur, F.; Ahmed, S.; Barbosa-Morais, N.L.; Tyrer, J.P.; Ponder, B.A.J.; Easton, D.F.; Pharoah, P.D.P.; Dunning, A.M.; et al. Association of Single-Nucleotide Polymorphisms in the Cell Cycle Genes with Breast Cancer in the British Population. Carcinogenesis 2008, 29, 333–341. [Google Scholar] [CrossRef]
- Campa, D.; Pastore, M.; Gentiluomo, M.; Talar-Wojnarowska, R.; Kupcinskas, J.; Malecka-Panas, E.; Neoptolemos, J.P.; Niesen, W.; Vodicka, P.; Delle Fave, G.; et al. Functional Single Nucleotide Polymorphisms within the Cyclin-Dependent Kinase Inhibitor 2A/2B Region Affect Pancreatic Cancer Risk. Oncotarget 2016, 7, 57011–57020. [Google Scholar] [CrossRef]
- Lesseur, C.; Ferreiro-Iglesias, A.; McKay, J.D.; Bossé, Y.; Johansson, M.; Gaborieau, V.; Landi, M.T.; Christiani, D.C.; Caporaso, N.C.; Bojesen, S.E.; et al. Genome-Wide Association Meta-Analysis Identifies Pleiotropic Risk Loci for Aerodigestive Squamous Cell Cancers. PLoS Genet. 2021, 17, e1009254. [Google Scholar] [CrossRef]
- Almontashiri, N.A.M.; Antoine, D.; Zhou, X.; Vilmundarson, R.O.; Zhang, S.X.; Hao, K.N.; Chen, H.-H.; Stewart, A.F.R. 9p21.3 Coronary Artery Disease Risk Variants Disrupt TEAD Transcription Factor-Dependent Transforming Growth Factor β Regulation of P16 Expression in Human Aortic Smooth Muscle Cells. Circulation 2015, 132, 1969–1978. [Google Scholar] [CrossRef]
- Zhu, B.; Zhu, Y.; Tian, J.; Shen, N.; Li, J.; Lou, J.; Ke, J.; Yang, Y.; Gong, Y.; Gong, J.; et al. A Functional Variant Rs1537373 in 9p21.3 Region Is Associated with Pancreatic Cancer Risk. Mol. Carcinog. 2019, 58, 760–766. [Google Scholar] [CrossRef]
- Al Olama, A.A.; Kote-Jarai, Z.; Berndt, S.I.; Conti, D.V.; Schumacher, F.; Han, Y.; Benlloch, S.; Hazelett, D.J.; Wang, Z.; Saunders, E.; et al. A Meta-Analysis of 87,040 Individuals Identifies 23 New Susceptibility Loci for Prostate Cancer. Nat. Genet. 2014, 46, 1103–1109. [Google Scholar] [CrossRef]
- Stein, M.M.; Thompson, E.E.; Schoettler, N.; Helling, B.A.; Magnaye, K.M.; Stanhope, C.; Igartua, C.; Morin, A.; Washington, C.; Nicolae, D.; et al. A Decade of Research on the 17q12-21 Asthma Locus: Piecing Together the Puzzle. J. Allergy Clin. Immunol. 2018, 142, 749–764.e3. [Google Scholar] [CrossRef]
- Cheng, J.; Cai, M.-Y.; Chen, Y.-N.; Li, Z.-C.; Tang, S.-S.; Yang, X.-L.; Chen, C.; Liu, X.; Xiong, X.-D. Variants in ANRIL Gene Correlated with Its Expression Contribute to Myocardial Infarction Risk. Oncotarget 2017, 8, 12607–12619. [Google Scholar] [CrossRef]
- He, S.; Cao, R.; Mao, Y.; Li, N.; Wang, Y.; Ma, H.; Tian, K. Alternative Splicing of PSMD13 Mediated by Genetic Variants Is Significantly Associated with Endometrial Cancer Risk. J. Gynecol. Oncol. 2023, 34, e40. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qu, L.; Chen, F.; Zhu, X. Propofol Upregulates MiR-320a and Reduces HMGB1 by Downregulating ANRIL to Inhibit PTC Cell Malignant Behaviors. Pathol. Res. Pract. 2020, 216, 152856. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, P.; Wan, T.; Tu, B.; Li, J.; Huang, F. TIGIT/PVR and LncRNA ANRIL Dual-Targetable PAMAM Polymeric Nanoparticles Efficiently Inhibited the Hepatoma Carcinoma by Combination of Immunotherapy and Gene Therapy. J. Drug Target. 2021, 29, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yu, T.; Shang, J.; Xiao, D.; Wang, X. Long Non-Coding RNA CDKN2B-AS1 Facilitates Laryngeal Squamous Cell Cancer Through Regulating MiR-497/CDK6 Pathway. Onco. Targets Ther. 2019, 12, 8853–8862. [Google Scholar] [CrossRef]
- Zhao, B.; Lu, Y.-L.; Yang, Y.; Hu, L.-B.; Bai, Y.; Li, R.-Q.; Zhang, G.-Y.; Li, J.; Bi, C.-W.; Yang, L.-B.; et al. Overexpression of LncRNA ANRIL Promoted the Proliferation and Migration of Prostate Cancer Cells via Regulating Let-7a/TGF-Β1/Smad Signaling Pathway. Cancer Biomark. 2018, 21, 613–620. [Google Scholar] [CrossRef]
- Du, Z.; Zhang, F.; Liu, L.; Shen, H.; Liu, T.; Jin, J.; Yu, N.; Wan, Z.; Wang, H.; Hu, X.; et al. LncRNA ANRIL Promotes HR Repair through Regulating PARP1 Expression by Sponging MiR-7-5p in Lung Cancer. BMC Cancer 2023, 23, 130. [Google Scholar] [CrossRef]
- Ma, M.-L.; Zhang, H.-Y.; Zhang, S.-Y.; Yi, X.-L. LncRNA CDKN2B-AS1 Sponges MiR-28-5p to Regulate Proliferation and Inhibit Apoptosis in Colorectal Cancer. Oncol. Rep. 2021, 46, 213. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Cheng, Z.; Dai, L.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA ANRIL Inhibits the Development of Cisplatin Resistance by Upregulating MiR-98 in Lung Cancer Cells. Oncol. Rep. 2020, 44, 1025–1036. [Google Scholar] [CrossRef]
- Hang, C.; Zhang, Y.; Qu, Y.; Wang, X. LncRNA ANRIL Represses Proliferation of Oral Squamous Cell Carcinoma Cells through Targeting MiR-99a. Minerva Med. 2022, 113, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, T.; Han, X.; Yuan, H. Knockdown of LncRNA ANRIL Suppresses Cell Proliferation, Metastasis, and Invasion via Regulating MiR-122-5p Expression in Hepatocellular Carcinoma. J. Cancer Res. Clin. Oncol. 2018, 144, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Ning, M.; Liu, Q.; Ding, X.; Wang, Y.; Liu, Q. Knockdown of Long Non-Coding RNA CDKN2B-AS1 Suppresses the Progression of Breast Cancer by MiR-122-5p/STK39 Axis. Bioengineered 2021, 12, 5125–5137. [Google Scholar] [CrossRef]
- Chai, L.; Yuan, Y.; Chen, C.; Zhou, J.; Wu, Y. The Role of Long Non-Coding RNA ANRIL in the Carcinogenesis of Oral Cancer by Targeting MiR-125a. Biomed. Pharmacother. 2018, 103, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Jiang, H.; Jiang, X. Downregulation of LncRNA ANRIL Inhibits Proliferation, Induces Apoptosis, and Enhances Radiosensitivity in Nasopharyngeal Carcinoma Cells through Regulating MiR-125a. Cancer Biol. Ther. 2017, 18, 331–338. [Google Scholar] [CrossRef]
- Yin, Y.; Yang, W.; Zhang, L.; Liu, K.; Luo, Z. Long Non-Coding RNA ANRIL and Its Target MicroRNAs (MicroRNA-34a, MicroRNA-125a and MicroRNA-186) Relate to Risk Stratification and Prognosis in Multiple Myeloma. Hematology 2021, 26, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kong, S.; Xu, A. LncRNA ANRIL Suppresses Proliferation and Promotes Apoptosis of Ovarian Cancer Cells by Regulating MiR-125a-3p/MAPK Signaling Pathway. Minerva Med. 2022, 113, 581–582. [Google Scholar] [CrossRef]
- Li, C.; Zhai, W.; Wan, L.; Li, J.; Huang, A.; Xing, S.; Fan, K. MicroRNA-125a Attenuates the Chemoresistance against Ubenimex in Non-Small Cell Lung Carcinoma via Targeting the Aminopeptidase N Signaling Pathway. J. Cell. Biochem. 2020, 121, 1716–1727. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Li, G.; Hu, J.; Liu, X.; Lin, L. LncRNA ANRIL Promotes Cell Growth, Migration and Invasion of Hepatocellular Carcinoma Cells via Sponging MiR-144. Anticancer Drugs 2019, 30, 1013–1021. [Google Scholar] [CrossRef]
- Wang, L.; Bi, R.; Li, L.; Zhou, K.; Yin, H. LncRNA ANRIL Aggravates the Chemoresistance of Pancreatic Cancer Cells to Gemcitabine by Targeting Inhibition of MiR-181a and Targeting HMGB1-Induced Autophagy. Aging 2021, 13, 19272–19281. [Google Scholar] [CrossRef]
- Sun, C.; Shen, C.; Zhang, Y.; Hu, C. LncRNA ANRIL Negatively Regulated Chitooligosaccharide-Induced Radiosensitivity in Colon Cancer Cells by Sponging MiR-181a-5p. Adv. Clin. Exp. Med. 2021, 30, 55–65. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Q.; Li, S.; Jiang, S.; Cui, J.; Dang, G. Interference of the Long Noncoding RNA CDKN2B-AS1 Upregulates MiR-181a-5p/TGFβI Axis to Restrain the Metastasis and Promote Apoptosis and Senescence of Cervical Cancer Cells. Cancer Med. 2019, 8, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Khasanov, D.; Gareev, I.; Valitov, E.; Sokhatskii, A.; Wang, C.; Pavlov, V.; Khasanova, G.; Ahmad, A. Differential Non-Coding RNAs Expression Profiles of Invasive and Non-Invasive Pituitary Adenomas. Non-Coding RNA Res. 2021, 6, 115–122. [Google Scholar] [CrossRef]
- Wang, G.; Xu, G.; Wang, W. Long Noncoding RNA CDKN2B-AS1 Facilitates Lung Cancer Development Through Regulating MiR-378b/NR2C2. Onco. Targets Ther. 2020, 13, 10641–10649. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Sun, H.; Liang, H.; Wang, Y.; Lu, M.; Guo, Z.; Lv, Z.; Ren, W. Evaluation of LncRNA ANRIL Potential in Hepatic Cancer Progression. J. Environ. Pathol. Toxicol. Oncol. 2019, 38, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.; Cao, H. Long Non-Coding RNA CDKN2B-AS1 Promotes Osteosarcoma by Increasing the Expression of MAP3K3 via Sponging MiR-4458. Vitr. Cell. Dev. Biol. Anim. 2020, 56, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Feng, L.; Liu, P.; Duan, W. ANRIL Promotes Chemoresistance via Disturbing Expression of ABCC1 by Regulating the Expression of Let-7a in Colorectal Cancer. Biosci. Rep. 2018, 38, BSR20180620. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long Non-Coding RNA ANRIL Is Required for the PRC2 Recruitment to and Silencing of P15(INK4B) Tumor Suppressor Gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Muniz, L.; Lazorthes, S.; Delmas, M.; Ouvrard, J.; Aguirrebengoa, M.; Trouche, D.; Nicolas, E. Circular ANRIL Isoforms Switch from Repressors to Activators of P15/CDKN2B Expression during RAF1 Oncogene-Induced Senescence. RNA Biol. 2021, 18, 404–420. [Google Scholar] [CrossRef]
- El Messaoudi-Aubert, S.; Nicholls, J.; Maertens, G.N.; Brookes, S.; Bernstein, E.; Peters, G. Role for the MOV10 RNA Helicase in Polycomb-Mediated Repression of the INK4a Tumor Suppressor. Nat. Struct. Mol. Biol. 2010, 17, 862–868. [Google Scholar] [CrossRef]
- Maenner, S.; Müller, M.; Fröhlich, J.; Langer, D.; Becker, P.B. ATP-Dependent RoX RNA Remodeling by the Helicase Maleless Enables Specific Association of MSL Proteins. Mol. Cell. 2013, 51, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.; Li, X.; Zhao, S.; Cui, S.; Tu, J. Long Non-Coding RNA CDKN2B-AS1 Contributes to Atherosclerotic Plaque Formation by Forming RNA-DNA Triplex in the CDKN2B Promoter. EBioMedicine 2020, 55, 102694. [Google Scholar] [CrossRef]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and Abundance Analysis of Human LncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Lo Sardo, V.; Chubukov, P.; Ferguson, W.; Kumar, A.; Teng, E.L.; Duran, M.; Zhang, L.; Cost, G.; Engler, A.J.; Urnov, F.; et al. Unveiling the Role of the Most Impactful Cardiovascular Risk Locus through Haplotype Editing. Cell 2018, 175, 1796–1810.e20. [Google Scholar] [CrossRef]
- Nie, F.; Sun, M.; Yang, J.; Xie, M.; Xu, T.; Xia, R.; Liu, Y.; Liu, X.; Zhang, E.; Lu, K.; et al. Long Noncoding RNA ANRIL Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef]
- Liu, L.; Ning, S.-B.; Fu, S.; Mao, Y.; Xiao, M.; Guo, B. Effects of LncRNA ANRIL on Proliferation and Apoptosis of Oral Squamous Cell Carcinoma Cells by Regulating TGF-β/Smad Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6194–6201. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Z.; Mao, C.; Zhou, Y.; Yu, L.; Yin, Y.; Wu, S.; Mou, X.; Zhu, Y. ANRIL Inhibits P15(INK4b) through the TGFβ1 Signaling Pathway in Human Esophageal Squamous Cell Carcinoma. Cell. Immunol. 2014, 289, 91–96. [Google Scholar] [CrossRef]
- Zhang, C.; Ge, S.; Gong, W.; Xu, J.; Guo, Z.; Liu, Z.; Gao, X.; Wei, X.; Ge, S. LncRNA ANRIL Acts as a Modular Scaffold of WDR5 and HDAC3 Complexes and Promotes Alteration of the Vascular Smooth Muscle Cell Phenotype. Cell Death Dis. 2020, 11, 435. [Google Scholar] [CrossRef]
- Huang, D.; Bi, C.; Zhao, Q.; Ding, X.; Bian, C.; Wang, H.; Wang, T.; Liu, H. Knockdown Long Non-Coding RNA ANRIL Inhibits Proliferation, Migration and Invasion of HepG2 Cells by down-Regulation of MiR-191. BMC Cancer 2018, 18, 919. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological Organization of Multichromosomal Regions by the Long Intergenic Noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The Matrix Protein HnRNP U Is Required for Chromosomal Localization of Xist RNA. Dev. Cell. 2010, 19, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schäfer, A.; Grummt, I.; Niehrs, C. GADD45A Binds R-Loops and Recruits TET1 to CpG Island Promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F.; Aune, T.M. Divergent LncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef]
- Gonzalez, I.; Munita, R.; Agirre, E.; Dittmer, T.A.; Gysling, K.; Misteli, T.; Luco, R.F. A LncRNA Regulates Alternative Splicing via Establishment of a Splicing-Specific Chromatin Signature. Nat. Struct. Mol. Biol. 2015, 22, 370–376. [Google Scholar] [CrossRef]
- Amann, T.; Bataille, F.; Spruss, T.; Dettmer, K.; Wild, P.; Liedtke, C.; Mühlbauer, M.; Kiefer, P.; Oefner, P.J.; Trautwein, C.; et al. Reduced Expression of Fibroblast Growth Factor Receptor 2IIIb in Hepatocellular Carcinoma Induces a More Aggressive Growth. Am. J. Pathol. 2010, 176, 1433–1442. [Google Scholar] [CrossRef]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef]
- Urbanski, L.M.; Leclair, N.; Anczuków, O. Alternative-Splicing Defects in Cancer: Splicing Regulators and Their Downstream Targets, Guiding the Way to Novel Cancer Therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476. [Google Scholar] [CrossRef]
- Eperon, I.C.; Makarova, O.V.; Mayeda, A.; Munroe, S.H.; Cáceres, J.F.; Hayward, D.G.; Krainer, A.R. Selection of Alternative 5’ Splice Sites: Role of U1 SnRNP and Models for the Antagonistic Effects of SF2/ASF and HnRNP A1. Mol. Cell. Biol. 2000, 20, 8303–8318. [Google Scholar] [CrossRef]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thåström, A.; Field, Y.; Moore, I.K.; Wang, J.-P.Z.; Widom, J. A Genomic Code for Nucleosome Positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef]
- Schwartz, S.; Meshorer, E.; Ast, G. Chromatin Organization Marks Exon-Intron Structure. Nat. Struct. Mol. Biol. 2009, 16, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Enroth, S.; Rada-Iglesias, A.; Wadelius, C.; Komorowski, J. Nucleosomes Are Well Positioned in Exons and Carry Characteristic Histone Modifications. Genome Res. 2009, 19, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Tilgner, H.; Nikolaou, C.; Althammer, S.; Sammeth, M.; Beato, M.; Valcárcel, J.; Guigó, R. Nucleosome Positioning as a Determinant of Exon Recognition. Nat. Struct. Mol. Biol. 2009, 16, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Saint-André, V.; Batsché, E.; Rachez, C.; Muchardt, C. Histone H3 Lysine 9 Trimethylation and HP1γ Favor Inclusion of Alternative Exons. Nat. Struct. Mol. Biol. 2011, 18, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Spies, N.; Nielsen, C.B.; Padgett, R.A.; Burge, C.B. Biased Chromatin Signatures around Polyadenylation Sites and Exons. Mol. Cell. 2009, 36, 245–254. [Google Scholar] [CrossRef]
- Agirre, E.; Oldfield, A.J.; Bellora, N.; Segelle, A.; Luco, R.F. Splicing-Associated Chromatin Signatures: A Combinatorial and Position-Dependent Role for Histone Marks in Splicing Definition. Nat. Commun. 2021, 12, 682. [Google Scholar] [CrossRef] [PubMed]
- Hon, G.; Wang, W.; Ren, B. Discovery and Annotation of Functional Chromatin Signatures in the Human Genome. PLoS Comput. Biol. 2009, 5, e1000566. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long Non-Coding RNAs: From Disease Code to Drug Role. Acta. Pharm. Sin. B 2021, 11, 340–354. [Google Scholar] [CrossRef]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA Structures with Small Molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).