Circulating Biomarkers Associated with the Diagnosis and Prognosis of B-Cell Progenitor Acute Lymphoblastic Leukemia
Abstract
:Simple Summary
Abstract
1. Introduction
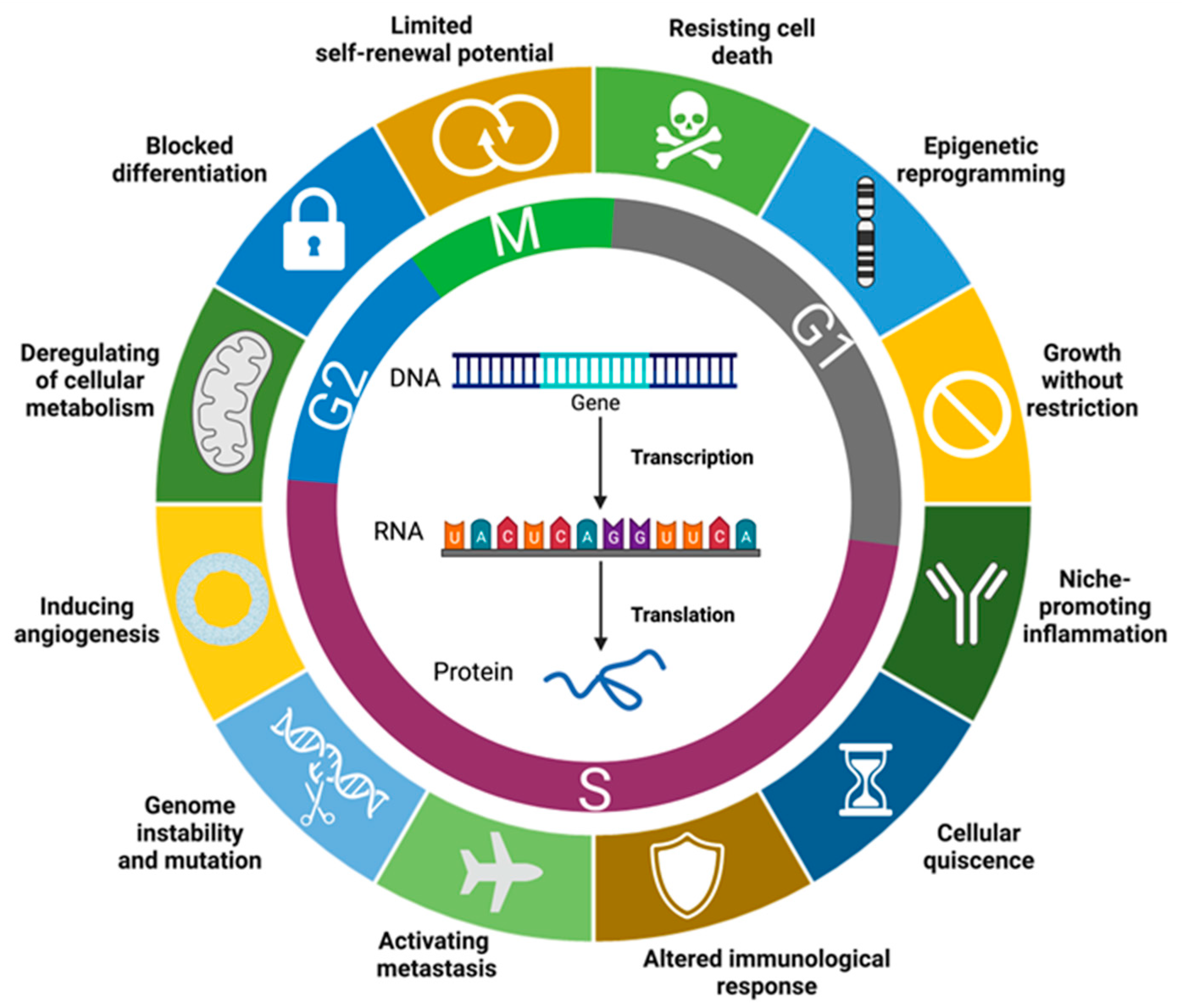
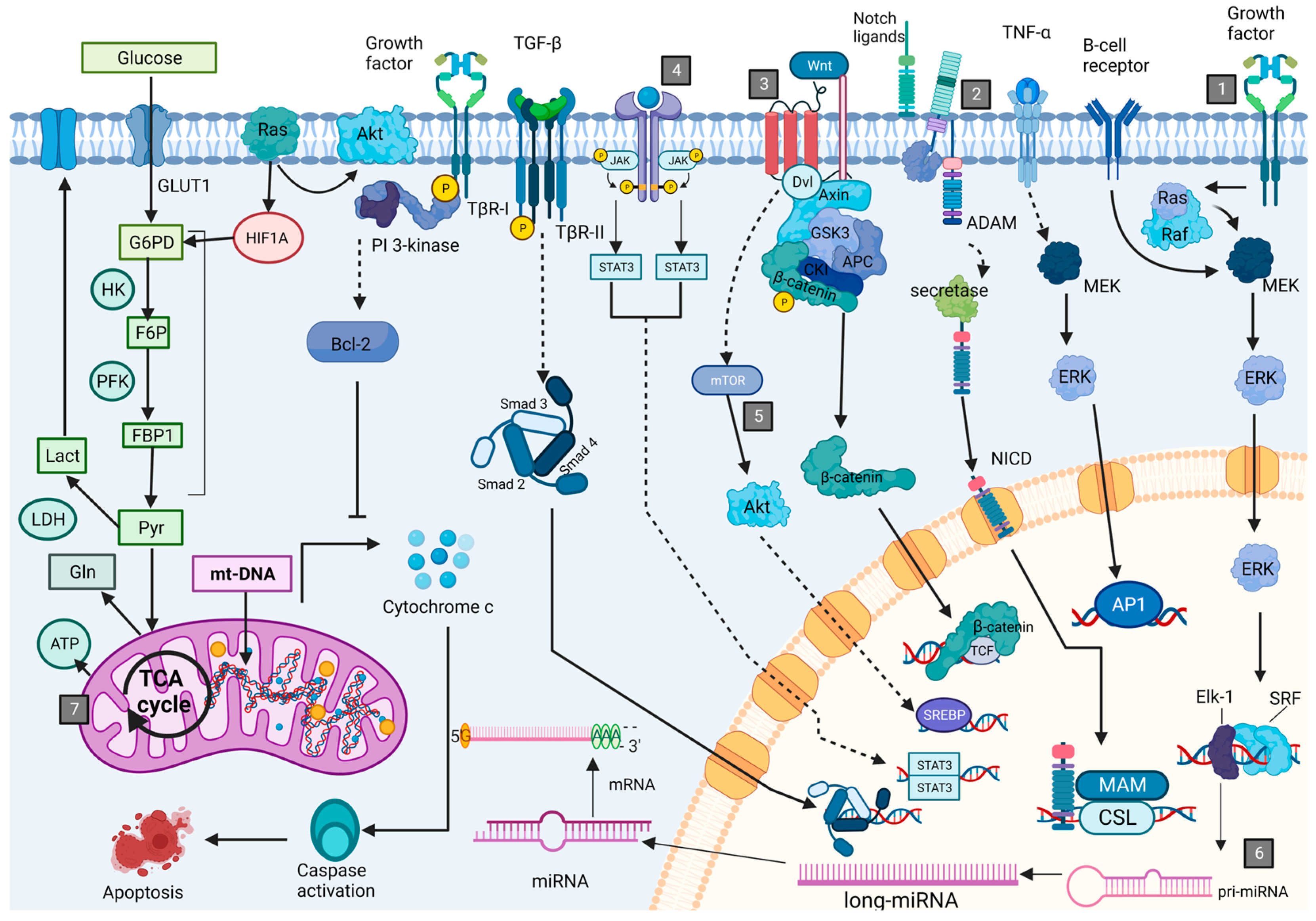
2. Leukemogenesis and Associated Cell Processes
General Description of Main Type of Molecules with Potential as Biomarkers for BCP-ALL
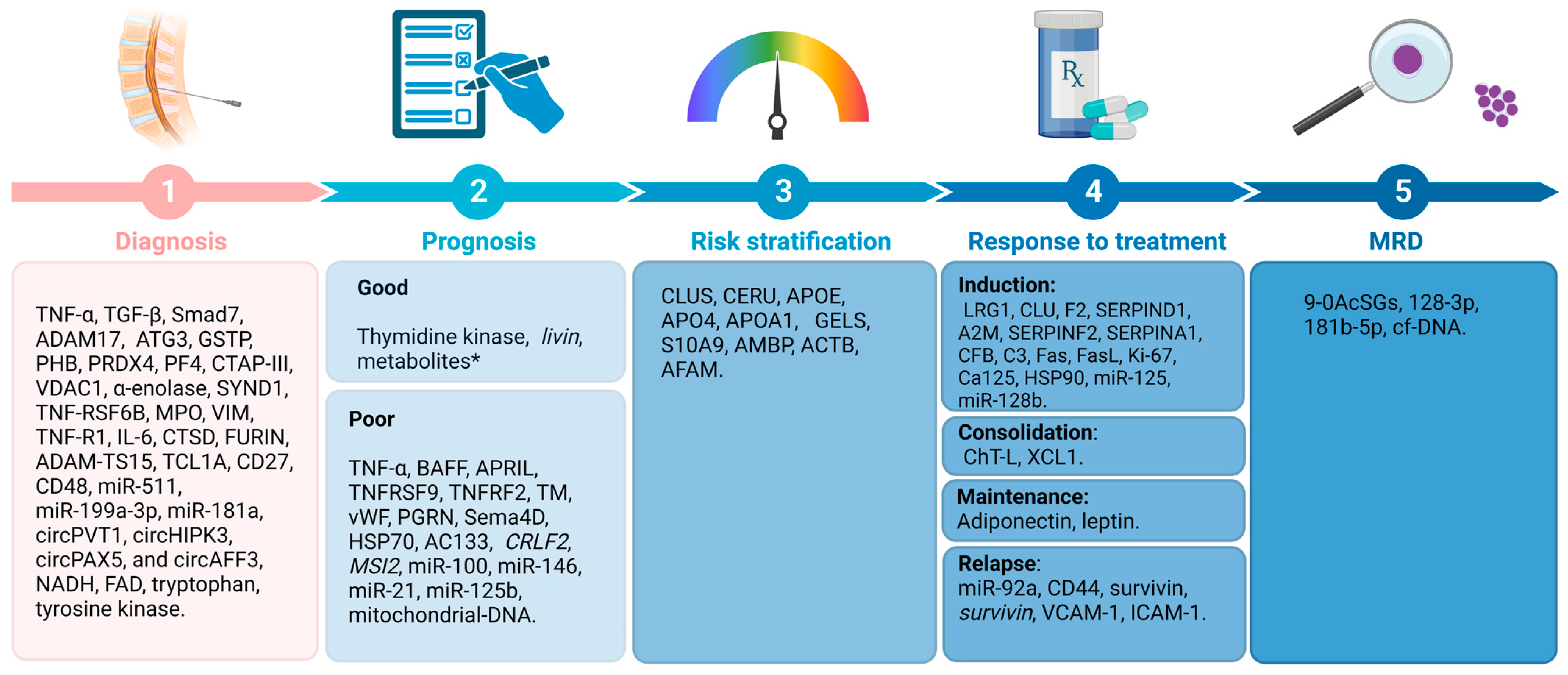
3. Diagnostic Biomarkers for BCP-ALL
3.1. Proteins Type BCP-ALL Biomarkers
3.2. RNA Type BCP-ALL Biomarkers
3.3. Metabolites Type BCP-ALL Biomarkers
4. Prognostic Biomarkers for BCP-ALL
4.1. BCP-ALL Biomarkers for Prognosis and Risk Stratification
4.1.1. Proteins
4.1.2. BCP-ALL RNA Biomarkers
- miRNAs
4.1.3. Mitochondrial DNA
4.1.4. Metabolites
4.2. Response to Treatment
4.2.1. Induction
4.2.2. Consolidation
4.2.3. Maintenance
4.2.4. Relapse
4.2.5. MRD
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fujita, T.C.; Sousa-Pereira, N.; Amarante, M.K.; Watanabe, M.A.E. Acute lymphoid leukemia etiopathogenesis. Mol. Biol. Rep. 2021, 48, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.A. Leukemia in Children. Pediatr. Rev. 2019, 40, 319–331. [Google Scholar] [CrossRef]
- Ahmed, M.B.; Shehata, H.H.; Moussa, M.; Ibrahim, T.M. Prognostic significance of survivin and tumor necrosis factor-alpha in adult acute lymphoblastic leukemia. Clin. Biochem. 2012, 45, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lv, Y.Q.; Liu, Y.F.; Du, X.J.; Li, B. Differential protein analysis of lymphocytes between children with acute lymphoblastic leukemia and healthy children. Leuk. Lymphoma 2013, 54, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S. WHO Classification of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2017. [Google Scholar]
- Fayed, D.; Donia, T.; El-Shanshory, M.; Ali, E.M.M.; Mohamed, T.M. Evaluation of MicroRNA92, MicroRNA638 in Acute Lymphoblastic Leukemia of Egyptian Children. Asian Pac. J. Cancer Prev. 2021, 22, 1567–1572. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, J.; Wu, P.; Feng, K.; Li, J.; Xie, Z.; Xue, P.; Cai, T.; Cui, Z.; Chen, X.; et al. Discovery and identification of potential biomarkers of pediatric acute lymphoblastic leukemia. Proteome Sci. 2009, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Masilamani, V.; Devanesan, S.; AlSalhi, M.S.; AlQahtany, F.S.; Farhat, K.H. Fluorescence spectral detection of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML): A novel photodiagnosis strategy. Photodiag. Photodyn. Ther. 2020, 29, 101634. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Shahid, W.; Shaheen, J.; Akhtar, M.W.; Sadaf, S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 22783. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.M.; Refaat, L.A.; Ismail, G.N.; Abdellateif, M.; Fadel, S.A.; AbdelAziz, R.S. Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 2020, 25, 405–413. [Google Scholar] [CrossRef]
- Brown, P.; Inaba, H.; Annesley, C.; Beck, J.; Colace, S.; Dallas, M.; DeSantes, K.; Kelly, K.; Kitko, C.; Lacayo, N.; et al. Pediatric Acute Lymphoblastic Leukemia, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2020, 18, 81–112. [Google Scholar] [CrossRef]
- Brown, P.A.; Shah, B.; Advani, A.; Aoun, P.; Boyer, M.W.; Burke, P.W.; DeAngelo, D.J.; Dinner, S.; Fathi, A.T.; Gauthier, J.; et al. Acute Lymphoblastic Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 1079–1109. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Yu, R.; Yang, S.; Liu, Y.; Zhu, Z. Identification and validation of serum autoantibodies in children with B-cell acute lymphoblastic leukemia by serological proteome analysis. Proteome Sci. 2022, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumour Biol. 2016, 37, 10571–10576. [Google Scholar] [CrossRef]
- Nabhan, M.; Louka, M.L.; Khairy, E.; Tash, F.; Ali-Labib, R.; El-Habashy, S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene 2017, 628, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, H.; Sun, X.; Sun, C.; Ren, L. Biomarker identification and pathway analysis by serum metabolomics of childhood acute lymphoblastic leukemia. Clin. Chim. Acta 2014, 436, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Carceles-Alvarez, A.; Ortega-Garcia, J.A.; Lopez-Hernandez, F.A.; Fuster-Soler, J.L.; Ramis, R.; Kloosterman, N.; Castillo, L.; Sanchez-Solis, M.; Claudio, L.; Ferris-Tortajada, J. Secondhand smoke: A new and modifiable prognostic factor in childhood acute lymphoblastic leukemias. Environ. Res. 2019, 178, 108689. [Google Scholar] [CrossRef]
- Vrooman, L.M.; Silverman, L.B. Treatment of Childhood Acute Lymphoblastic Leukemia: Prognostic Factors and Clinical Advances. Curr. Hematol. Malig. Rep. 2016, 11, 385–394. [Google Scholar] [CrossRef]
- Aly, R.M.; Yousef, A.B. Prognostic significance of lymphoid enhancer-binding factor-1 expression in egyptian adult B-acute lymphocytic leukemia patients. Turk. J. Haematol. 2015, 32, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Aguirre, C.H.; Flores-Jimenez, J.A.; Alatorre-Ricardo, J.; Cantu-Rodriguez, O.G.; Rosas-Taraco, A.; Salazar-Riojas, R.; Jaime-Perez, J.C.; Sanchez-Cardenas, M.; Lopez-Silva, L.; Martinez-Castilla, A.M.; et al. The prognostic significance of serum XCL1 concentration in patients with acute lymphoblastic leukemia: A pilot study. Ann. Hematol. 2017, 96, 2015–2024. [Google Scholar] [CrossRef]
- Kruse, A.; Abdel-Azim, N.; Kim, H.N.; Ruan, Y.; Phan, V.; Ogana, H.; Wang, W.; Lee, R.; Gang, E.J.; Khazal, S.; et al. Minimal Residual Disease Detection in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, C.; Han, L.; Jain, N.; Roberts, K.G.; Ma, H.; Cai, T.; Cavazos, A.; Tabe, Y.; Jacamo, R.O.; et al. Inhibition of mTORC1/C2 signaling improves anti-leukemia efficacy of JAK/STAT blockade in CRLF2 rearranged and/or JAK driven Philadelphia chromosome-like acute B-cell lymphoblastic leukemia. Oncotarget 2018, 9, 8027–8041. [Google Scholar] [CrossRef]
- Cante-Barrett, K.; Spijkers-Hagelstein, J.A.; Buijs-Gladdines, J.G.; Uitdehaag, J.C.; Smits, W.K.; van der Zwet, J.; Buijsman, R.C.; Zaman, G.J.; Pieters, R.; Meijerink, J.P. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016, 30, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Tovar, C.F.; Zeron, H.M.; Romero, M.D.; Sanchez, Y.V.; Romero, I.T. Glycogen Synthase Kinase-3beta (GSK-3beta) and Nuclear Factor Kappa-B (NFKB) in Childhood Acute Lymphoblastic Leukemia. Adv. Clin. Exp. Med. 2016, 25, 1139–1147. [Google Scholar] [CrossRef]
- Vrooman, L.M.; Silverman, L.B. Childhood acute lymphoblastic leukemia: Update on prognostic factors. Curr. Opin. Pediatr. 2009, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Aly, R.M.; Ghazy, H.F. Prognostic significance of MSI2 predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int. J. Lab. Hematol. 2015, 37, 272–278. [Google Scholar] [CrossRef]
- Aguirre-Guillen, W.A.; Angeles-Floriano, T.; Lopez-Martinez, B.; Reyes-Morales, H.; Zlotnik, A.; Valle-Rios, R. Omics techniques and biobanks to find new biomarkers for the early detection of acute lymphoblastic leukemia in middle-income countries: A perspective from Mexico. Bol. Med. Hosp. Infant. Mex. 2017, 74, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Liu, H.; Xu, J.; Cen, X.; Wang, Q.; Xu, W.; Wang, W.; Qiu, Z.; Ou, J.; Dong, Y.; et al. Expression of human Krüppel-like factor 3 in peripheral blood as a promising biomarker for acute leukemia. Cancer Med. 2020, 9, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Morad, H.M.; Abou-Elzahab, M.M.; Aref, S.; El-Sokkary, A.M.A. Diagnostic Value of (1)H NMR-Based Metabolomics in Acute Lymphoblastic Leukemia, Acute Myeloid Leukemia, and Breast Cancer. ACS Omega 2022, 7, 8128–8140. [Google Scholar] [CrossRef]
- Hussan, S.S.; Maqsood, N.; Wang, Q.; Tao, S.; Sadaf, S. A panel of epigenetically dysregulated Wnt signaling pathway genes for non-invasive diagnosis of pediatric acute lymphoblastic leukemia. Cancer Biomark. 2021, 32, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hu, J.; Hu, G. Biomarker Studies in Early Detection and Prognosis of Breast Cancer. Adv. Exp. Med. Biol. 2017, 1026, 27–39. [Google Scholar] [CrossRef]
- Damanti, C.C.; Gaffo, E.; Lovisa, F.; Garbin, A.; Di Battista, P.; Gallingani, I.; Tosato, A.; Pillon, M.; Carraro, E.; Mascarin, M.; et al. MiR-26a-5p as a Reference to Normalize MicroRNA qRT-PCR Levels in Plasma Exosomes of Pediatric Hematological Malignancies. Cells 2021, 10, 101. [Google Scholar] [CrossRef]
- Lee, R.D.; Munro, S.A.; Knutson, T.P.; LaRue, R.S.; Heltemes-Harris, L.M.; Farrar, M.A. Single-cell analysis identifies dynamic gene expression networks that govern B cell development and transformation. Nat. Commun. 2021, 12, 6843. [Google Scholar] [CrossRef]
- Simioni, C.; Conti, I.; Varano, G.; Brenna, C.; Costanzi, E.; Neri, L.M. The Complexity of the Tumor Microenvironment and Its Role in Acute Lymphoblastic Leukemia: Implications for Therapies. Front. Oncol. 2021, 11, 673506. [Google Scholar] [CrossRef] [PubMed]
- Tsiftsoglou, A.S.; Bonovolias, I.D.; Tsiftsoglou, S.A. Multilevel targeting of hematopoietic stem cell self-renewal, differentiation and apoptosis for leukemia therapy. Pharmacol. Ther. 2009, 122, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J.E. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef]
- Koschade, S.E.; Brandts, C.H. Selective Autophagy in Normal and Malignant Hematopoiesis. J. Mol. Biol. 2020, 432, 261–282. [Google Scholar] [CrossRef]
- Mizia-Malarz, A.; Sobol-Milejska, G. Assessment of Angiogenesis in Children with Acute Lymphoblastic Leukemia Based on Serum Vascular Endothelial Growth Factor Assay. Indian J. Med. Paediatr. Oncol. 2017, 38, 321–325. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, B.; Sun, J.; He, Q.; Zhao, Y. Knockdown of GPSM1 Inhibits the Proliferation and Promotes the Apoptosis of B-Cell Acute Lymphoblastic Leukemia Cells by Suppressing the ADCY6-RAPGEF3-JNK Signaling Pathway. Pathol. Oncol. Res. 2021, 27, 643376. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Zhu, X.; Radich, J.P.; Ao, P. From molecular interaction to acute promyelocytic leukemia: Calculating leukemogenesis and remission from endogenous molecular-cellular network. Sci. Rep. 2016, 6, 24307. [Google Scholar] [CrossRef]
- Alexander, S. Clinically defining and managing high-risk pediatric patients with acute lymphoblastic leukemia. Hematol. Am. Soc. Hematol. Educ. Program. 2014, 2014, 181–189. [Google Scholar] [CrossRef]
- Liang, T.; Wang, N.; Li, W.; Li, A.; Wang, J.; Cui, J.; Liu, N.; Li, Y.; Li, L.; Yang, G.; et al. Identification of complement C3f-desArg and its derivative for acute leukemia diagnosis and minimal residual disease assessment. Proteomics 2010, 10, 90–98. [Google Scholar] [CrossRef]
- Volm, M.; Hecker, S.; Sauerbrey, A.; Mattern, J. Predictive value of statin, a G0-associated cell cycle protein, in childhood acute lymphoblastic leukemia. Int. J. Cancer 1995, 64, 166–170. [Google Scholar] [CrossRef]
- Weiss, A.; Saw, S.; Aiken, A.; Aliar, K.; Shao, Y.W.; Fang, H.; Narala, S.; Shetty, R.; Waterhouse, P.D.; Khokha, R. Abnormal B-cell development in TIMP-deficient bone marrow. Blood Adv. 2021, 5, 3960–3974. [Google Scholar] [CrossRef]
- Cristea, I.M.; Gaskell, S.J.; Whetton, A.D. Proteomics techniques and their application to hematology. Blood 2004, 103, 3624–3634. [Google Scholar] [CrossRef] [PubMed]
- Miljkovic-Licina, M.; Arraud, N.; Zahra, A.D.; Ropraz, P.; Matthes, T. Quantification and Phenotypic Characterization of Extracellular Vesicles from Patients with Acute Myeloid and B-Cell Lymphoblastic Leukemia. Cancers 2021, 14, 56. [Google Scholar] [CrossRef]
- Gao, Y.J.; He, Y.J.; Yang, Z.L.; Shao, H.Y.; Zuo, Y.; Bai, Y.; Chen, H.; Chen, X.C.; Qin, F.X.; Tan, S.; et al. Increased integrity of circulating cell-free DNA in plasma of patients with acute leukemia. Clin. Chem. Lab. Med. 2010, 48, 1651–1656. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef]
- Staal, F.J.; Famili, F.; Garcia Perez, L.; Pike-Overzet, K. Aberrant Wnt Signaling in Leukemia. Cancers 2016, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Garis, M.; Garrett-Sinha, L.A. Notch Signaling in B Cell Immune Responses. Front. Immunol. 2020, 11, 609324. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Philips, R.L.; Wang, Y.; Cheon, H.; Kanno, Y.; Gadina, M.; Sartorelli, V.; Horvath, C.M.; Darnell, J.E., Jr.; Stark, G.R.; O’Shea, J.J. The JAK-STAT pathway at 30: Much learned, much more to do. Cell 2022, 185, 3857–3876. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.W. Biomarker classification, validation, and what to look for in 2017 and beyond. BJU Int 2017, 119, 812–814. [Google Scholar] [CrossRef] [PubMed]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, Endpoints, and Other Tools) Resource; FDA-NIH Biomarker Working Group: Silver Spring, MD, USA, 2016. [Google Scholar]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; International Agency for Research on Cancer: Lyon, France, 2008; Volume 2. [Google Scholar]
- Cui, J.W.; Wang, J.; He, K.; Jin, B.F.; Wang, H.X.; Li, W.; Kang, L.H.; Hu, M.R.; Li, H.Y.; Yu, M.; et al. Proteomic analysis of human acute leukemia cells: Insight into their classification. Clin. Cancer Res. 2004, 10, 6887–6896. [Google Scholar] [CrossRef]
- Malcles, M.H.; Wang, H.W.; Koumi, A.; Tsai, Y.H.; Yu, M.; Godfrey, A.; Boshoff, C. Characterisation of the anti-apoptotic function of survivin-DeltaEx3 during TNFalpha-mediated cell death. Br. J. Cancer 2007, 96, 1659–1666. [Google Scholar] [CrossRef]
- Aref, S.; Salama, O.; Shamaa, S.; El-Refaie, M.; Mourkos, H. Angiogenesis factor pattern differs in acute lymphoblastic leukemia and chronic lymphocytic leukemia. Hematology 2007, 12, 319–324. [Google Scholar] [CrossRef]
- Potapnev, M.P.; Petyovka, N.V.; Belevtsev, M.V.; Savitskiy, V.P.; Migal, N.V. Plasma level of tumor necrosis factor-alpha (TNF-alpha) correlates with leukocytosis and biological features of leukemic cells, but not treatment response of children with acute lymphoblastic leukemia. Leuk. Lymphoma 2003, 44, 1077–1079. [Google Scholar] [CrossRef]
- Zakhary, N.I.; Boshra, S.A.; El-Sawalhi, M.M.; Fahim, A.T.; Ebeid, E.N. Insulin-like growth factor system in Egyptian children with acute lymphoblastic leukemia. Genet. Test. Mol. Biomark. 2012, 16, 1067–1072. [Google Scholar] [CrossRef]
- Pal, S.; Bandyopadhyay, S.; Chatterjee, M.; Bhattacharya, D.K.; Minto, L.; Hall, A.G.; Mandal, C. Antibodies against 9-O-acetylated sialoglycans: A potent marker to monitor clinical status in childhood acute lymphoblastic leukemia. Clin. Biochem. 2004, 37, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, M.M. Clinical significance of serum p53 and epidermal growth factor receptor in patients with acute leukemia. Asian Pac. J. Cancer Prev. 2013, 14, 4295–4299. [Google Scholar] [CrossRef] [PubMed]
- Pane, F.; Savoia, M.; Fortunato, G.; Camera, A.; Rotoli, B.; Salvatore, F.; Sacchetti, L. Serum pseudouridine in the diagnosis of acute leukaemias and as a novel prognostic indicator in acute lymphoblastic leukaemia. Clin. Biochem. 1993, 26, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Xing, C.; Li, R.; Cheng, Z.; Deng, M.; Luo, Y.; Li, H.; Zhang, G.; Sheng, Y.; Peng, H.; et al. Proteomic profiling of plasma exosomes from patients with B-cell acute lymphoblastic leukemia. Sci. Rep. 2022, 12, 11975. [Google Scholar] [CrossRef]
- Elgendi, H.M.; Mekawy, M.A.; Abdel Wahab, S.E.; Tawfik, L.M.; Ismail, E.A.; Adly, A.A. AC133 expression in egyptian children with acute leukemia: Impact on treatment response and disease outcome. J. Pediatr. Hematol. Oncol. 2010, 32, 286–293. [Google Scholar] [CrossRef]
- Mohammadi, M.; Amirmahani, F.; Goharrizi, K.J.; Pakzad, R.; Dolat, H. Evaluating the expression level of Survivin gene in different groups of B-cell acute lymphoblastic leukemia patients of Iran. Mol. Biol. Rep. 2019, 46, 2679–2684. [Google Scholar] [CrossRef]
- Alkhouly, N.; Shehata, I.; Ahmed, M.B.; Shehata, H.; Hassan, S.; Ibrahim, T. HLA-G expression in acute lymphoblastic leukemia: A significant prognostic tumor biomarker. Med. Oncol. 2013, 30, 460. [Google Scholar] [CrossRef]
- Swellam, M.; Hashim, M.; Mahmoud, M.S.; Ramadan, A.; Hassan, N.M. Aberrant Expression of Some Circulating miRNAs in Childhood Acute Lymphoblastic Leukemia. Biochem. Genet. 2018, 56, 283–294. [Google Scholar] [CrossRef]
- Luna-Aguirre, C.M.; de la Luz Martinez-Fierro, M.; Mar-Aguilar, F.; Garza-Veloz, I.; Trevino-Alvarado, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark. 2015, 15, 299–310. [Google Scholar] [CrossRef]
- El-maadawy, E.A.; Bakry, R.M.; Moussa, M.M.; El-Naby, S.; Talaat, R.M.J.C. Alteration in miRNAs expression in paediatric acute lymphocyticleukaemia: Insight into patients’ therapeutic response. Pharmacol. Pharm. 2021, 48, 35–43. [Google Scholar] [CrossRef]
- Ohyashiki, J.H.; Umezu, T.; Kobayashi, C.; Hamamura, R.S.; Tanaka, M.; Kuroda, M.; Ohyashiki, K. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: In vivo assessment of cell to plasma ratio of miR-92a. BMC Res. Notes 2010, 3, 347. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Elshimy, A.A.; Hassan, S.S.; Matbouly, S.; Safwat, G.; Zannoun, M.; Riad, R.A. Dysregulation of miR-125b predicts poor response to therapy in pediatric acute lymphoblastic leukemia. J. Cell Biochem. 2018, 120, 7428–7438. [Google Scholar] [CrossRef]
- Nemes, K.; Csoka, M.; Nagy, N.; Mark, A.; Varadi, Z.; Danko, T.; Kovacs, G.; Kopper, L.; Sebestyen, A. Expression of certain leukemia/lymphoma related microRNAs and its correlation with prognosis in childhood acute lymphoblastic leukemia. Pathol. Oncol. Res. 2015, 21, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Abu Sabaa, A.; Shen, Q.; Lennmyr, E.B.; Enblad, A.P.; Gammelgard, G.; Molin, D.; Hein, A.; Freyhult, E.; Kamali-Moghaddam, M.; Hoglund, M.; et al. Plasma protein biomarker profiling reveals major differences between acute leukaemia, lymphoma patients and controls. N Biotechnol. 2022, 71, 21–29. [Google Scholar] [CrossRef]
- Wik, L.; Nordberg, N.; Broberg, J.; Bjorkesten, J.; Assarsson, E.; Henriksson, S.; Grundberg, I.; Pettersson, E.; Westerberg, C.; Liljeroth, E.; et al. Proximity Extension Assay in Combination with Next-Generation Sequencing for High-throughput Proteome-wide Analysis. Mol. Cell. Proteom. MCP 2021, 20, 100168. [Google Scholar] [CrossRef]
- Limijadi, E.K.S.; Budiwijono, I.; Samsuria, I.K.; Adhipireno, P.; Devi, W.R. Coagulation and Fibrinolysis Profiles of Acute Myeloblastic Leukemia: Preliminary Assessment of Hypercoagulability. Eur. J. Mol. Clin. Med. 2021, 8, 607–615. [Google Scholar]
- Seftalioglu, A.; Karakus, S. Syndecan-1/CD138 expression in normal myeloid, acute lymphoblastic and myeloblastic leukemia cells. Acta Histochem. 2003, 105, 213–221. [Google Scholar] [CrossRef]
- Lenting, P.J.; Christophe, O.D.; Denis, C.V. von Willebrand factor biosynthesis, secretion, and clearance: Connecting the far ends. Blood 2015, 125, 2019–2028. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef]
- Maimaitiyiming, Y.; Ye, L.; Yang, T.; Yu, W.; Naranmandura, H. Linear and Circular Long Non-Coding RNAs in Acute Lymphoblastic Leukemia: From Pathogenesis to Classification and Treatment. Int. J. Mol. Sci. 2022, 23, 4442. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, J.; Shao, T.; Zhang, Y.; Chen, H.; Li, X. RNA Function Prediction. Methods Mol. Biol. 2017, 1654, 17–28. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Perez de Acha, O.; Rossi, M.; Gorospe, M. Circular RNAs in Blood Malignancies. Front. Mol. Biosci. 2020, 7, 109. [Google Scholar] [CrossRef]
- Gaffo, E.; Boldrin, E.; Dal Molin, A.; Bresolin, S.; Bonizzato, A.; Trentin, L.; Frasson, C.; Debatin, K.-M.; Meyer, L.H.; te Kronnie, G.; et al. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci. Rep. 2019, 9, 14670. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Siddiqui, A.J.; Shamsi, T.; Choudhary, M.I.; Rahman, A.U. Serum metabonomics of acute leukemia using nuclear magnetic resonance spectroscopy. Sci. Rep. 2016, 6, 30693. [Google Scholar] [CrossRef]
- Bannur, Z.; Teh, L.K.; Hennesy, T.; Rosli, W.R.; Mohamad, N.; Nasir, A.; Ankathil, R.; Zakaria, Z.A.; Baba, A.; Salleh, M.Z. The differential metabolite profiles of acute lymphoblastic leukaemic patients treated with 6-mercaptopurine using untargeted metabolomics approach. Clin. Biochem. 2014, 47, 427–431. [Google Scholar] [CrossRef]
- Bolkun, L.; Lemancewicz, D.; Jablonska, E.; Szumowska, A.; Bolkun-Skornicka, U.; Moniuszko, M.; Dzieciol, J.; Kloczko, J. Prognostic significance of ligands belonging to tumour necrosis factor superfamily in acute lymphoblastic leukaemia. Leuk. Res. 2015, 39, 290–295. [Google Scholar] [CrossRef]
- Sun, B.; Wu, Y.; Wu, L.; Cui, M.; Ni, H.; Yang, Z.; Xu, M.; Wang, H. Raised expression of APRIL in Chinese children with acute lymphoblastic leukemia and its clinical implications. J. Pediatr. Hematol. Oncol. 2014, 36, 276–280. [Google Scholar] [CrossRef]
- El Kramani, N.; Elsherbiny, N.M.; El-Gayar, A.M.; Ebrahim, M.A.; Al-Gayyar, M.M.H. Clinical significance of the TNF-alpha receptors, TNFRSF2 and TNFRSF9, on cell migration molecules Fascin-1 and Versican in acute leukemia. Cytokine 2018, 111, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Giordano, P.; Muggeo, P.; Delvecchio, M.; Carbonara, S.; Romano, A.; Altomare, M.; Ricci, G.; Valente, F.; Zito, A.; Scicchitano, P.; et al. Endothelial dysfunction and cardiovascular risk factors in childhood acute lymphoblastic leukemia survivors. Int. J. Cardiol. 2017, 228, 621–627. [Google Scholar] [CrossRef]
- Hatzipantelis, E.S.; Athanassiou-Metaxa, M.; Gombakis, N.; Tzimouli, V.; Taparkou, A.; Sidi-Fragandrea, V.; Garipidou, V.; Papageorgiou, T.; Kleta, D.; Koliouskas, D.E.; et al. Thrombomodulin and von Willebrand factor: Relation to endothelial dysfunction and disease outcome in children with acute lymphoblastic leukemia. Acta Haematol. 2011, 125, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Hagag, A.A.; Abdel-Lateef, A.E.; Aly, R. Prognostic value of plasma levels of thrombomodulin and von Willebrand factor in Egyptian children with acute lymphoblastic leukemia. J. Oncol. Pharm. Pract. 2014, 20, 356–361. [Google Scholar] [CrossRef]
- Qin, J.; Huang, S.; Qian, J.; Xu, C.; Li, S.; Yu, S.; Yan, H.; Wu, M.; Chen, J.; Ren, H.; et al. The prognostic relevance and expression of progranulin in adult patients with acute myeloid leukemia. Medicine 2020, 99, e18574. [Google Scholar] [CrossRef] [PubMed]
- El-Ghammaz, A.M.S.; Azzazi, M.O.; Mostafa, N.; Hegab, H.M.; Mahmoud, A.A. Prognostic significance of serum progranulin level in de novo adult acute lymphoblastic leukemia patients. Clin. Exp. Med. 2020, 20, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zhu, L. Semaphorins and Their Receptors: From Axonal Guidance to Atherosclerosis. Front. Physiol. 2018, 9, 1236. [Google Scholar] [CrossRef]
- Wei, L.; Li, H.; Tamagnone, L.; You, H. Semaphorins and Their Receptors in Hematological Malignancies. Front. Oncol. 2019, 9, 382. [Google Scholar] [CrossRef]
- Stelzer Gil, R.N.; Inbar, P.; Shahar, Z.; Michal, T.; Simon, F.; TsippiIny, S.; Ron, N.; Iris, L.; Yaron, M.; Sergey, K.; et al. GeneCards—The Human Gene Database. Available online: https://www.genecards.org (accessed on 25 May 2023).
- Jiang, H.; Tang, J.; Qiu, L.; Zhang, Z.; Shi, S.; Xue, L.; Kui, L.; Huang, T.; Nan, W.; Zhou, B.; et al. Semaphorin 4D is a potential biomarker in pediatric leukemia and promotes leukemogenesis by activating PI3K/AKT and ERK signaling pathways. Oncol. Rep. 2021, 45, 1. [Google Scholar] [CrossRef]
- Xue, L.; Shi, S.; Lei, H.; Zhang, Z.; Tian, X.; Qiu, L.; Tang, J.; Kui, L.; Nan, W.; Cao, X.; et al. Soluble Sema4D Level Is Positively Correlated with Sema4D Expression in PBMCs and Peripheral Blast Number in Acute Leukemia. Dis. Markers 2022, 2022, 1384471. [Google Scholar] [CrossRef]
- Yeh, C.H.; Tseng, R.; Hannah, A.; Estrov, Z.; Estey, E.; Kantarjian, H.; Albitar, M. Clinical correlation of circulating heat shock protein 70 in acute leukemia. Leuk. Res. 2010, 34, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Barreto, A.; Gonzalez, J.M.; Kabingu, E.; Asea, A.; Fiorentino, S. Stress-induced release of HSC70 from human tumors. Cell Immunol. 2003, 222, 97–104. [Google Scholar] [CrossRef]
- Cox, C.V.; Diamanti, P.; Evely, R.S.; Kearns, P.R.; Blair, A. Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood 2009, 113, 3287–3296. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Lambrou, G.I.; Vougas, K.; Karamolegou, K.; Tsangaris, G.T.; Tzortzatou-Stathopoulou, F. Protein biomarkers distinguish between high- and low-risk pediatric acute lymphoblastic leukemia in a tissue specific manner. J. Hematol. Oncol. 2013, 6, 52. [Google Scholar] [CrossRef]
- Ibrahim, L.; Aladle, D.; Mansour, A.; Hammad, A.; Al Wakeel, A.A.; Abd El-Hameed, S.A. Expression and prognostic significance of livin/BIRC7 in childhood acute lymphoblastic leukemia. Med. Oncol. 2014, 31, 941. [Google Scholar] [CrossRef] [PubMed]
- Chiaretti, S.; Brugnoletti, F.; Messina, M.; Paoloni, F.; Fedullo, A.L.; Piciocchi, A.; Elia, L.; Vitale, A.; Mauro, E.; Ferrara, F.; et al. CRLF2 overexpression identifies an unfavourable subgroup of adult B-cell precursor acute lymphoblastic leukemia lacking recurrent genetic abnormalities. Leuk. Res. 2016, 41, 36–42. [Google Scholar] [CrossRef]
- Russell, L.J.; Capasso, M.; Vater, I.; Akasaka, T.; Bernard, O.A.; Calasanz, M.J.; Chandrasekaran, T.; Chapiro, E.; Gesk, S.; Griffiths, M.; et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood 2009, 114, 2688–2698. [Google Scholar] [CrossRef]
- Palacios, F.; Yan, X.-J.; Ferrer, G.; Chen, S.-S.; Vergani, S.; Yang, X.; Gardner, J.; Barrientos, J.C.; Rock, P.; Burack, R.; et al. Musashi 2 influences chronic lymphocytic leukemia cell survival and growth making it a potential therapeutic target. Leukemia 2021, 35, 1037–1052. [Google Scholar] [CrossRef]
- Akpinar, F.; Polat, A.; Balci, Y.I.; Akça, H.; Şenol, H.; Tokgun, O. Microrna Expression Profiles and Changes with Treatment on Childhood Leukemias. Int. J. Hematol. Oncol. 2020, 30, 72–80. [Google Scholar] [CrossRef]
- Ramani, R.; Megason, G.; Schallheim, J.; Karlson, C.; Vijayakumar, V.; Vijayakumar, S.; Hicks, C. Integrative Analysis of MicroRNA-Mediated Gene Signatures and Pathways Modulating White Blood Cell Count in Childhood Acute Lymphoblastic Leukemia. Biomark. Insights 2017, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yang, X.; Hu, S.; Kang, M.; Chen, J.; Fang, Y. A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. Cancer Med. 2019, 8, 2553–2560. [Google Scholar] [CrossRef] [PubMed]
- Schotte, D.; De Menezes, R.X.; Akbari Moqadam, F.; Khankahdani, L.M.; Lange-Turenhout, E.; Chen, C.; Pieters, R.; Den Boer, M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Bakhshi, S.; Thakkar, H.; Gerards, M.; Singh, A. Elevated mitochondrial DNA copy numbers in pediatric acute lymphoblastic leukemia: A potential biomarker for predicting inferior survival. Pediatr. Blood Cancer 2018, 65, 1–7. [Google Scholar] [CrossRef]
- Kwok, C.S.; Quah, T.C.; Ariffin, H.; Tay, S.K.; Yeoh, A.E. Mitochondrial D-loop polymorphisms and mitochondrial DNA content in childhood acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2011, 33, e239–e244. [Google Scholar] [CrossRef]
- Kodron, A.; Ghanim, M.; Krawczyk, K.K.; Stelmaszczyk-Emmel, A.; Tonska, K.; Demkow, U.; Bartnik, E. Mitochondrial DNA in pediatric leukemia patients. Acta Biochim. Pol. 2017, 64, 183–187. [Google Scholar] [CrossRef]
- Oh, B.L.Z.; Lee, S.H.R.; Yeoh, A.E.J. Curing the Curable: Managing Low-Risk Acute Lymphoblastic Leukemia in Resource Limited Countries. J. Clin. Med. 2021, 10, 4728. [Google Scholar] [CrossRef]
- Brandalise, S.; Odone, V.; Pereira, W.; Andrea, M.; Zanichelli, M.; Aranega, V. Treatment results of three consecutive Brazilian cooperative childhood ALL protocols: GBTLI-80, GBTLI-82 and -85. ALL Brazilian Group. Leukemia 1993, 7 (Suppl. 2), S142–S145. [Google Scholar]
- Cavalcante Mde, S.; Torres-Romero, J.C.; Lobo, M.D.; Moreno, F.B.; Bezerra, L.P.; Lima, D.S.; Matos, J.C.; Moreira Rde, A.; Monteiro-Moreira, A.C. A panel of glycoproteins as candidate biomarkers for early diagnosis and treatment evaluation of B-cell acute lymphoblastic leukemia. Biomark. Res. 2016, 4, 1. [Google Scholar] [CrossRef]
- Fathi, M.; Amirghofran, Z.; Shahriari, M. Soluble Fas and Fas ligand and prognosis in children with acute lymphoblastic leukemia. Med. Oncol. 2012, 29, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Bruey, J.M.; Kantarjian, H.; Estrov, Z.; Zhang, Z.; Ma, W.; Albitar, F.; Abdool, A.; Thomas, D.; Yeh, C.; O’Brien, S.; et al. Circulating Ki-67 protein in plasma as a biomarker and prognostic indicator of acute lymphoblastic leukemia. Leuk. Res. 2010, 34, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Birgen, D.; Ertem, U.; Duru, F.; Sahin, G.; Yuksek, N.; Bozkurt, C.; Karacan, C.D.; Aksoy, C. Serum Ca 125 levels in children with acute leukemia and lymphoma. Leuk. Lymphoma 2005, 46, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Klikova, K.; Pilchova, I.; Stefanikova, A.; Hatok, J.; Dobrota, D.; Racay, P. The Role of Heat Shock Proteins in Leukemia. Klin. Onkol. 2016, 29, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Conter, V.; Bartram, C.R.; Valsecchi, M.G.; Schrauder, A.; Panzer-Grumayer, R.; Moricke, A.; Arico, M.; Zimmermann, M.; Mann, G.; De Rossi, G.; et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: Results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 2010, 115, 3206–3214. [Google Scholar] [CrossRef]
- Pawlik-Gwozdecka, D.; Gorska-Ponikowska, M.; Adamkiewicz-Drozynska, E.; Niedzwiecki, M. Serum heat shock protein 90 as a future predictive biomarker in childhood acute lymphoblastic leukemia. Cent. Eur. J. Immunol. 2021, 46, 63–67. [Google Scholar] [CrossRef]
- Stary, J.; Zimmermann, M.; Campbell, M.; Castillo, L.; Dibar, E.; Donska, S.; Gonzalez, A.; Izraeli, S.; Janic, D.; Jazbec, J.; et al. Intensive chemotherapy for childhood acute lymphoblastic leukemia: Results of the randomized intercontinental trial ALL IC-BFM 2002. J. Clin. Oncol. 2014, 32, 174–184. [Google Scholar] [CrossRef]
- Krawczuk-Rybak, M.; Leszczynska, E.; Malinowska, I.; Matysiak, M.; Ostrowska, H. Proteasome chymotrypsin-like activity in plasma as a useful marker for children with acute lymphoblastic leukemia. Scand. J. Clin. Lab. Investig. 2012, 72, 67–72. [Google Scholar] [CrossRef]
- Zinngrebe, J.; Debatin, K.M.; Fischer-Posovszky, P. Adipocytes in hematopoiesis and acute leukemia: Friends, enemies, or innocent bystanders? Leukemia 2020, 34, 2305–2316. [Google Scholar] [CrossRef]
- Moschovi, M.; Trimis, G.; Vounatsou, M.; Katsibardi, K.; Margeli, A.; Damianos, A.; Chrousos, G.; Papassotiriou, I. Serial plasma concentrations of adiponectin, leptin, and resistin during therapy in children with acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2010, 32, e8–e13. [Google Scholar] [CrossRef]
- Aref, S.; Ibrahim, L.; Azmy, E.; Al Ashary, R. Impact of serum adiponectin and leptin levels in acute leukemia. Hematology 2013, 18, 198–203. [Google Scholar] [CrossRef]
- Buchmann, S.; Schrappe, M.; Baruchel, A.; Biondi, A.; Borowitz, M.; Campbell, M.; Cario, G.; Cazzaniga, G.; Escherich, G.; Harrison, C.J.; et al. Remission, treatment failure, and relapse in pediatric ALL: An international consensus of the Ponte-di-Legno Consortium. Blood 2022, 139, 1785–1793. [Google Scholar] [CrossRef]
- O’Neill, K.L.; Zhang, F.; Li, H.; Fuja, D.G.; Murray, B.K. Thymidine kinase 1--a prognostic and diagnostic indicator in ALL and AML patients. Leukemia 2007, 21, 560–563. [Google Scholar] [CrossRef]
- Zhang, F.; Shao, X.; Li, H.; Robison, J.G.; Murray, B.K.; O’Neill, K.L. A monoclonal antibody specific for human thymidine kinase 1. Hybridoma 2001, 20, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Martinez, B.; Vilchis Ordonez, A.; Salazar Garcia, M.; Klunder-Klunder, M.; Parra-Ortega, I.; Dorantes-Acosta, E.; Angeles-Floriano, T. Thymidine Kinase: A Biomarker for Recently Diagnosed Acute Leukemia in Pediatric Patients According to the Cell Line Involved. Arch. Med. Res. 2015, 46, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Ishii, G.; Sugaya, Y.; Nishimura, M.; Saito, Y.; Harigaya, K. Potential use of serum CD44 as an indicator of tumour progression in acute leukemia. Hematol. Oncol. 1999, 17, 161–168. [Google Scholar] [CrossRef]
- Takeuchi, M.; Tanizawa, A.; Fukumoto, Y.; Kikawa, Y.; Mayumi, M. Serum soluble CD44 in pediatric patients with acute leukemia. J. Pediatr. Hematol. Oncol. 1999, 21, 384–388. [Google Scholar] [CrossRef]
- Hatzistilianou, M.; Athanassiadou, F.; Agguridaki, C.; Catriu, D. Circulating soluble adhesion molecule levels in children with acute lymphoblastic leukaemia. Eur. J. Pediatr. 1997, 156, 537–540. [Google Scholar] [CrossRef]
- Neaga, A.; Jimbu, L.; Mesaros, O.; Bota, M.; Lazar, D.; Cainap, S.; Blag, C.; Zdrenghea, M. Why Do Children with Acute Lymphoblastic Leukemia Fare Better Than Adults? Cancers 2021, 13, 3886. [Google Scholar] [CrossRef]
- Contreras Yametti, G.P.; Ostrow, T.H.; Jasinski, S.; Raetz, E.A.; Carroll, W.L.; Evensen, N.A. Minimal Residual Disease in Acute Lymphoblastic Leukemia: Current Practice and Future Directions. Cancers 2021, 13, 1847. [Google Scholar] [CrossRef]
- Campana, D.; Coustan-Smith, E. Measurements of treatment response in childhood acute leukemia. Korean J. Hematol. 2012, 47, 245–254. [Google Scholar] [CrossRef]
- Rzepiel, A.; Kutszegi, N.; Gezsi, A.; Sagi, J.C.; Egyed, B.; Peter, G.; Butz, H.; Nyiro, G.; Muller, J.; Kovacs, G.T.; et al. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y.; Hu, T.; He, X.; Zou, Y.; Deng, Q.; Ke, J.; Lian, L.; He, X.; Zhao, D.; et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol. Oncol. 2021, 15, 2702–2714. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, H.; Huang, Y.; Xu, S.; Zhou, Y.; Zhang, W.; Li, J.; Ming, Y.; Wang, X.; Zhao, S.; et al. Genome-wide cell-free DNA methylation analyses improve accuracy of non-invasive diagnostic imaging for early-stage breast cancer. Mol. Cancer 2021, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Uehiro, N.; Sato, F.; Pu, F.; Tanaka, S.; Kawashima, M.; Kawaguchi, K.; Sugimoto, M.; Saji, S.; Toi, M. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 2016, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Li, L.; Xu, M.; Wang, X.; Wang, F.; Ni, H. Significance of BAFF/APRIL Expression and Their Receptors in Pediatric Patients With Acute Lymphoblastic Leukemia. J. Pediatr. Hematol./Oncol. 2016, 38, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Meena, R.; Nangia, A.; Sharma, S.; Chandra, J. Serum Levels of Vascular Endothelial Growth Factor and Its Receptor in Newly Diagnosed Paediatric Acute Lymphoblastic Leukemia. Indian J. Hematol. Blood Transfus. 2021, 37, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Olejnik, I. Serum soluble L-selectin in childhood acute lymphoblastic leukemia. Pediatr. Int. 1999, 41, 246–248. [Google Scholar] [CrossRef]
- Ebrahimi-Rad, M.; Khatami, S.; Ansari, S.; Jalylfar, S.; Valadbeigi, S.; Saghiri, R. Adenosine Deaminase 1 as a Biomarker for Diagnosis and Monitoring of Patients with Acute Lymphoblastic Leukemia. J. Med. Biochem. 2018, 37, 128–133. [Google Scholar] [CrossRef]
- Abdool, A.; Yeh, C.H.; Kantarjian, H.; O’Brien, S.; Bruey, J.; Giles, F.; Albitar, M. Circulating CD33 and its clinical value in acute leukemia. Exp. Hematol. 2010, 38, 462–471. [Google Scholar] [CrossRef]
- Aref, S.; Azmy, E.; El-Bakry, K.; Ibrahim, L.; Mabed, M. Prognostic impact of CD200 and CD56 expression in adult acute lymphoblastic leukemia patients. Hematology 2018, 23, 263–270. [Google Scholar] [CrossRef]
- Bhatia, P.; Masih, S.; Varma, N.; Bansal, D.; Trehan, A. High Expression of Lung Resistance Protein mRNA at Diagnosis Predicts Poor Early Response to Induction Chemotherapy in Childhood Acute Lymphoblastic Leukemia. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 6663–6668. [Google Scholar] [CrossRef] [PubMed]
- Duyu, M.; Durmaz, B.; Gunduz, C.; Vergin, C.; Yilmaz Karapinar, D.; Aksoylar, S.; Kavakli, K.; Cetingul, N.; Irken, G.; Yaman, Y.; et al. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res. Int. 2014, 2014, 967585. [Google Scholar] [CrossRef] [PubMed]
- Al Nakeeb, R.H.; Alrubaye, D.J.I.J.o.S. The Expression of Different Micrornas in Iraqi Patients with Childhood Acute Leukemia and Their Association to C/EBP-Î’Serum Level. Iraqi J. Sci. 2020, 2879–2887. [Google Scholar] [CrossRef]
- Labib, H.A.; Elantouny, N.G.; Ibrahim, N.F.; Alnagar, A.A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology 2017, 22, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Asnafi, A.A.; Khodadi, E.; Golchin, N.; Alghasi, A.; Tavakolifar, Y.; Saki, N.J.F.i.B. Association between microRNA-21, microRNA-150, and micro-RNA-451 expression and clinical outcome of patients with acute lymphoblastic leukemia. Front. Biol 2017, 12, 63–70. [Google Scholar] [CrossRef]
- Anagnostopoulou, E.; Papanastasopoulou, C.; Papastamataki, M.; Kotsiou, A.; Topouzoglou, Z.; Anagnostopoulos, N.; Sitaras, N. Serum Hyaluronic Acid Levels Are Altered in Acute Leukemia Patients: Potential Prognostic Implications. Acta Haematol. 2017, 138, 44–51. [Google Scholar] [CrossRef]
- Rautonen, J.; Siimes, M.A.J.C. Elevated serum transaminase activity at diagnosis is associated with rapidly progressing disease in children with acute lymphoblastic leukemia. Cancer 1988, 61, 754–757. [Google Scholar] [CrossRef]

| Biomarker | Blood Sample | Leukemia Type | Area under the ROC Curve | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Smad 7 | Serum | ALL | 0.81 | 63 | 100 | 100 | 73 | |
| TGF-β1 | Serum | ALL | 0.79 | 57 | 93 | 89.5 | 68 | [16] |
| Smad 7 TGF-β1 miR-181a | Serum | ALL | - | 100 | 93 | 93.7 | 100 | |
| IGF-I | Serum | ALL | - | 60.6 | 73.3 | - | - | [64] |
| IGF-II | Serum | ALL | - | 72.2 | 73.3 | - | - | |
| IGFBP-2 | Serum | ALL | - | 72.2 | 86.7 | - | - | |
| IGFBP-3 | Serum | ALL | - | 93.9 | 93.9 | - | - | |
| Anti-9-0AcSGs | Serum | ALL-GI | - | 98.9 | 92.1 | 96.8 | 97.2 | [65] |
| Anti-9-0AcSGs | Serum | ALL-GI | - | 96.8 | 95.9 | 96.8 | 95.9 | |
| PF4 CTAP-II | Serum | ALL | - | 91.8 | 90 | - | - | [7] |
| C3f | Serum | AL | 0.99 | 97 | 100 | - | - | [45] |
| TNF-α | Serum | ALL | 0.94 | 91.7 | 100 | - | - | [3] |
| Survivin | Serum | ALL | 0.98 | 90 | 80 | - | - | |
| p53 | Serum | AL | 0.8 | 52 | 100 | - | - | [66] |
| EGFR | Serum | AL | 0.93 | 73.9 | 95.8 | - | - | |
| Pseudouridine | Serum | ALL | - | 90 | 97.5 | - | - | [67] |
| ADAM 17 | Plasma | BCP-ALL | 0.98 | 100 | 100 | - | - | [68] |
| ATG3 | Plasma | BCP-ALL | 0.95 | 100 | 100 | - | - | |
| AC133 * | Whole blood | ALL | - | 100 | 100 | 100 | 100 | [69] |
| miR-181a | Serum | ALL | 0.93 | 86.5 | 93.3 | 92.8 | 87.5 | [16] |
| miR-146a | Plasma | BCP-ALL | 1 | 100 | 100 | - | - | [9] |
| mRNA Survivin | Whole blood | BCP-ALL | 0.85 | 95 | 95 | - | - | [70] |
| mRNA HLA-G | PBMC | ALL | - | 74 | 100 | - | - | [71] |
| miR-125b-1 | Serum | ALL | 0.85 | 83.7 | 100 | - | - | [72] |
| miR-203 | Serum | ALL | 0.87 | 97.7 | 87 | - | - | |
| miR-100 | PBMC | ALL | 0.87 | 82.7 | 100 | - | - | [15] |
| miR-196a | PBMC | ALL | 0.537 | 46.6 | 100 | - | - | |
| miR-146a | PBMC | ALL | 1 | 100 | 100 | - | - | |
| miR-511 | Plasma | BCP-ALL | 1 | 100 | 100 | 1 | 1 | [73] |
| miR-34a | Plasma | BCP-ALL | 0.98 | 92 | 100 | 1 | 0.70 | |
| miR-22 | Plasma | BCP-ALL | 0.91 | 79 | 100 | 1 | 0.54 | |
| miR-26a | Plasma | BCP-ALL | 0.91 | 79 | 100 | 1 | 0.47 | |
| miR-221 | Plasma | BCP-ALL | 0.92 | 83 | 100 | 1 | 0.54 | |
| miR-223 | Plasma | BCP-ALL | 0.93 | 89 | 100 | 1 | 0.64 | |
| miR-21 | PBMC | ALL | 0.565 | 44 | 55 | - | - | [74] |
| miR-26 | PBMC | ALL | 0.464 | 54 | 50 | - | - | |
| miR-148a | PBMC | ALL | 0.719 | 74 | 79 | - | - | |
| miR-133b | PBMC | ALL | 0.669 | 70 | 60 | - | - | |
| miR-24 | PBMC | ALL | 0.785 | 72 | 81 | - | - | |
| miR-92a | PBMC and plasma | ALL | 0.99 | - | - | - | - | [75] |
| miR-92a | Plasma | ALL | 0.755 | 41.5 | 100 | 100 | 36.7 | [6] |
| miR-638 | Plasma | ALL | 0.86 | 54.7 | 100 | 100 | 42.9 | |
| miR-125b | PBMC | ALL | 0.99 | 98 | 96.7 | - | - | [76] |
| mRNA-Bcl-2 | PBMC | ALL | 0.9 | 96.7 | 70 | - | - | |
| miR-128b | PBMC | ALL | - | 75 | 87.5 | - | - | [77] |
| cf-DNA levels | Plasma | ALL, AML | 0.79 | 65 | 100 | - | - | [50] |
| cf-DNA integrity | Plasma | ALL, AML | 88 | 78 | 90 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Zúñiga, C.D.; Garza-Veloz, I.; Martínez-Rendón, J.; Ureño-Segura, M.; Delgado-Enciso, I.; Martinez-Fierro, M.L. Circulating Biomarkers Associated with the Diagnosis and Prognosis of B-Cell Progenitor Acute Lymphoblastic Leukemia. Cancers 2023, 15, 4186. https://doi.org/10.3390/cancers15164186
Álvarez-Zúñiga CD, Garza-Veloz I, Martínez-Rendón J, Ureño-Segura M, Delgado-Enciso I, Martinez-Fierro ML. Circulating Biomarkers Associated with the Diagnosis and Prognosis of B-Cell Progenitor Acute Lymphoblastic Leukemia. Cancers. 2023; 15(16):4186. https://doi.org/10.3390/cancers15164186
Chicago/Turabian StyleÁlvarez-Zúñiga, Claudia Daniela, Idalia Garza-Veloz, Jacqueline Martínez-Rendón, Misael Ureño-Segura, Iván Delgado-Enciso, and Margarita L. Martinez-Fierro. 2023. "Circulating Biomarkers Associated with the Diagnosis and Prognosis of B-Cell Progenitor Acute Lymphoblastic Leukemia" Cancers 15, no. 16: 4186. https://doi.org/10.3390/cancers15164186







