Predicting Severe Haematological Toxicity in Gastrointestinal Cancer Patients Undergoing 5-FU-Based Chemotherapy: A Bayesian Network Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Clinical Information
2.3. Model Development
2.4. Data Processing
- Consolidation of databases: We combined individual patient databases into a single database, compiling all pertinent information for each patient.
- Standardization of variable formats: We ensured that all variables were standardized in terms of format, facilitating further analysis and processing.
- Removal of variables with excessive missing values: We excluded variables with missing values exceeding 10%. Those variables underwent thorough examination to ensure that their removal would not introduce bias or compromise the predictive capability of our model. This threshold was selected to strike a balance between retaining important information and ensuring the reliability of the model. Prior to their elimination, we verified the lack of a significant correlation with our target variable in order to preserve the integrity of our model.
- Imputation of missing values: For the remaining variables with missing values, we imputed the median value of the respective variable. This step allowed for the preservation of the overall structure and relationships within the data while accounting for missing information.
- Conversion of date variables into numerical variables: We transformed date variables into numerical variables by measuring the time elapsed between events of interest. This step enabled the incorporation of time-related information into the model.
- Identification of the target variable (Toxicity): We selected relevant toxicities for the study and recategorized the target variable as binary, where 1 represents the presence of severe toxicity of interest, and 0 represents its absence.
- Splitting the dataset into training and testing sets: We divided the cleaned database into a training set (80%) and a testing set (20%) to create and train the predictive model using the training set and perform an independent validation using the testing set. This approach allowed us to assess the model’s performance on unseen data and ensure its generalizability to new cases. Furthermore, the division of the datasets was performed in such a way that each set maintained the same proportion of the target variable, toxicity, ensuring a balanced distribution for a more accurate analysis.
2.5. Importance of Variables
- A random forest [34] model was constructed using 1000 decision trees, with toxicity as the target variable and all available variables serving as predictors.
- The MeanDecreaseGini coefficient for each variable was recorded, reflecting their importance in the model.
2.6. Bayesian Network Model Design
- Database Augmentation: Due to the imbalanced nature of the original dataset, with more cases of individuals without toxicity compared with those with toxicity, we decided to augment the TRAIN dataset using the SMOTE (Synthetic Minority Over-sampling Technique) function from the performanceEstimation library [35,36] This approach aimed to balance the dataset by generating synthetic samples for the under-represented class, thereby enhancing the model’s ability to accurately predict severe toxicity.
- Dataset Partitioning: The TRAIN dataset was partitioned into 10 subgroups to facilitate the application of a 10-fold cross-validation scheme. This partitioning maintained a representative distribution of the target variable (toxicity) categories in each subgroup comparable to the overall TRAIN dataset.
- Optimization Strategy: In the analysis of mixed Bayesian networks using the bnlearn R library [37], the aic-cg method was employed for configuring the network structure. This method utilizes the Akaike Information Criterion (AIC) score [38,39] to select the optimal network structure, considering both numerical and categorical variables. By utilizing the aic-cg method, it is possible to obtain a network structure that balances model complexity and goodness of fit, leading to more accurate predictions and insights in the analysis of mixed data.
- Adhering to the variable order determined by the previously obtained importance ranking, one variable at a time was incorporated into the model’s variable set, which initially contained only the target variable, toxicity.
- Cross-validation was performed on the updated variable set. In each iteration, a network structure was designed and its parameters determined using K-1 groups, while the remaining K group was employed to assess the predictive capacity of the preceding model.
- The cross-validation yielded a list of 10 estimations of the model’s predictive capacity. If the inclusion of a variable resulted in a higher set of estimations compared with the current model’s estimations, the variable’s incorporation was deemed successful.
- The iterative process continued until all variables were evaluated.
- Model Validation: The predictive capability of the Bayesian network structure, which was established in the previous phase, was evaluated using both the TRAIN dataset for training and the TEST dataset for validation.
3. Results
3.1. Data Processing
3.2. Importance of Variables
3.3. Bayesian Network Model Design
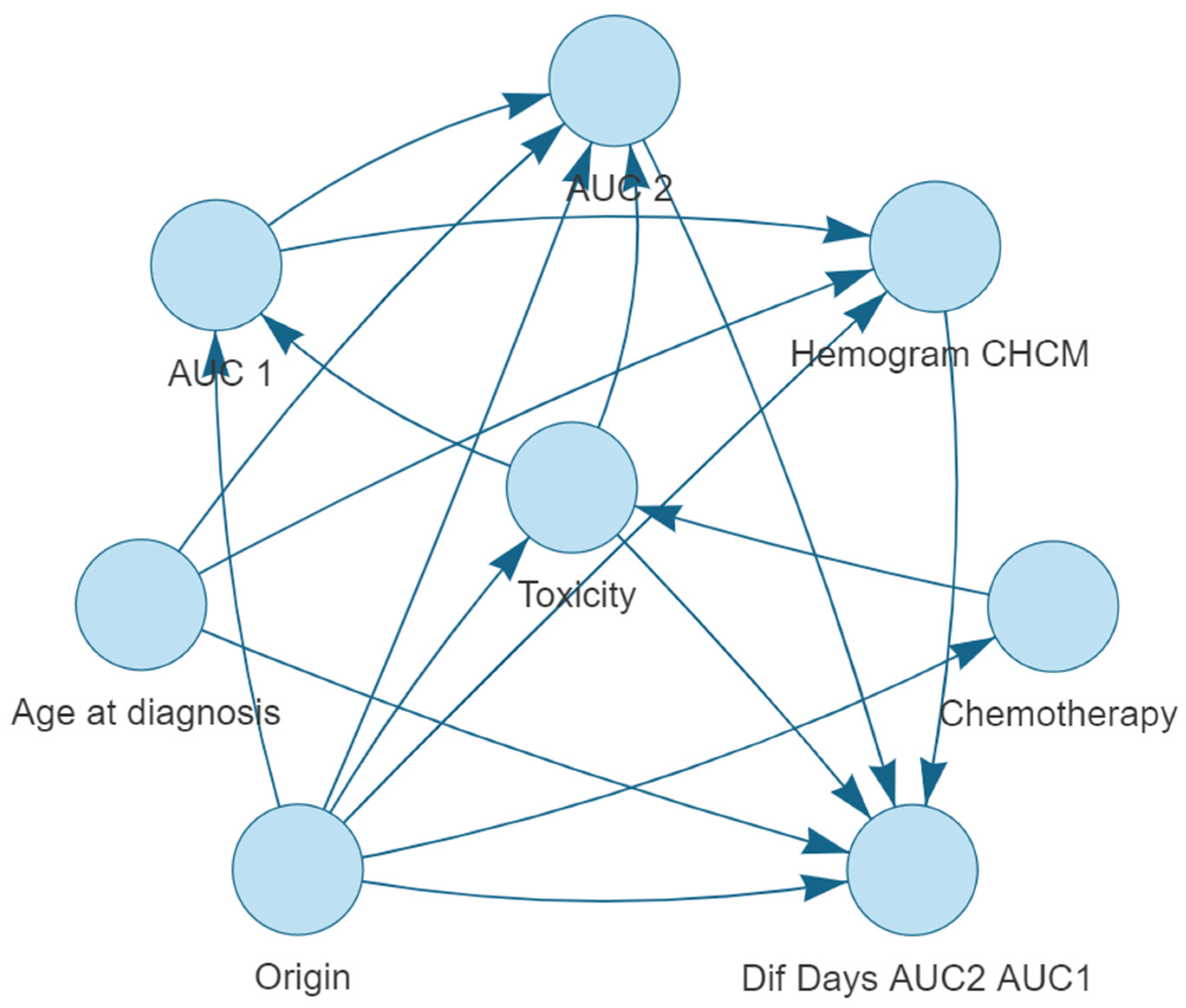
Bayesian Network Structure
- Origin is connected to: Dif Days AUC2 AUC1, Chemotherapy, CBC MCHC, Toxicity, AUC 1 and AUC 2.
- Toxicity is connected to: Dif Days AUC2 AUC1, AUC 1, and AUC 2.
- Age at diagnosis is connected to: CBC MCHC, AUC 2, and Dif Days AUC2 AUC1.
- AUC 1 is connected to: AUC 2 and CBC MCHC.
- AUC 2 is connected to: Dif Days AUC2 AUC1.
- CBC MCHC is connected to: Dif Days AUC2 AUC1.
- Chemotherapy is connected to: Toxicity.
3.4. Model Performance Metrics
3.4.1. Cross-Validation on TRAIN Dataset
- Accuracy: The average accuracy of the model, obtained from the 10-fold cross-validation, indicates the proportion of correct predictions out of the total number of predictions made. In our study, the Bayesian network model achieved an average accuracy of 0.85 with a standard deviation of 0.05.
- Sensitivity: The average sensitivity measures the model’s ability to correctly identify patients who experience severe toxicity. Our model demonstrated an average sensitivity of 0.82 with a standard deviation of 0.14.
- Specificity: The average specificity assesses the model’s ability to correctly identify patients who do not experience severe toxicity. In this study, the Bayesian network model displayed an average specificity of 0.87 with a standard deviation of 0.07.
3.4.2. Validation on TEST Dataset
- Accuracy: The accuracy of the model on the TEST dataset was 0.80.
- Sensitivity: The sensitivity of the model on the TEST dataset was 0.71.
- Specificity: The specificity of the model on the TEST dataset was 0.83.
3.5. Model Implementation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barin-Le Guellec, C.; Lafay-Chebassier, C.; Ingrand, I.; Tournamille, J.F.; Boudet, A.; Lanoue, M.C.; Defos-sez, G.; Ingrand, P.; Perault-Pochat, M.C.; Etienne-Grimaldi, M.C. Toxicities associated with chemotherapy regimens containing a fluoropyrimidine: A real-life evaluation in France. Eur. J. Cancer 2020, 124, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.; Piedbois, P.; Buyse, M.; Pignon, J.P.; Rougier, P.; Ryan, L.; Hansen, R.; Zee, B.; Weinerman, B.; Pater, J.; et al. Toxicity of fluorouracil in patients with advanced colorectal cancer: Effect of administra-tion schedule and prognostic factors. J. Clin. Oncol. 1998, 16, 3537–3541. [Google Scholar] [PubMed]
- Breton, C.; Aparicio, T.; Le Malicot, K.; Ducreux, M.; Lecomte, T.; Bachet, J.B.; Taieb, J.; Legoux, J.L.; Gramont, A.D.; Bennouna, J.; et al. Predictive factors of severe early treatment-related toxicity in patients receiving first-line treatment for metastatic colorectal cancer: Pooled analysis of 2190 patients enrolled in Fédération Francophone de Cancérologie Digestive (FFCD) trials. Eur. J. Cancer 2021, 153, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, O.; Ahmed, O. Predictors of toxicity-related hospitalization in four randomized studies of 5-fluorouracil-based chemotherapy in metastatic colorectal cancer. Int. J. Color. Dis. 2019, 34, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Al-Batran, S.E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar]
- Zhang, B.; Zhou, F.; Hong, J.; Ng, D.M.; Yang, T.; Zhou, X.; Jin, J.; Zhou, F.; Chen, P.; Xu, Y. The role of FOLFIRINOX in metastatic pancreatic cancer: A meta-analysis. World J. Surg. Oncol. 2021, 19, 182. [Google Scholar] [CrossRef]
- Khali, K.A.; Musalm, H.S.; Hassan, M.A.; Mahmoud, I.A. Triplet (FOLFOXIRI) Versus Doublet (FOLFOX or FOLFIRI) Regimen as First Line Treatment in Metastatic Colorectal Carcinoma, a Prospective Phase II, Randomized Controlled Trial. Asian Pac. J. Cancer Prev. APJCP 2022, 23, 3421. [Google Scholar] [CrossRef]
- Afzal, S.; Gusella, M.; Vainer, B.; Vogel, U.B.; Andersen, J.T.; Broedbaek, K.; Petersen, M.; Jiminez-SOlem, E.; Bertolaso, L.; Barile, C.; et al. Combinations of polymorphisms in genes involved in the 5-Fluorouracil metabolism pathway are associated with gastrointestinal toxicity in chemotherapy-treated colorectal cancer patients. Clin. Cancer Res. 2011, 17, 3822–3829. [Google Scholar] [CrossRef]
- Pullarkat, S.T.; Stoehlmacher, J.; Ghaderi, V.; Xiong, Y.P.; Ingles, S.A.; Sherrod, A.; Warren, R.; Tsao-Wei, D.; Groshen, S.; Lenz, H.J. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenom. J. 2001, 1, 65–70. [Google Scholar] [CrossRef]
- Lee, A.M.; Shi, Q.; Alberts, S.R.; Sargent, D.J.; Sinicrope, F.A.; Berenberg, J.L.; Grothey, A.; Polite, B.; Chan, E.; Gill, S.; et al. Association between DPYD c. 1129-5923 C > G/hapB3 and severe toxicity to 5-fluorouracil-based chemotherapy in stage III colon cancer patients: NCCTG N0147 (Alliance). Pharmacogenom. Genom. 2016, 26, 133. [Google Scholar] [CrossRef] [PubMed]
- De Luca, O.; Salerno, G.; De Bernardini, D.; Torre, M.S.; Simmaco, M.; Lionetto, L.; Gentile, G.; Borro, M. Predicting dihydropyrimidine dehydrogenase deficiency and related 5-fluorouracil toxicity: Opportunities and challenges of DPYD exon sequencing and the role of phenotyping assays. Int. J. Mol. Sci. 2022, 23, 13923. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.M.; Shi, Q.; Pavey, E.; Alberts, S.R.; Sargent, D.J.; Sinicrope, F.A.; Berenberg, J.L.; Goldberg, R.M.; Diasio, R.B. DPYD variants as predictors of 5-fluorouracil toxicity in adjuvant colon cancer treatment (NCCTG N0147). J. Natl. Cancer Inst. 2014, 106, dju298. [Google Scholar] [CrossRef] [PubMed]
- Gusella, M.; Frigo, A.C.; Bolzonella, C.; Marinelli, R.; Barile, C.; Bononi, A.; Crepaldi, G.; Menon, D.; Stievano, L.; Toso, S.; et al. Predictors of survival and toxicity in patients on adjuvant therapy with 5-fluorouracil for colorectal cancer. Br. J. Cancer 2009, 100, 1549–1557. [Google Scholar] [CrossRef]
- Oyaga-Iriarte, E.; Insausti, A.; Sayar, O.; Aldaz, A. Prediction of irinotecan toxicity in metastatic colorectal cancer patients based on machine learning models with pharmacokinetic parameters. J. Pharmacol. Sci. 2019, 140, 20–25. [Google Scholar] [CrossRef]
- Bae, J.H.; Kim, M.; Lim, J.S.; Geem, Z.W. Feature selection for colon cancer detection using k-means clustering and modified harmony search algorithm. Mathematics 2021, 9, 570. [Google Scholar] [CrossRef]
- Birks, J.; Bankhead, C.; Holt, T.A.; Fuller, A.; Patnick, J. Evaluation of a prediction model for colorectal cancer: Retrospective analysis of 2.5 million patient records. Cancer Med. 2017, 6, 2453–2460. [Google Scholar] [CrossRef]
- Song, Z.; Yu, C.; Zou, S.; Wang, W.; Huang, Y.; Ding, X.; Liu, J.; Shao, L.; Yuan, J.; Gou, X.; et al. Automatic deep learning-based colorectal adenoma detection system and its similarities with pathologists. BMJ Open 2020, 10, e036423. [Google Scholar] [CrossRef]
- Alboaneen, D.; Alqarni, R.; Alqahtani, S.; Alrashidi, M.; Alhuda, R.; Alyahyan, E.; Alshammari, T. Predicting Colorectal Cancer Using Machine and Deep Learning Algorithms: Challenges and Opportunities. Big Data Cogn. Comput. 2023, 7, 74. [Google Scholar] [CrossRef]
- Isci, S.; Dogan, H.; Ozturk, C.; Otu, H.H. Bayesian network prior: Network analysis of biological data using external knowledge. Bioinformatics 2014, 30, 860–867. [Google Scholar] [CrossRef]
- Angelopoulos, N.; Chatzipli, A.; Nangalia, J.; Maura, F.; Campbell, P.J. Bayesian networks elucidate complex genomic landscapes in cancer. Commun. Biol. 2022, 5, 306. [Google Scholar] [CrossRef] [PubMed]
- Kalet, A.M.; Doctor, J.N.; Gennari, J.H.; Phillips, M.H. Developing Bayesian networks from a dependency-layered ontology: A proof-of-concept in radiation oncology. Med. Phys. 2017, 44, 4350–4359. [Google Scholar] [CrossRef] [PubMed]
- van der Gaag, L.C.; Bodlaender, H.L.; Feelders, A. Monotonicity in Bayesian networks. arXiv 2012, arXiv:1207.4160. [Google Scholar]
- Arora, P.; Boyne, D.; Slater, J.J.; Gupta, A.; Brenner, D.R.; Druzdzel, M.J. Bayesian networks for risk prediction using real-world data: A tool for precision medicine. Value Health 2019, 22, 439–445. [Google Scholar] [CrossRef]
- Zihni, E.; Madai, V.I.; Livne, M.; Galinovic, I.; Khalil, A.A.; Fiebach, J.B.; Frey, D. Opening the black box of artificial intelligence for clinical decision support: A study predicting stroke outcome. PLoS ONE 2020, 15, e0231166. [Google Scholar] [CrossRef]
- Vásquez-Morales, G.R.; Martinez-Monterrubio, S.M.; Moreno-Ger, P.; Recio-Garcia, J.A. Explainable prediction of chronic renal disease in the colombian population using neural networks and case-based reasoning. IEEE Access 2019, 7, 152900–152910. [Google Scholar] [CrossRef]
- Pintelas, E.; Liaskos, M.; Livieris, I.E.; Kotsiantis, S.; Pintelas, P. Explainable machine learning framework for image classification problems: Case study on glioma cancer prediction. J. Imaging 2020, 6, 37. [Google Scholar] [CrossRef]
- Saraswat, D.; Bhattacharya, P.; Verma, A.; Prasad, V.K.; Tanwar, S.; Sharma, G.; Bokoro, P.N.; Sharma, R. Explainable AI for healthcare 5.0: Opportunities and challenges. IEEE Access 2022, 10, 84486–84517. [Google Scholar] [CrossRef]
- Gusella, M.; Ferrazzi, E.; Ferrari, M.; Padrini, R. New limited sampling strategy for determining 5-fluorouracil area under the concentration-time curve after rapid intravenous bolus. Ther. Drug Monit. 2002, 24, 425–431. [Google Scholar] [CrossRef]
- Etienne, M.C.; Chatelut, E.; Pivot, X.; Lavit, M.; Pujol, A.; Canal, P.; Milano, G. Co-variables influencing 5-fluorouracil clearance during continuous venous infusion. A NONMEM analysis. Eur. J. Cancer 1998, 34, 92–97. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. (n.d.). Available online: https://www.r-project.org (accessed on 29 June 2023).
- Han, H.; Guo, X.; Yu, H. Variable selection using mean decrease accuracy and mean decrease gini based on random forest. In Proceedings of the 2016 7th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 26–28 August 2016; IEEE: New York, NY, USA, 2016; pp. 219–224. [Google Scholar]
- Calle, M.L.; Urrea, V. Stability of Random Forest importance measures. Brief. Bioinform. 2011, 12, 86–89. [Google Scholar] [CrossRef]
- Cutler, A.; Cutler, D.R.; Stevens, J.R. Random forests. In Ensemble Machine Learning: Methods and Applications; Springer: New York, NY, USA, 2012; pp. 157–175. [Google Scholar]
- Torgo, M.L. Package “PerformanceEstimation”. R-project.org. 2022. Available online: https://cran.r-project.org/web/packages/performanceEstimation/performanceEstimation.pdf (accessed on 29 June 2023).
- Dablain, D.; Krawczyk, B.; Chawla, N.V. DeepSMOTE: Fusing deep learning and SMOTE for imbalanced data. IEEE Trans. Neural Netw. Learn. Syst. 2022. [Google Scholar] [CrossRef]
- Bnlearn—Bayesian Network Structure Learning. (n.d.). Bnlearn.com. Available online: https://www.bnlearn.com/ (accessed on 27 March 2023).
- Scutari, M. Learning Bayesian networks with the bnlearn R package. arXiv 2009, arXiv:0908.3817. [Google Scholar]
- Cavanaugh, J.E.; Neath, A.A. The Akaike information criterion: Background, derivation, properties, application, interpretation, and refinements. Wiley Interdiscip. Rev. Comput. Stat. 2019, 11, e1460. [Google Scholar] [CrossRef]
- Tsalic, M.; Bar-Sela, G.; Beny, A.; Visel, B.; Haim, N. Severe toxicity related to the 5-fluorouracil/leucovorin combination (the Mayo Clinic regimen): A prospective study in colorectal cancer patients. Am. J. Clin. Oncol. 2003, 26, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Anthoney, D.A.; Crellin, A.M.; Sebag-Montefiore, D.; Messruther, J.; Seymour, M.T. Weekly 5-fluorouracil and leucovorin: Achieving lower toxicity with higher dose-intensity in adjuvant chemotherapy after colorectal cancer resection. Ann. Oncol. 2004, 15, 568–573. [Google Scholar] [CrossRef]
- Garg, M.B.; Lincz, L.F.; Adler, K.; Scorgie, F.E.; Ackland, S.P.; Sakoff, J.A. Predicting 5-fluorouracil toxicity in colorectal cancer patients from peripheral blood cell telomere length: A multivariate analysis. Br. J. Cancer 2012, 107, 1525–1533. [Google Scholar] [CrossRef]
- Sharma, R.; Hoskins, J.M.; Rivory, L.P.; Zucknick, M.; London, R.; Liddle, C.; Clarke, S.J. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms and toxicity to capecitabine in advanced colorectal cancer patients. Clin. Cancer Res. 2008, 14, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Meulendijks, D.; Henricks, L.M.; Jacobs, B.A.; Aliev, A.; Deenen, M.J.; de Vries, N.; Rosing, H.; van Werkhoven, E.; de Boer, A.; Beijnen, J.H.; et al. Pretreatment serum uracil concentration as a predictor of severe and fatal fluoropyrimidine-associated toxicity. Br. J. Cancer 2017, 116, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Wiberg, H.; Yu, P.; Montanaro, P.; Mather, J.; Birz, S.; Schneider, M.; Bertsimas, D. Prediction of neutropenic events in chemotherapy patients: A machine learning approach. JCO Clin. Cancer Inform. 2021, 5, 904–911. [Google Scholar] [CrossRef]


| Variable | Number of Imputed Values |
|---|---|
| Area under the curve of the second cycle (AUC_2) | 2 |
| CBC Platelet amplitude distribution (PDW) | 4 |
| CBC PTC | 4 |
| CBC Red Cell Distribution Width (RDW) | 1 |
| CBC Mean Platelet Volume (MPV) | 1 |
| Plasma creatinine | 23 |
| MDRD (GFR algorithm) | 24 |
| Variable | Min | 1st.Qu. | Median | Mean | 3rd.Qu. | Max | SD |
|---|---|---|---|---|---|---|---|
| Age at diagnosis | 27.04 | 57.092 | 64.819 | 63.714 | 71.712 | 90.808 | 10.762 |
| BMI | 15.800 | 23.400 | 26.000 | 26.068 | 28.300 | 44.200 | 4.250 |
| Dif_Days_AUC1_Analy1 | −12.000 | 2.000 | 2.000 | 3.974 | 5.000 | 23.000 | 3.740 |
| Dif_Days_AUC2_Analy1 | 2.000 | 16.000 | 18.000 | 21.401 | 22.000 | 381.000 | 25.574 |
| Dif_Days_AUC2_AUC1 | 11.000 | 14.000 | 14.000 | 17.464 | 16.000 | 379.000 | 25.475 |
| AUC-1 | 13.500 | 25.000 | 29.000 | 29.536 | 32.500 | 59.700 | 6.762 |
| AUC-2 | 13.700 | 26.000 | 28.700 | 28.704 | 31.000 | 54.000 | 5.043 |
| CBC Bas (109/L) | 0 | 0.020 | 0.040 | 0.043 | 0.050 | 0.190 | 0.026 |
| CBC Basophils (%) | 0 | 0.300 | 0.500 | 0.542 | 0.700 | 1.700 | 0.288 |
| CBC MCHC (g/dL) | 27.200 | 31.700 | 32.600 | 32.472 | 33.400 | 51.800 | 2.017 |
| CBC Eos (109/L) | 0 | 0.080 | 0.130 | 0.192 | 0.235 | 2.260 | 0.244 |
| CBC Eosinophils (109/L) | 0 | 1.100 | 1.800 | 2.324 | 3.100 | 21.000 | 2.242 |
| CBC Hb (g/dL) | 7.400 | 11.400 | 13.300 | 12.880 | 14.400 | 17.700 | 1.905 |
| CBC HCM (pg) | 17.600 | 27.400 | 29.300 | 28.855 | 30.700 | 52.500 | 3.548 |
| CBC Hematies (1012/L) | 2.440 | 4.145 | 4.520 | 4.480 | 4.870 | 6.070 | 0.55 |
| CBC Hto (%) | 24.200 | 35.900 | 40.500 | 39.625 | 43.350 | 53.900 | 5.233 |
| CBC Leukocytes (109/L) | 3320 | 6.425 | 7.640 | 8.216 | 9.645 | 30.850 | 2.554 |
| CBC Lin (109/L) | 0.460 | 1.395 | 1.750 | 1.792 | 2.085 | 4.080 | 0.621 |
| CBC Lymphocytes (%) | 2.700 | 17.650 | 23.500 | 23.420 | 28.250 | 49.500 | 8.234 |
| CBC Mon (109/L) | 0.190 | 0.485 | 0.610 | 0.652 | 0.770 | 1.650 | 0.239 |
| CBC Monocytes (%) | 1.500 | 6.600 | 8.100 | 8.175 | 9.550 | 18.500 | 2.295 |
| CBC Neu (109/L) | 1.360 | 3.965 | 5.050 | 5.531 | 6.545 | 28.150 | 2.208 |
| CBC Neutrophils (%) | 35.600 | 60.500 | 65.600 | 65.494 | 71.400 | 92.800 | 9.430 |
| CBC PDW (fL) | 8.700 | 11.500 | 12.900 | 13.889 | 15.250 | 66.700 | 4.157 |
| CBC Platelet (109/L) | 85.000 | 207.000 | 267.000 | 278.798 | 343.000 | 595.000 | 96.363 |
| CBC PTC (%) | 0.080 | 0.200 | 0.280 | 0.281 | 0.350 | 0.640 | 0.094 |
| CBC RDW (%) | 11.600 | 12.850 | 13.600 | 14.564 | 15.150 | 30.300 | 2.928 |
| CBC VCM (fL) | 64.300 | 85.950 | 89.300 | 88.656 | 92.900 | 108.000 | 7.233 |
| CBC VPM (fL) | 6.500 | 9.500 | 10.200 | 10.193 | 10.950 | 13.500 | 1.225 |
| Blood Creatinine (mg/dL) | 0.400 | 0.700 | 0.800 | 0.872 | 1.000 | 2.900 | 0.234 |
| MDRD(GFR)(mL/min/1.73 m2) | 23.000 | 80.000 | 92.000 | 92.581 | 105.000 | 171.000 | 21.637 |
| Variable | Category | Number |
|---|---|---|
| Sex | Female | 79 |
| Male | 188 | |
| ECOG | 0 | 111 |
| 1 | 156 | |
| Histology | Adenocarcinoma | 263 |
| Carcinoma | 4 | |
| Origin | Colon | 81 |
| Gastric | 45 | |
| Pancreas | 82 | |
| Rectum | 59 | |
| Stage | Disseminated | 76 |
| Localized | 21 | |
| Regional | 170 | |
| Type of | FLOT | 18 |
| Chemotherapy | FOLFOX | 134 |
| FOLFOXIRI | 115 | |
| Pyrimidines | Altered | 11 |
| Metabolism | Normal | 256 |
| Patient Status | Dead with disease | 108 |
| Dead without disease | 5 | |
| Alive with disease | 34 | |
| Alive without disease | 120 |
| Toxicities | Patients |
|---|---|
| Anemia | 6/267 |
| Asthenia | 6/267 |
| Diarrhoea | 11/267 |
| Hyporexia | 1/267 |
| Leukopenia | 8/267 |
| Lymphopenia | 13/267 |
| Mucositis | 3/267 |
| Neutropenia | 66/267 |
| Nausea/vomiting | 2/267 |
| Rash | 1/267 |
| Neurological toxicity | 1/267 |
| Cardiac toxicity | 1/267 |
| Thrombocytopenia | 5/267 |
| Variable | MeanDecreaseGini |
|---|---|
| Dif_Days_AUC2_AUC1 | 7.68418463 |
| Chemotherapy | 6.63702847 |
| AUC-1 | 5.98405905 |
| Origin | 3.66820382 |
| Dif_Days_AUC2_Analy1 | 3.12617860 |
| MDRD (GFR algorithm) | 2.75232908 |
| AUC-2 | 2.64474938 |
| CBC Eos | 2.51249340 |
| CBC PDW | 2.51066431 |
| CBC Platelet | 2.35070235 |
| CBC Leukocytes | 2.20782943 |
| BMI | 2.16055155 |
| CBC RDW | 2.11807334 |
| Age at diagnosis | 2.09749240 |
| CBC Eosinophils | 2.06942951 |
| CBC Mon | 2.04356005 |
| CBC Neutrophils | 2.03195574 |
| CBC Hematies | 1.99520591 |
| CBC Neu | 1.94319862 |
| CBC Lymphocytes | 1.92257386 |
| CBC Lin | 1.90860498 |
| CBC Hb | 1.79924777 |
| CBC MCHC | 1.75612465 |
| CBC Hto | 1.72569409 |
| CBC PTC | 1.67414576 |
| CBC Monocytes | 1.60481585 |
| CBC VCM | 1.58960997 |
| CBC VPM | 1.57007868 |
| CBC HCM | 1.53288421 |
| Blood Creatinine | 1.33964382 |
| CBC Basophils | 1.00624322 |
| Dif_Days_AUC1_Analy1 | 0.95062264 |
| CBC Bas | 0.91029812 |
| Status | 0.80868056 |
| Stage | 0.54241400 |
| ECOG | 0.50190054 |
| Sex | 0.19311684 |
| Pyrimidines * | 0.05256130 |
| Histology | 0.02240322 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz Sarrias, O.; Gónzalez Deza, C.; Rodríguez Rodríguez, J.; Arrizibita Iriarte, O.; Vizcay Atienza, A.; Zumárraga Lizundia, T.; Sayar Beristain, O.; Aldaz Pastor, A. Predicting Severe Haematological Toxicity in Gastrointestinal Cancer Patients Undergoing 5-FU-Based Chemotherapy: A Bayesian Network Approach. Cancers 2023, 15, 4206. https://doi.org/10.3390/cancers15174206
Ruiz Sarrias O, Gónzalez Deza C, Rodríguez Rodríguez J, Arrizibita Iriarte O, Vizcay Atienza A, Zumárraga Lizundia T, Sayar Beristain O, Aldaz Pastor A. Predicting Severe Haematological Toxicity in Gastrointestinal Cancer Patients Undergoing 5-FU-Based Chemotherapy: A Bayesian Network Approach. Cancers. 2023; 15(17):4206. https://doi.org/10.3390/cancers15174206
Chicago/Turabian StyleRuiz Sarrias, Oskitz, Cristina Gónzalez Deza, Javier Rodríguez Rodríguez, Olast Arrizibita Iriarte, Angel Vizcay Atienza, Teresa Zumárraga Lizundia, Onintza Sayar Beristain, and Azucena Aldaz Pastor. 2023. "Predicting Severe Haematological Toxicity in Gastrointestinal Cancer Patients Undergoing 5-FU-Based Chemotherapy: A Bayesian Network Approach" Cancers 15, no. 17: 4206. https://doi.org/10.3390/cancers15174206






