Mutational Landscape and Precision Medicine in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Mutational Landscape of HCC
2.1. Microsatellite Instability
2.2. BRCA and BRCAness Mutations
2.3. Gene Fusions
2.4. Omics Signature
2.5. Mutational Burden
2.6. TERT
2.7. PT53
2.8. WNT-ß-Catenin
2.9. ARID1A
2.10. CDKN2A
2.11. CCND1
2.12. Angiogenesis Pathways
3. Precision Medicine to Guide Therapy
3.1. Arterially Directed Therapies
3.2. Curative-Intent Interventions
3.3. Systemic Therapies
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Serraino, D.; Fratino, L.; Piselli, P. Epidemiological Aspects of Hepatocellular Carcinoma. In Hepatocellular Carcinoma; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Global Burden of Disease Liver Cancer Collaboration; Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Waghray, A.; Murali, A.R.; Menon, K.N. Hepatocellular carcinoma: From diagnosis to treatment. World J. Hepatol. 2015, 7, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-C.; Wang, J.-J.; Chen, Y.-S.; Chen, J.-J.; Tsai, T.-C.; Lai, C.-C.; Sun, D.-P.; Shi, H.-Y. Trends and predictors of outcomes after surgery for hepatocellular carcinoma: A nationwide population-based study in Taiwan. Eur. J. Surg. Oncol. 2015, 41, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today/online-analysis-map?v=2020&mode=population&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=11&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&nb_items=10&group_cancer=1&include_nmsc=0&include_nmsc_other=0&projection=natural-earth&color_palette=default&map_scale=quantile&map_nb_colors=5&continent=0&show_ranking=0&rotate=%255B10%252C0%255D (accessed on 17 May 2023).
- Tian, Z.; Xu, C.; Yang, P.; Lin, Z.; Wu, W.; Zhang, W.; Ding, J.; Ding, R.; Zhang, X.; Dou, K. Molecular pathogenesis: Connections between viral hepatitis-induced and non-alcoholic steatohepatitis-induced hepatocellular carcinoma. Front. Immunol. 2022, 13, 984728. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Han, T.; Muskhelishvili, L.; Fuscoe, J.C.; Ross, S.A.; Beland, F.A.; Pogribny, I. Coupling global methylation and gene expression profiles reveal key pathophysiological events in liver injury induced by a methyl-deficient diet. Mol. Nutr. Food Res. 2011, 55, 411–418. [Google Scholar] [CrossRef]
- Yang, B.; Guo, M.; Herman, J.G.; Clark, D.P. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am. J. Pathol. 2003, 163, 1101–1107. [Google Scholar] [CrossRef]
- Zhang, D.; Guo, S.; Schrodi, S.J. Mechanisms of DNA Methylation in Virus-Host Interaction in Hepatitis B Infection: Pathogenesis and Oncogenetic Properties. Int. J. Mol. Sci. 2021, 22, 9858. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelly, R.; Galle, P.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef]
- Arvanitakis, K.; Mitroulis, I.; Chatzigeorgiou, A.; Elefsiniotis, I.; Germanidis, G. The Liver Cancer Immune Microenvironment: Emerging Concepts for Myeloid Cell Profiling with Diagnostic and Therapeutic Implications. Cancers 2023, 15, 1522. [Google Scholar] [CrossRef]
- Akhoon, N. Precision Medicine: A New Paradigm in Therapeutics. Int. J. Prev. Med. 2021, 12, 12. [Google Scholar] [CrossRef]
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef]
- Brown, Z.J.; Patwardhan, S.; Bean, J.; Pawlik, T.M. Molecular diagnostics and biomarkers in cholangiocarcinoma. Surg. Oncol. 2022, 44, 101851. [Google Scholar] [CrossRef] [PubMed]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An Introduction to the Performance of Immunohistochemistry. Methods Mol. Biol. 2019, 1897, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Yang, P.-C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Mettman, D.; Saeed, A.; Shold, J.; Laury, R.; Ly, A.; Khan, I.; Golem, S.; Olyaee, M.; O’Neil, M. Refined pancreatobiliary UroVysion criteria and an approach for further optimization. Cancer Med. 2021, 10, 5725–5738. [Google Scholar] [CrossRef]
- Barr Fritcher, E.G.; Voss, J.S.; Brankley, S.M.; Campion, M.B.; Jenkins, S.M.; Keeney, M.E.; Henry, M.; Kerr, S.; Chaiteerakij, R.; Pestova, E.; et al. An Optimized Set of Fluorescence In Situ Hybridization Probes for Detection of Pancreatobiliary Tract Cancer in Cytology Brush Samples. Gastroenterology 2015, 149, 1813–1824.e1. [Google Scholar] [CrossRef]
- Patrinos, G.P.; Danielson, P.B.; Ansorge, W.J. Molecular Diagnostics: Past, Present, and Future. In Molecular Diagnostics, 3rd ed.; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Khemlina, G.; Ikeda, S.; Kurzrock, R. The biology of Hepatocellular carcinoma: Implications for genomic and immune therapies. Mol. Cancer 2017, 16, 149. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.-C.; Villanueva, A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin. Cancer. Res. 2015, 21, 1786–1788. [Google Scholar] [CrossRef]
- Bruix, J.; Han, K.-H.; Gores, G.; Llovet, J.M.; Mazzaferro, V. Liver cancer: Approaching a personalized care. J. Hepatol. 2015, 62, S144–S156. [Google Scholar] [CrossRef]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Ahn, S.-M.; Jang, S.J.; Shim, J.H.; Kim, D.; Hong, S.-M.; Sung, C.O.; Baek, D.; Haq, F.; Ansari, A.; Lee, S.; et al. Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014, 60, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Takai, A.; Dang, H.T.; Wang, X.W. Identification of drivers from cancer genome diversity in hepatocellular carcinoma. Int. J. Mol. Sci. 2014, 15, 11142–11160. [Google Scholar] [CrossRef]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-source consolidation of human biological pathways. Database 2015, 2015, bav006. [Google Scholar] [CrossRef]
- Schwaederlé, M.; Lazar, V.; Validire, P.; Hansson, J.; Lacroix, L.; Soria, J.-C.; Pawitan, Y.; Kurzrock, R. VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Antiangiogenesis Therapy. Cancer Res. 2015, 75, 1187–1190. [Google Scholar] [CrossRef]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 436. [Google Scholar] [CrossRef]
- Said, R.; Hong, D.S.; Warneke, C.L.; Lee, J.J.; Wheler, J.J.; Janku, F.; Naing, A.; Falchook, G.; Fu, S.; Piha-Paul, S.; et al. P53 mutations in advanced cancers: Clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget 2013, 4, 705–714. [Google Scholar] [CrossRef]
- Wong, C.H.; Wong, C.S.C.; Chan, S.L. Targeting angiogenic genes as a therapeutic approach for hepatocellular carcinoma. Curr. Gene. Ther. 2015, 15, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Gopal, P.; Singal, A.G. The changing landscape of hepatocellular carcinoma: Etiology, genetics, and therapy. Am. J. Pathol. 2014, 184, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Kogiso, T.; Nagahara, H.; Hashimoto, E.; Ariizumi, S.; Yamamoto, M.; Shiratori, K. Efficient induction of apoptosis by wee1 kinase inhibition in hepatocellular carcinoma cells. PLoS ONE 2014, 9, e100495. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.gov—Bethesda MNLoM: Clinicaltrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 20 July 2023).
- Boon, E.M.J.; Keller, J.J.; Wormhoudt, T.A.M.; Giardiello, F.M.; Offerhaus, G.J.A.; van der Neut, R.; Pals, S.T. Sulindac targets nuclear beta-catenin accumulation and Wnt signalling in adenomas of patients with familial adenomatous polyposis and in human colorectal cancer cell lines. Br. J. Cancer. 2004, 90, 224–229. [Google Scholar] [CrossRef]
- Huang, S.-M.A.; Mishina, Y.M.; Liu, S.; Cheung, A.; Stegmeier, F.; Michaud, G.A.; Charlat, O.; Wiellette, E.; Zhang, Y.; Wiessner, S.; et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009, 461, 614–620. [Google Scholar] [CrossRef]
- Llovet, J.M.; Decaens, T.; Raoul, J.-L.; Boucher, E.; Kudo, M.; Chang, C.; Kang, Y.; Assenat, E.; Lim, H.; Boige, V.; et al. Brivanib in patients with advanced hepatocellular carcinoma who were intolerant to sorafenib or for whom sorafenib failed: Results from the randomized phase III BRISK-PS study. J. Clin. Oncol. 2013, 31, 3509–3516. [Google Scholar] [CrossRef]
- Ho, H.K.; Yeo, A.H.L.; Kang, T.S.; Chua, B.T. Current strategies for inhibiting FGFR activities in clinical applications: Opportunities, challenges and toxicological considerations. Drug Discov. Today 2014, 19, 51–62. [Google Scholar] [CrossRef]
- Mukai, S.; Kanzaki, H.; Ogasawara, S.; Ishino, T.; Ogawa, K.; Nakagawa, M.; Fujiwara, K.; Unozawa, H.; Iwanaga, T.; Sakuma, T.; et al. Exploring microsatellite instability in patients with advanced hepatocellular carcinoma and its tumor microenvironment. JGH Open 2021, 5, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.E.; Puccini, A.; Grothey, A.; Raghavan, D.; Goldberg, R.M.; Xiu, J.; Korn, M.; Weinberg, B.; Hwang, J.; Shields, A.; et al. Landscape of Tumor Mutation Load, Mismatch Repair Deficiency, and PD-L1 Expression in a Large Patient Cohort of Gastrointestinal Cancers. Mol. Cancer Res. 2018, 16, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017, 8, 15180. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol. 2017, 2017, 1–15. [Google Scholar] [CrossRef]
- Mei, J.; Wang, R.; Xia, D.; Yang, X.; Zhou, W.; Wang, H.; Liu, C. BRCA1 Is a Novel Prognostic Indicator and Associates with Immune Cell Infiltration in Hepatocellular Carcinoma. DNA Cell Biol. 2020, 39, 1838–1849. [Google Scholar] [CrossRef]
- Chen, Z.-H.; Yu, Y.P.; Tao, J.; Liu, S.; Tseng, G.; Nalesnik, M.; Hamilton, R.; Bhargava, R.; Nelson, J.; Pennathur, A.; et al. MAN2A1-FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice. Gastroenterology 2017, 153, 1120–1132.e15. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, J.; Song, Y.; Gao, P.; Wang, X.; Chen, M.; Li, Y.; Zu, W. Roles of fusion genes in digestive system cancers: Dawn for cancer precision therapy. Crit. Rev. Oncol. Hematol. 2022, 171, 103622. [Google Scholar] [CrossRef]
- Toyota, A.; Goto, M.; Miyamoto, M.; Nagashima, Y.; Iwasaki, S.; Komatsu, T.; Momose, T.; Yoshida, K.; Tsukada, T.; Matsufuji, T.; et al. Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model. Biochem. Biophys. Res. Commun. 2022, 621, 157–161. [Google Scholar] [CrossRef]
- Pilarczyk, M.; Fazel-Najafabadi, M.; Kouril, M.; Shamsaei, B.; Vasiliauskas, J.; Niu, W.; Mahi, N.; Zhang, L.; Clark, N.; Ren, Y.; et al. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nat. Commun. 2022, 13, 4678. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, X.; Leung, K.-S.; Wong, M.-H.; Tsui, S.K.-W.; Cheng, L. meGPS: A multi-omics signature for hepatocellular carcinoma detection integrating methylome and transcriptome data. Bioinformatics 2022, 38, 3513–3522. [Google Scholar] [CrossRef]
- Gabbia, D.; De Martin, S. Tumor Mutational Burden for Predicting Prognosis and Therapy Outcome of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 3441. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wu, H.; Pan, T.; Zheng, X.; Yang, X.; Zhang, G.; Lian, Y.; Lin, J.; Peng, L. A novel panel based on immune infiltration and tumor mutational burden for prognostic prediction in hepatocellular carcinoma. Aging 2021, 13, 8563–8587. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.-X.; Zhu, Q.-G.; Zhang, S.-M.; Guan, L.; Li, T.; Zhang, L.; Wang, S.; Ren, W.; Chen, X.; Zhao, J.; et al. Precision medicine for hepatocellular carcinoma: Driver mutations and targeted therapy. Oncotarget 2017, 8, 55715–55730. [Google Scholar] [CrossRef]
- Low, K.C.; Tergaonkar, V. Telomerase: Central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013, 38, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Dratwa, M.; Wysoczańska, B.; Łacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929. [Google Scholar] [CrossRef]
- Ambrozkiewicz, F.; Trailin, A.; Červenková, L.; Vaclavikova, R.; Hanicinec, V.; Allah, M.A.O.; Palek, R.; Treska, V.; Daum, O.; Tonar, Z.; et al. CTNNB1 mutations, TERT polymorphism and CD8+ cell densities in resected hepatocellular carcinoma are associated with longer time to recurrence. BMC Cancer 2022, 22, 884. [Google Scholar] [CrossRef]
- Saretzki, G. Extra-telomeric functions of human telomerase: Cancer, mitochondria and oxidative stress. Curr. Pharm. Des. 2014, 20, 6386–6403. [Google Scholar] [CrossRef]
- Romaniuk, A.; Paszel-Jaworska, A.; Totoń, E.; Lisiak, N.; Hołysz, H.; Królak, A.; Grodecka-Fazdecka, S.; Rubis, B. The non-canonical functions of telomerase: To turn off or not to turn off. Mol. Biol. Rep. 2019, 46, 1401–1411. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Yu, J. Integrated Analysis of Altered lncRNA, circRNA, microRNA, and mRNA Expression in Hepatocellular Carcinoma Carrying TERT Promoter Mutations. J. Hepatocell. Carcinoma 2022, 9, 1201–1215. [Google Scholar] [CrossRef]
- Trung, N.T.; Hoan, N.X.; Trung, P.Q.; Binh, M.T.; Van Tong, H.; Toan, N.L.; Bang, M.; Song, L. Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci. Rep. 2020, 10, 8181. [Google Scholar] [CrossRef]
- Brosh, R.; Rotter, V. When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer 2009, 9, 701–713. [Google Scholar] [CrossRef]
- Woo, H.G.; Wang, X.W.; Budhu, A.; Kim, Y.H.; Kwon, S.M.; Tang, Z.-Y.; Sun, Z.; Harris, C.; Thorgeirsson, S. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011, 140, 1063–1070. [Google Scholar] [CrossRef]
- Liu, J.; Ma, Q.; Zhang, M.; Wang, X.; Zhang, D.; Li, W.; Wang, F.; Wu, E. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: Evidence from a systematic review and meta-analysis. Eur. J. Cancer 2012, 48, 2328–2338. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-C.; Liu, T.-P.; Andriani, V.; Athoillah, M.; Wang, C.-Y.; Yang, P.-M. Bioinformatics Analysis Identifies Precision Treatment with Paclitaxel for Hepatocellular Carcinoma Patients Harboring Mutant TP53 or Wild-Type CTNNB1 Gene. J. Pers. Med. 2021, 11, 1199. [Google Scholar] [CrossRef]
- Yang, C.; Huang, X.; Li, Y.; Chen, J.; Lv, Y.; Dai, S. Prognosis and personalized treatment prediction in TP53-mutant hepatocellular carcinoma: An in silico strategy towards precision oncology. Brief. Bioinform. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Corda, G.; Sala, A. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis 2017, 6, e364. [Google Scholar] [CrossRef]
- Daniels, D.L.; Weis, W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005, 12, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xu, Z.; Zhang, Y.; Evert, M.; Calvisi, D.F.; Chen, X. β-Catenin signaling in hepatocellular carcinoma. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Monga, S.P. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef]
- Bugter, J.M.; Fenderico, N.; Maurice, M.M. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat. Rev. Cancer 2021, 21, 5–21. [Google Scholar] [CrossRef]
- Lu, D.; Choi, M.Y.; Yu, J.; Castro, J.E.; Kipps, T.J.; Carson, D.A. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13253–13257. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; He, L.; Dai, W.-Q.; Xu, Y.-P.; Wu, D.; Lin, C.-L.; Wu, S.; Cheng, P.; Zhang, Y.; Shen, M.; et al. Salinomycin inhibits proliferation and induces apoptosis of human hepatocellular carcinoma cells in vitro and in vivo. PLoS ONE 2012, 7, e50638. [Google Scholar] [CrossRef] [PubMed]
- Mullen, J.; Kato, S.; Sicklick, J.K.; Kurzrock, R. Targeting ARID1A mutations in cancer. Cancer Treat. Rev. 2021, 100, 102287. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Ui, A.; Kanno, S.-I.; Ogiwara, H.; Nagase, T.; Kohno, T.; Yasui, A. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014, 74, 2465–2475. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Peng, Y.; Wei, L.; Zhang, W.; Yang, L.; Lan, L.; Kapoor, P.; Ju, Z.; Mo, Q.; Shih, I.; et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov. 2015, 5, 752–767. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, G.; Ouyang, X.; Zai, D.; Zhou, J.; Li, X.; Zhang, Q.; Zhao, J. Loss of ARID1A Promotes Hepatocellular Carcinoma Progression via Up-regulation of MYC Transcription. J. Clin. Transl. Hepatol. 2021, 9, 528–536. [Google Scholar] [CrossRef]
- Abdel-Moety, A.; Baddour, N.; Salem, P.; Rady, A.; El-Shendidi, A. ARID1A expression in hepatocellular carcinoma and relation to tumor recurrence after microwave ablation. Clin. Exp. Hepatol. 2022, 8, 49–59. [Google Scholar] [CrossRef]
- Luo, J.-P.; Wang, J.; Huang, J.-H. CDKN2A is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20211103. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, M.; Fu, B.M. Inhibition of endothelial nitric oxide synthase decreases breast cancer cell MDA-MB-231 adhesion to intact microvessels under physiological flows. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H1735–H1747. [Google Scholar] [CrossRef]
- Qin, L.-X. Inflammatory immune responses in tumor microenvironment and metastasis of hepatocellular carcinoma. Cancer Microenviron. 2012, 5, 203–209. [Google Scholar] [CrossRef]
- Keenan, B.P.; Fong, L.; Kelley, R.K. Immunotherapy in hepatocellular carcinoma: The complex interface between inflammation, fibrosis, and the immune response. J. Immunother. Cancer 2019, 7, 267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hong, R.; Li, L.; Wang, Y.; Du, P.; Ou, Y.; Zhao, Z.; Liu, X.; Xiao, W.; Dong, D.; et al. The chromosome 11q13.3 amplification associated lymph node metastasis is driven by miR-548k through modulating tumor microenvironment. Mol. Cancer 2018, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Alao, J.P. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol. Cancer 2007, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Wang, Y.; Zhang, H. CCND1 silencing suppresses liver cancer stem cell differentiation and overcomes 5-Fluorouracil resistance in hepatocellular carcinoma. J. Pharmacol. Sci. 2020, 143, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Duda, D.G.; Sahani, D.V.; Jain, R.K. HCC and angiogenesis: Possible targets and future directions. Nat. Rev. Clin. Oncol. 2011, 8, 292–301. [Google Scholar] [CrossRef]
- Zhu, A.X.; Park, J.O.; Ryoo, B.-Y.; Yen, C.-J.; Poon, R.; Pastorelli, D.; Blanc, J.; Chung, H.; Baron, A.; Pfiffer, T.; et al. Ramucirumab versus placebo as second-line treatment in patients with advanced hepatocellular carcinoma following first-line therapy with sorafenib (REACH): A randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2015, 16, 859–870. [Google Scholar] [CrossRef]
- Xiang, Q.; Chen, W.; Ren, M.; Wang, J.; Zhang, H.; Deng, D.Y.B.; Zhang, L.; Shang, C.; Chen, Y. Cabozantinib suppresses tumor growth and metastasis in hepatocellular carcinoma by a dual blockade of VEGFR2 and MET. Clin. Cancer Res. 2014, 20, 2959–2970. [Google Scholar] [CrossRef]
- Morse, M.A.; Sun, W.; Kim, R.; He, A.R.; Abada, P.B.; Mynderse, M.; Finn, R. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 912–920. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Z.; Peng, Y.; Li, D.; Jiang, Y.; Pan, F.; Li, Y.; Lai, Y.; Cui, Z.; Zhang, K.; et al. Correlationship between Ki67, VEGF, and p53 and Hepatocellular Carcinoma Recurrence in Liver Transplant Patients. Biomed. Res. Int. 2021, 2021, 6651397. [Google Scholar] [CrossRef]
- Lacin, S.; Yalcin, S. The Prognostic Value of Circulating VEGF-A Level in Patients with Hepatocellular Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820971677. [Google Scholar] [CrossRef]
- Yu, J.-H.; Kim, J.M.; Kim, J.K.; Choi, S.J.; Lee, K.S.; Lee, J.-W.; Chang, H.; Lee, J. Platelet-derived growth factor receptor α in hepatocellular carcinoma is a prognostic marker independent of underlying liver cirrhosis. Oncotarget 2017, 8, 39534–39546. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.S.; Hughes, S.D.; Gilbertson, D.G.; Palmer, T.E.; Holdren, M.S.; Haran, A.C.; Odell, M.; Bauer, R.; Ren, H.; Haugen, H.; et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 3389–3394. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Yamakuchi, M.; Shimizu, T.; Kadono, J.; Furoi, A.; Gejima, K.; Komokata, T.; Koriyama, C.; Hashiguchi, T.; Imoto, Y. Predictive Value of Diminished Serum PDGF-BB after Curative Resection of Hepatocellular Cancer. J. Oncol. 2019, 2019, 1925315. [Google Scholar] [CrossRef]
- Tacher, V.; Lin, M.; Duran, R.; Yarmohammadi, H.; Lee, H.; Chapiro, J.; Chao, M.; Wang, Z.; Frangakis, C.; Sohn, J.; et al. Comparison of Existing Response Criteria in Patients with Hepatocellular Carcinoma Treated with Transarterial Chemoembolization Using a 3D Quantitative Approach. Radiology 2016, 278, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Galun, D.; Mijac, D.; Filipovic, A.; Bogdanovic, A.; Zivanovic, M.; Masulovic, D. Precision Medicine for Hepatocellular Carcinoma: Clinical Perspective. J. Pers. Med. 2022, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Ziv, E.; Zhang, Y.; Kelly, L.; Nikolovski, I.; Boas, F.E.; Erinjeri, J.P.; Cai, L.; Petre, E.; Brody, L.; Covey, A.; et al. NRF2 Dysregulation in Hepatocellular Carcinoma and Ischemia: A Cohort Study and Laboratory Investigation. Radiology 2020, 297, 225–234. [Google Scholar] [CrossRef]
- Martin, S.P.; Fako, V.; Dang, H.; Dominguez, D.A.; Khatib, S.; Ma, L.; Wang, H.; Zheng, W.; Wang, X. PKM2 inhibition may reverse therapeutic resistance to transarterial chemoembolization in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 99. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.; Schreiber, S.; Cantley, L. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef]
- Allaire, M.; Goumard, C.; Lim, C.; Le Cleach, A.; Wagner, M.; Scatton, O. New frontiers in liver resection for hepatocellular carcinoma. JHEP Rep. 2020, 2, 100134. [Google Scholar] [CrossRef]
- European Association For The Study Of The Liver, European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef]
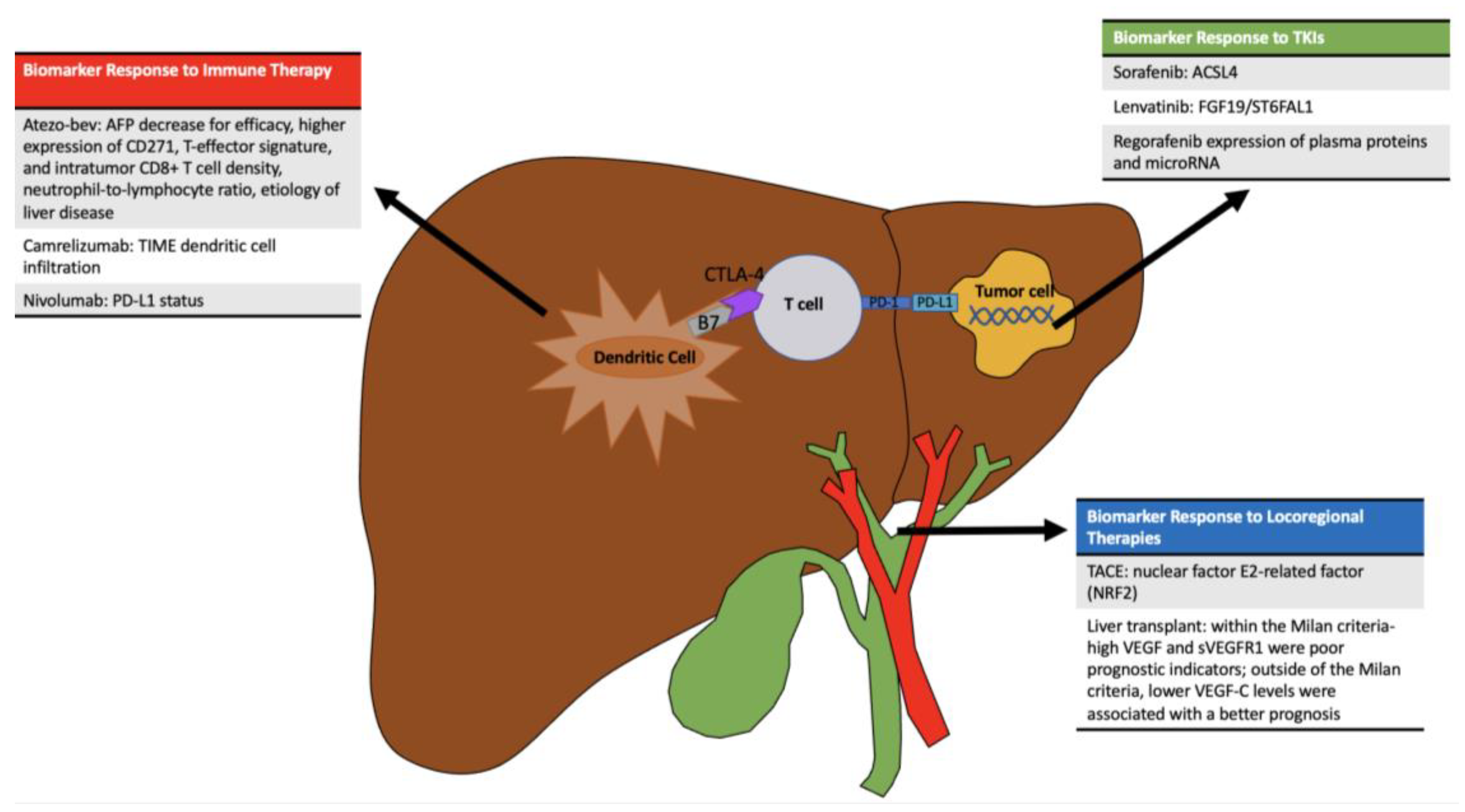
- Duda, D.G.; Dima, S.O.; Cucu, D.; Sorop, A.; Klein, S.; Ancukiewicz, M.; Kitahara, S.; Iacob, S.; Bacalbasa, N.; Tomescu, D.; et al. Potential Circulating Biomarkers of Recurrence after Hepatic Resection or Liver Transplantation in Hepatocellular Carcinoma Patients. Cancers 2020, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Pommergaard, H.C.; Yde, C.W.; Ahlborn, L.B.; Andersen, C.L.; Henriksen, T.V.; Hasselby, J.P.; Rostved, A.; Sorensen, C.; Rohrberg, K.; Nielsen, F.; et al. Personalized circulating tumor DNA in patients with hepatocellular carcinoma: A pilot study. Mol. Biol. Rep. 2022, 49, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Pinyol, R.; Montal, R.; Bassaganyas, L.; Sia, D.; Takayama, T.; Chau, G.-Y.; Mazzaferro, V.; Roayaie, S.; Lee, H.; Kokudo, N.; et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut 2019, 68, 1065–1075. [Google Scholar] [CrossRef]
- Huang, A.; Yang, X.-R.; Chung, W.-Y.; Dennison, A.R.; Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target. Ther. 2020, 5, 146. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Lim, H.; Kudo, M.; Breder, V.; Merle, P.; et al. Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J. Hepatol. 2022, 76, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Chan, S.L.; Kudo, M.; Lau, G.; Kelley, R.K.; Furuse, J.; Sukeepaisarnjaroen, W.; Kan, Y.; Dao, T.; De Toni, E.; et al. Phase 3 randomized, open-label, multicenter study of tremelimumab (T) and durvalumab (D) as first-line therapy in patients (pts) with unresectable hepatocellular carcinoma (uHCC): HIMALAYA. J. Clin. Oncol. 2022, 40, 379. [Google Scholar] [CrossRef]
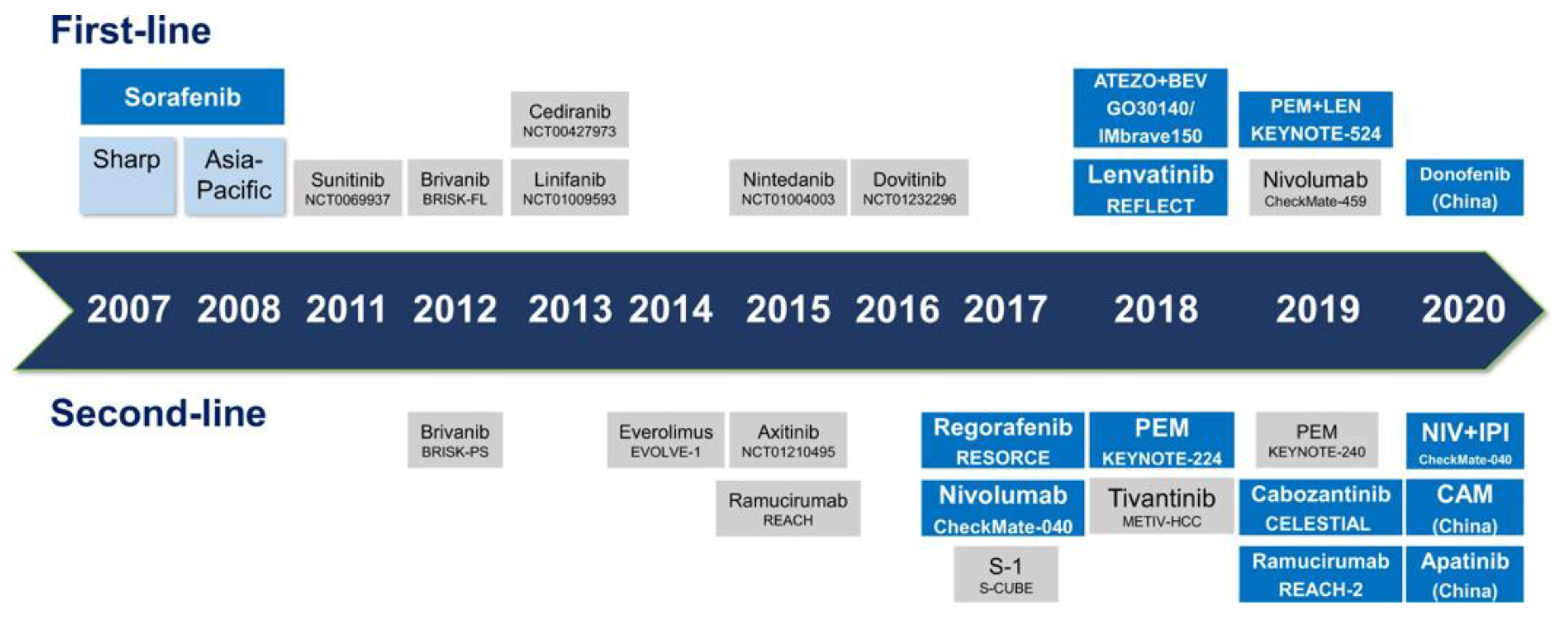
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.; Santoro, A.; Raoul, J.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Kang, Y.-K.; Chen, Z.; Tsao, C.-J.; Qin, S.; Kim, J.S.; Luo, R.; Feng, J.; Ye, S.; Yang, T.; et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009, 10, 25–34. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Alsina, A.; Kudo, M.; Vogel, A.; Cheng, A.-L.; Tak, W.Y.; Ryoo, B.-Y.; Evans, T.; Lopez Lopez, C.; Daniele, C.; Misir, R.; et al. Effects of Subsequent Systemic Anticancer Medication Following First-Line Lenvatinib: A Post Hoc Responder Analysis from the Phase 3 REFLECT Study in Unresectable Hepatocellular Carcinoma. Liver Cancer 2020, 9, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Verset, G.; Borbath, I.; Karwal, M.; Verslype, C.; Van Vlierberghe, H.; Kardosh, A.; Zagonel, V.; Stal, P.; Sarker, D.; Palmer, D.; et al. Pembrolizumab Monotherapy for Previously Untreated Advanced Hepatocellular Carcinoma: Data from the Open-Label, Phase II KEYNOTE-224 Trial. Clin. Cancer Res. 2022, 28, 2547–2554. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, A.; Fulgenzi, C.A.M.; Nishida, N.; Schönlein, M.; von Felden, J.; Schulze, K.; Wege, H.; Gaillard, V.; Saeed, A.; Wietharn, B.; et al. Preliminary evidence of safety and tolerability of atezolizumab plus bevacizumab in patients with hepatocellular carcinoma and Child-Pugh A and B cirrhosis: A real-world study. Hepatology 2022, 76, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Schenker, M.; Burotto, M.; Richardet, M.; Ciuleanu, T.; Goncalves, A.; Steeghs, N.; Schoffski, P.; Ascierto, P.; Maio, M.; Lugowska, I.; et al. Abstract CT022: CheckMate 848: A randomized, open-label, phase 2 study of nivolumab in combination with ipilimumab or nivolumab monotherapy in patients with advanced or metastatic solid tumors of high tumor mutational burden. Cancer Res. 2022, 82, CT022. [Google Scholar] [CrossRef]
- Bruix, J.; Qin, S.; Merle, P.; Granito, A.; Huang, Y.-H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Breder, V.; et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 389, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Meyer, T.; Cheng, A.-L.; El-Khoueiry, A.B.; Rimassa, L.; Ryoo, B.-Y.; Cicin, I.; Merle, P.; Chen, Y.; Park, J.; et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N. Engl. J. Med. 2018, 379, 54–63. [Google Scholar] [CrossRef]
- Yau, T.; Kang, Y.-K.; Kim, T.-Y.; El-Khoueiry, A.B.; Santoro, A.; Sangro, B.; Melero, I.; Kudo, M.; Hou, M.; Matilla, A.; et al. Efficacy and Safety of Nivolumab Plus Ipilimumab in Patients with Advanced Hepatocellular Carcinoma Previously Treated With Sorafenib: The CheckMate 040 Randomized Clinical Trial. JAMA Oncol. 2020, 6, e204564. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.-Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients with Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Qin, S.; Chen, Z.; Fang, W.; Ren, Z.; Xu, R.; Ryoo, B.-Y.; Meng, Z.; Bahi, Y.; Chen, X.; Liu, X.; et al. Pembrolizumab Versus Placebo as Second-Line Therapy in Patients from Asia With Advanced Hepatocellular Carcinoma: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2023, 41, 1434–1443. [Google Scholar] [CrossRef]
- Berton, D.; Banerjee, S.; Curigliano, G.; Cresta, S.; Arkenau, H.T.; Abdeddaim, C.; Kristeleit, R.; Redondo, A.; Leath, C.; Torres, A.; et al. Antitumor activity of dostarlimab in patients with mismatch repair-deficient/microsatellite instability-high tumors: A combined analysis of two cohorts in the GARNET study [abstract]. J. Linical Oncol. 2021, 39, 2564. [Google Scholar] [CrossRef]
- Subbiah, V.; Wolf, J.; Konda, B.; Kang, H.; Spira, A.; Weiss, J.; Takeda, M.; Ohe, Y.; Khan, S.; Ohashi, K.; et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): A phase 1/2, open-label, basket trial. Lancet Oncol. 2022, 23, 1261–1273. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Dayyani, F.; Yen, C.-J.; Ren, Z.; Bai, Y.; Meng, Z.; Pan, H.; Dillon, P.; Mhatre, S.; Gaillard, V.; et al. Alpha-Fetoprotein as a Potential Surrogate Biomarker for Atezolizumab + Bevacizumab Treatment of Hepatocellular Carcinoma. Clin. Cancer Res. 2022, 28, 3537–3545. [Google Scholar] [CrossRef]
- Zhu, A.X.; Abbas, A.R.; de Galarreta, M.R.; Guan, Y.; Lu, S.; Koeppen, H.; Zhang, W.; Hsu, C.; He, A.; Ryoo, B.; et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat. Med. 2022, 28, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Chon, Y.E.; Cheon, J.; Kim, H.; Kang, B.; Ha, Y.; Kim, D.Y.; Hwang, S.; Chon, H.; Kim, B. Predictive biomarkers of survival in patients with advanced hepatocellular carcinoma receiving atezolizumab plus bevacizumab treatment. Cancer Med. 2023, 12, 2731–2738. [Google Scholar] [CrossRef]
- Myojin, Y.; Kodama, T.; Maesaka, K.; Motooka, D.; Sato, Y.; Tanaka, S.; Abe, Y.; Ohkawa, K.; Mita, E.; Hayashi, Y.; et al. ST6GAL1 Is a Novel Serum Biomarker for Lenvatinib-Susceptible FGF19-Driven Hepatocellular Carcinoma. Clin. Cancer Res. 2021, 27, 1150–1161. [Google Scholar] [CrossRef]
- Kim, C.M.; Hwang, S.; Keam, B.; Yu, Y.S.; Kim, J.H.; Kim, D.-S.; Bae, S.; Kim, G.; Lee, J.; Seo, Y.; et al. Gene Signature for Sorafenib Susceptibility in Hepatocellular Carcinoma: Different Approach with a Predictive Biomarker. Liver Cancer 2020, 9, 182–192. [Google Scholar] [CrossRef]
- Feng, J.; Lu, P.-Z.; Zhu, G.-Z.; Hooi, S.C.; Wu, Y.; Huang, X.-W.; Dai, H.; Chen, P.; Li, Z.; Su, W.; et al. ACSL4 is a predictive biomarker of sorafenib sensitivity in hepatocellular carcinoma. Acta Pharmacol. Sin. 2021, 42, 160–170. [Google Scholar] [CrossRef]
- Rimassa, L.; Kelley, R.K.; Meyer, T.; Ryoo, B.-Y.; Merle, P.; Park, J.-W.; Blanc, J.; Lim, H.; Tran, A.; Chan, Y.; et al. Outcomes Based on Plasma Biomarkers for the Phase 3 CELESTIAL Trial of Cabozantinib versus Placebo in Advanced Hepatocellular Carcinoma. Liver Cancer 2022, 11, 38–47. [Google Scholar] [CrossRef]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schütz, G.; Thierauch, K.; Zopf, D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef]
- Teufel, M.; Seidel, H.; Köchert, K.; Meinhardt, G.; Finn, R.S.; Llovet, J.M.; Bruix, J. Biomarkers Associated with Response to Regorafenib in Patients With Hepatocellular Carcinoma. Gastroenterology 2019, 156, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Tang, W.; Qian, X.; Li, X.; Cheng, F.; Wang, K.; Zhang, F.; Zhang, C.; Li, D.; Song, J.; et al. Efficacy and safety of camrelizumab plus apatinib during the perioperative period in resectable hepatocellular carcinoma: A single-arm, open label, phase II clinical trial. J. Immunother. Cancer 2022, 10, e004656. [Google Scholar] [CrossRef] [PubMed]
- Sangro, B.; Melero, I.; Wadhawan, S.; Finn, R.S.; Abou-Alfa, G.K.; Cheng, A.L.; Yau, T.; Furuse, J.; Park, J.; Boyd, Z.; et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J. Hepatol. 2020, 73, 1460–1469. [Google Scholar] [CrossRef]
- Sun, J.; Althoff, K.N.; Jing, Y.; Horberg, M.A.; Buchacz, K.; Gill, M.J.; Justica, A.; Rabkin, C.; Goedert, J.; Sigel, K.; et al. Trends in Hepatocellular Carcinoma Incidence and Risk Among Persons With HIV in the US and Canada, 1996–2015. JAMA Netw. Open 2021, 4, e2037512. [Google Scholar] [CrossRef] [PubMed]
| Material | Methods | Examples |
|---|---|---|
| DNA | Sequencing—Process of determination of the consistent nucleotides of the DNA. First popularized by Fred Sanger, latest techniques called next-generation sequencing (NGS) run millions of these reactions simultaneously, making sequencing faster and cheaper. | Used in exploratory studies and miRNA detection |
| DNA probes—Detect specific DNA sequences. They are often tagged with fluorescent markers, which transmit a signal. | Tailored as per need | |
| DNA microarray—Consists of numerous DNA probes arranged in rows and columns on a small glass surface. Allows for detection of multiple sequences at the same time—so-called ‘high-throughput’ analysis. This allows chip-based detection of multiple variations of the same mutation. | GeneChip® | |
| Fluorescence in situ hybridization (FISH)—Allows for visualization of the presence and location of specific NDA mutations. These are seen under a fluorescent microscope. | Pancreatobiliary FISH by UroVision | |
| Polymerase chain reaction (PCR)—Revolutionary technique that produces millions of copies of the desired DNA fragment, which can be detected; nowadays, real-time PCR involves simultaneous amplification and detection, making the entire process faster. | The Cobas® KRAS Mutation Test | |
| Comparative genome hybridization—Provides an overall picture of chromosomal gains and losses throughout the whole genome of the tumor. | Array-based CGH | |
| Liquid biopsy—Laboratory testing of bodily fluid samples, including blood or urine, allowing for detection of circulating tumor cells, circulating tumor DNA, cell-free DNA, circulating miRNA, and exosomes. Multiple non-invasive samples may be taken over time, allowing for potential detection, treatment response, and surveillance for disease recurrence. | ||
| RNA | Gene expression testing—These tests study mRNA in the cells to determine activity of different genes. | MammaPrint®, Oncotype DX® Breast |
| Reverse transcriptase PCR—Reverse transcriptase is an enzyme that converts RNA into DNA, which is then detected by conventional PCR. | Detection of specific miRNAs | |
| Protein | Immunohistochemistry—Uses antibodies to identify specific proteins. Can provide quantitative and qualitative results. | Pathway Anti-Her2/NEU (4B5) Rabbit Monoclonal Primary Antibody |
| Mass spectrometry (MS)—Process of volatilization and ionization of proteins and peptides followed by their detection based on their mass/charge ratio using a mass analyzer. MS may be coupled with liquid or gas chromatography to achieve better separation. | ||
| Nuclear magnetic resonance (NMR) spectroscopy uses a magnetic field and a radiofrequency pulse to measure organic and some inorganic compounds inside biological samples (as solid tissue or extracted metabolite). | ||
| Western blot (WB)—Proteins are separated based on molecular weight through gel electrophoresis, then transferred to a band-producing membrane, and the protein of interest is identified through labeled antibodies. |
| Gene | Aberration Frequency | Pathway | Function | Examples of Potential Targeted Agents |
|---|---|---|---|---|
| TERT promoter | 60% | Telomerase maintenance | Add telomere repeats (TTAFFF) onto chromosome ends, compensating for the erosion of protective telomeric ends that is a normal part of cell division. | |
| TP53 | Mutation: 3–40%; Loss: 2–15% | P53 pathway | Tumor suppressor TP53 gene regulates the expression of VEGF-A. Antiangiogenic agents were correlated with longer PFS in patients harboring PT53-mutant tumors. | Bevacizumab, ramucirumab, sorafenib, and Wee-1 inhibitors |
| CTNNB1 | Mutation: 11–41% | Wnt pathway | Regulates cell adhesion, growth, and differentiation. | BBI608, a potent small molecule inhibitor; PRI-724; and Sulindac |
| AXIN1 | 5–19% | Wnt pathway | Regulates cell adhesion, growth, and differentiation. | Small molecular inhibitor XAV939 |
| ARID1A | Mutation: 4–17% | Chromatin remodeling | Transcriptional activation and repression of selected genes via chromatin remodeling. | CDK4/6 inhibitor palbociclib |
| CDKN2A | Deletion: 7–8% | Cell cycle | Tumor suppressor gene promotes cell cycle arrest in G1 and G2 phases. Suppresses MDM2. | CDK4/6 inhibitor palbociclib |
| ARID2 | Mutation: 5–7% | Chromatin remodeling | Tumor suppressor gene with a role in the transcription, activation, and repression of selected genes. | CDK4/6 inhibitor palbociclib |
| RPS6KA3 | Mutation: 4–7% | Dual-function regulation of MPAK/ERK and mTOR signaling | Mediates stress-induced and mitogenic activation of transcription factors and cellular differentiation, proliferation, and survival. | CDK4/6 inhibitor palbociclib |
| CCND1 | Alterations (focal amplications or deletions): 4.7–7% | P53 pathway cell cycle | Functions as a regulatory subunit of CDK4 or CDK6, the activity of which is required for cell cycle progression. | Palbociclib |
| FGF3, FGF4, or FGF19 | Alterations (focal amplications or deletions): 4–5.6% | FGF pathway | FGF family members possess broad mitogenic and cell survival activities and are operative in tumor growth and invasion, as well as tissue repair. | Brivanib, BIBF 1120, dovitinib, and lenvatinib |
| Setting | Mechanism of Action | Evidence | |
|---|---|---|---|
| Atezolizumab + Bevacizumab | Preferred regimen (child class A only) Certain circumstances (child class B only) | Atezolizumab is a monoclonal antibody that binds PD-L1 Bevacizumab is a monoclonal antibody that inhibits angiogenesis by binding to circulating VEGF and interrupting its ability to bind to VEGFR | Atezolizumab + bevacizumab vs. sorafenib: median OS, 19.2 mo vs. 13.4 mo. (95% CI); PFS, 6.9 mo vs. 4.2 mo (95% CI) Atezolizumab + bevacizumab: median OS, 14.9 mo; median PFS, 6.8 mo (95% CI) |
| Tremelimumab-actl + Durvalumab | Preferred regimen | Tremelimumab is a monoclonal antibody that targets the activity of CTLA-4 Durvalumab is a monoclonal antibody that blocks the interaction of PD-L1 and CD80 | Tremelimumab + durvalumab vs. sorafenib: median OS, 16.43 mo vs. 16.56 mo (95% CI) |
| Sorafenib | Other recommended (child class A or B7 only) | A multikinase inhibitor that works to decrease angiogenesis through inhibition of VEGF receptors, PDGF, and raf kinase | Sorafenib vs. placebo: median OS, 10.7 mo vs. 7.9 (95% CI); TTRP, 5.5 mo vs. 2.8 mo Sorafenib vs. placebo in Asia-Pacific population: median OS, 6.5 mo vs. 4.2 mo (95% CI); PFS, 2.8 mo vs. 1.4 mo |
| Lenvatinib | Other recommended (child class A only) | A multikinase inhibitor including VEGF, fibroblast growth factor receptor (FGFR), PDGR, KIT, and RET | Lenvatinib vs. sorafenib: median OS, 13.6 mo vs. 12.3 mo (95% CI) Lenvatinib + subsequent anticancer rx. vs. sorafenib + subsequent anticancer rx: median OS, 25.7 mo vs. 22.3 mo (95% CI) |
| Durvalumab | Other recommended | A monoclonal antibody that blocks the interaction of PD-L1 and CD80 | |
| Pembrolizumab | Other recommended | A monoclonal antibody that binds PD-L1 | Monotherapy: median OS, 17 mo (95% CI); median PFS, 4 mo (95% CI) Pembrolizumab vs. placebo in pts previously treated with sorafenib: median OS, 13.9 mo vs. 10.6 mo (95% CI); median PFS, 3.0 mo vs. 2.8 mo (95% CI) Pembrolizumab vs. Placebo in pts previously treated with sorafenib or oxaliplatin-based chemotherapy: median OS, 14.6 mo vs. 13.0 mo (95% CI); median PFS, 2.6 vs. 2.3 mo (95% CI) |
| Nivolumab | Certain circumstances (child class B only) | A monoclonal antibody that binds PD-L1 | Nivolumab vs. sorafenib: median OS, 16.4 mo vs. 14.7 (95% CI) |
| Nivolumab + Ipilimumab | Certain circumstances (TMB-H tumors) | Nivolumab is a monoclonal antibody that binds PD-L1 Ipilimumab is a monoclonal antibody that binds CTLA-4 | Nivolumab + ipilimumab tTMB-H vs. bTMB-H: median OS, 14.5 mo vs. 8.5 mo (95% CI); median PFS, 4.1 mo vs. 2.8 mo (95% CI) Nivolumab + ipilimumab in pts previously treated with sorafenib + N Q2wks (arm A), N + I Q3wks (arm B), N Q3wks + I Q6wks, OR 32% arm A, 27% arm B, and 29% arm C |
| Second-Line Therapy | |||
| Regorafenib | Child class A only | A multikinase inhibitor including VEGF1/2/3, PDGFR, FGFR1, c-KIT, RAF1, BRAF, and RET. | Regorafenib vs. placebo after sorafenib use: median OS, 10.6 mo vs. 7.8 mo (95% CI) |
| Cabozantinib | Child class A only | A multikinase inhibitor including tyrosine kinase, c-MET, VEGFR, AXL, and RET | Cabozantinib vs. placebo: median OS, 10.2 mo vs. 8.0 mo (95% CI); median PFS, 5.2 mo vs. 1.9 mo (95% CI) |
| Lenvatinib | Child class A only | A multikinase inhibitor including VEGF1/2/3, PDGFR, FGFR1/2/3/4, c-KIT, and RET | Lenvatinib vs. sorafenib: median OS, 13.6 mo vs. 12.3 mo (95% CI) Lenvatinib + subsequent anticancer rx. vs. sorafenib + subsequent anticancer rx: median OS, 25.7 mo vs. 22.3 mo (95% CI) |
| Nivolumab + ipilimumab | Child class A only TMB-H tumors | Nivolumab is a monoclonal antibody that binds PD-L1 Ipilimumab is a monoclonal antibody that binds CTLA-4 | Nivolumab + ipilimumab tTMB-H vs. bTMB-H: median OS, 14.5 mo vs. 8.5 mo (95% CI); median PFS, 4.1 mo vs. 2.8 mo (95% CI) Nivolumab + ipilimumab in pts previously treated with sorafenib + N Q2wks (arm A), N + I Q3wks (arm B), N Q3wks + I Q6wks, OR 32% arm A, 27% arm B, and 29% arm C |
| Pembrolizumab | Child class A only | Immunoglobulin G1 monoclonal antibody that binds to VEGFR and inhibits angiogenesis by decreasing endothelial cell permeability, migration, and proliferation | Monotherapy: median OS, 17 mo (95% CI); median PFS, 4 mo (95% CI) Pembrolizumab vs. placebo in pts previously treated with sorafenib: median OS, 13.9 mo vs. 10.6 mo (95% CI); median PFS, 3.0 mo vs. 2.8 mo (95% CI) Pembrolizumab vs. placebo in pts previously treated with sorafenib or oxaliplatin-based chemotherapy: median OS, 14.6 mo vs. 13.0 mo (95% CI); median PFS, 2.6 vs. 2.3 mo (95% CI) |
| Ramucirumab | AFP>400 ng/mL and Child class A only | A VEGFR2 antagonist | Ramucirumab vs. placebo in pts previously treated with sorafenib: median OS, 8.5 vs. 7.3 mo (95% CI); median PFS, 3.7 mo vs. 2.8 mo (95% CI) |
| Nivolumab | Child class B only | A monoclonal antibody that binds PD-L1 | Nivolumab vs. sorafenib: median OS, 16.4 mo vs. 14.7 (95% CI) |
| Dostarlimab-gxly | MSI-H/dMMR tumors | A monoclonal antibody that binds PD-L1 | In pts with solid tumors and dMMR/MSI-H: ORR, 87% (95% CI) |
| Selpercatinib | RET gene-fusion-positive tumors | A kinase inhibitor including wild-type RET and mutated RET isoforms | In pts with RET fusion-positive advanced solid tumors in solid tumors other than non-small cell lung cancer and thyroid cancer: ORR, 43.9% (95% CI) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorji, L.; Brown, Z.J.; Pawlik, T.M. Mutational Landscape and Precision Medicine in Hepatocellular Carcinoma. Cancers 2023, 15, 4221. https://doi.org/10.3390/cancers15174221
Gorji L, Brown ZJ, Pawlik TM. Mutational Landscape and Precision Medicine in Hepatocellular Carcinoma. Cancers. 2023; 15(17):4221. https://doi.org/10.3390/cancers15174221
Chicago/Turabian StyleGorji, Leva, Zachary J. Brown, and Timothy M. Pawlik. 2023. "Mutational Landscape and Precision Medicine in Hepatocellular Carcinoma" Cancers 15, no. 17: 4221. https://doi.org/10.3390/cancers15174221
APA StyleGorji, L., Brown, Z. J., & Pawlik, T. M. (2023). Mutational Landscape and Precision Medicine in Hepatocellular Carcinoma. Cancers, 15(17), 4221. https://doi.org/10.3390/cancers15174221







