The Prognostic Value of ASPHD1 and ZBTB12 in Colorectal Cancer: A Machine Learning-Based Integrated Bioinformatics Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Data Processing
2.2. Patient’s Samples
2.3. DNA-Seq and Whole Exome Sequencing
2.4. Differential Gene Expression Analysis
2.5. Gene Set, Ontology, and Pathway Enrichment Analysis
2.6. Survival Analysis
2.7. Machine Learning Method
2.8. Computational Workflow
2.9. Protein–Protein Interaction (PPI) Network
2.10. Kaplan–Meier Survival Curve
2.11. Receiver Operating Characteristic (ROC) Curve Analysis
2.12. Quantitative Real-Time-PCR Validation
3. Results
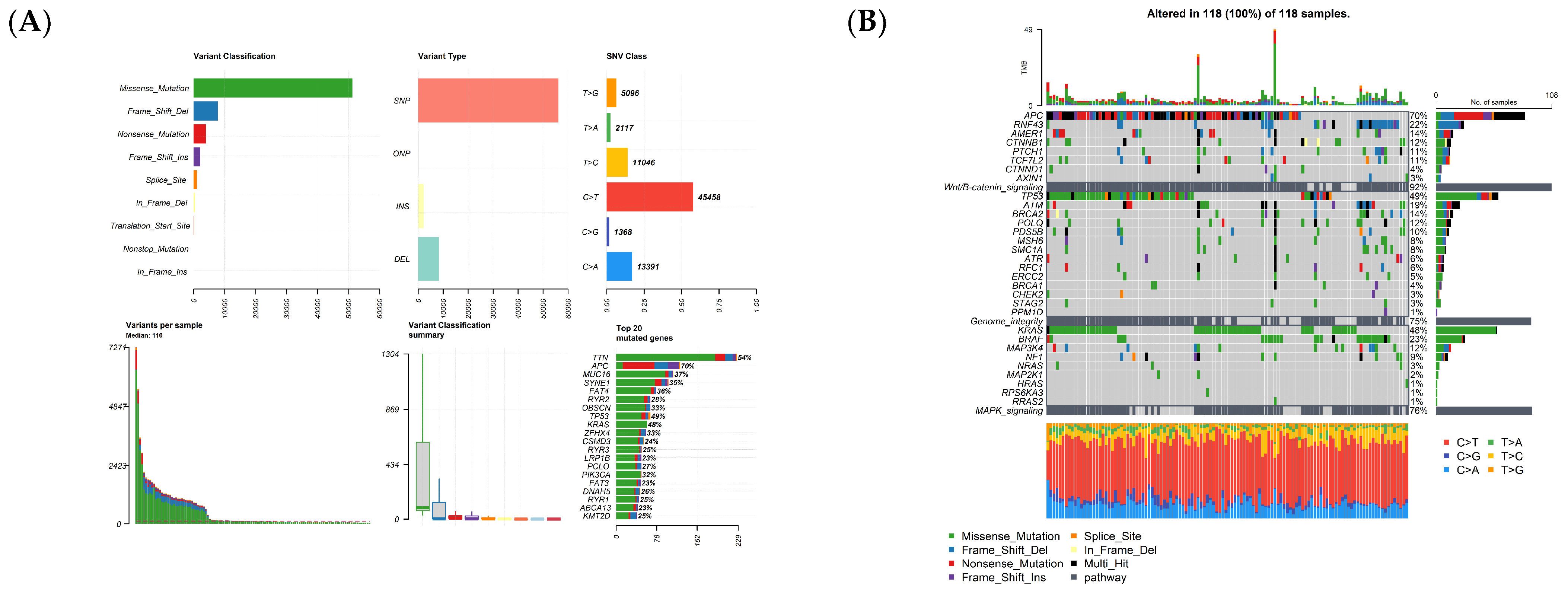
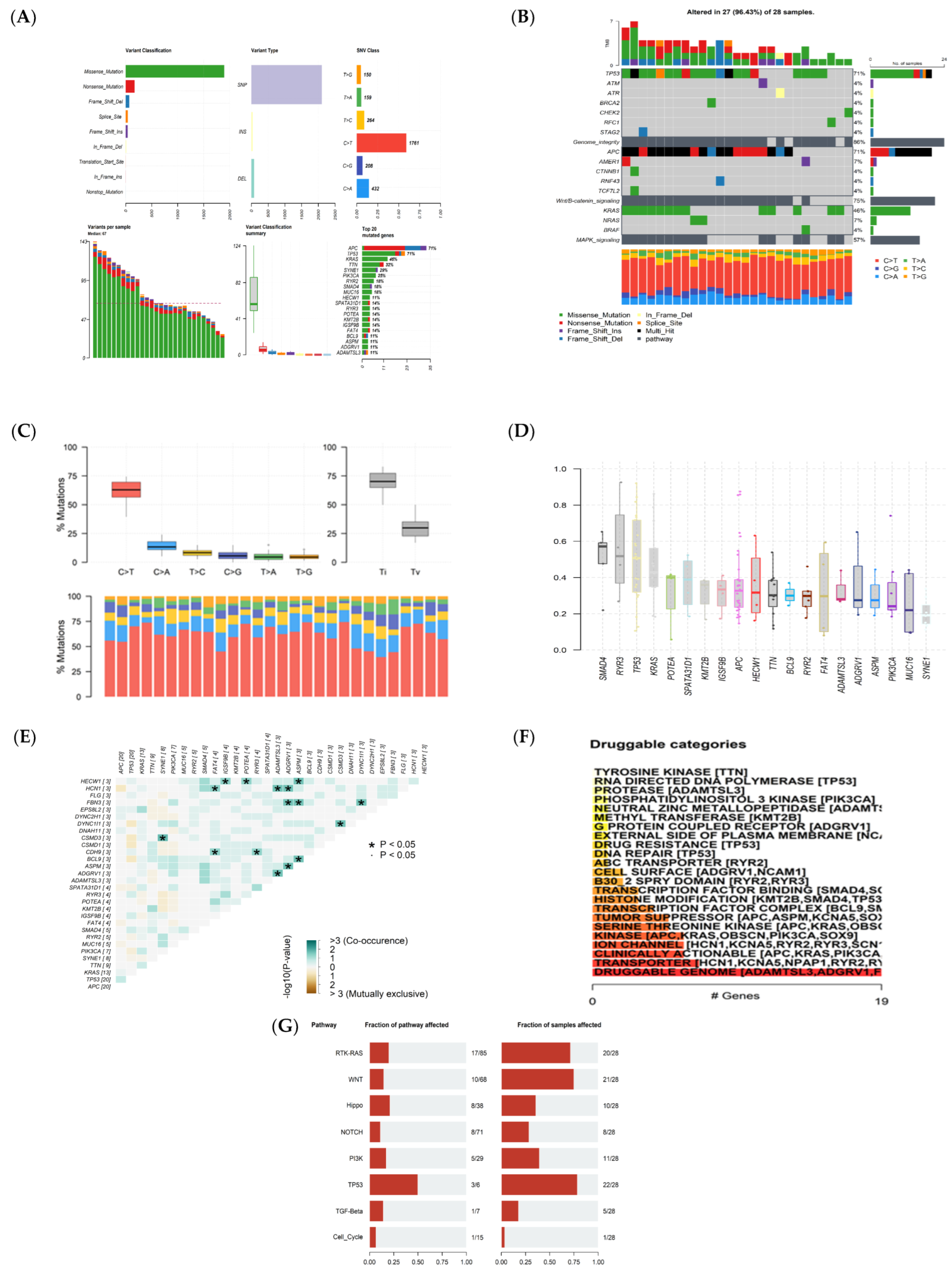
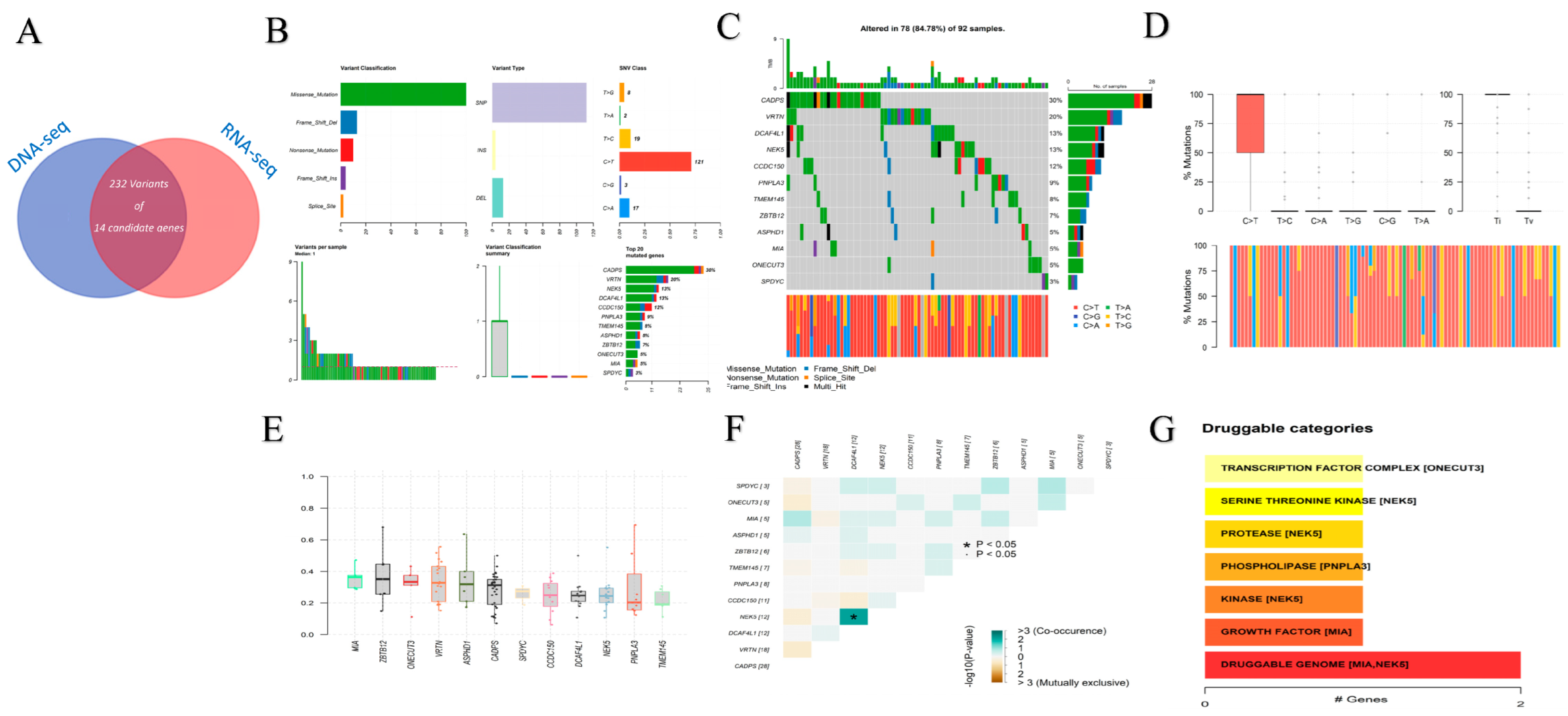
3.1. Whole Exome Sequencing
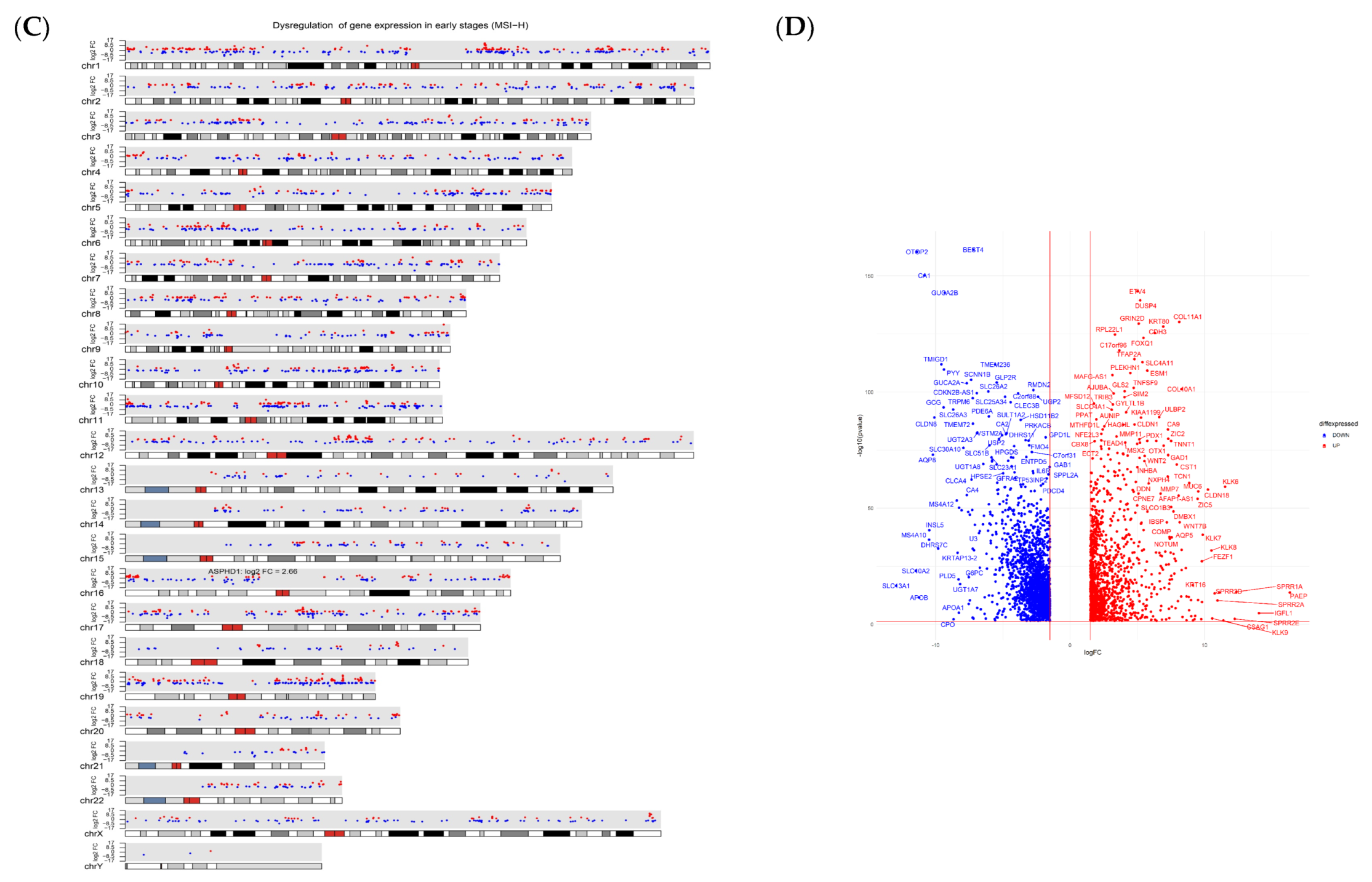
3.2. Gene Expression Profiling, Identification of DEGs, and Pathway Enrichment Analysis
3.3. Machine Learning Analysis
3.4. The Prognostic Value of ZBTB12 and ASPHD1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Yin, Z.; Yao, C.; Zhang, L.; Qi, S. Application of artificial intelligence in diagnosis and treatment of colorectal cancer: A novel Prospect. Front. Med. 2023, 10, 1128084. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Hammad, A.; Elshaer, M.; Tang, X. Identification of potential biomarkers with colorectal cancer based on bioinformatics analysis and machine learning. Math. Biosci. Eng. 2021, 18, 8997–9015. [Google Scholar] [CrossRef]
- Maurya, N.S.; Kushwaha, S.; Chawade, A.; Mani, A. Transcriptome profiling by combined machine learning and statistical R analysis identifies TMEM236 as a potential novel diagnostic biomarker for colorectal cancer. Sci. Rep. 2021, 11, 14304. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, L.; Weng, S.; Guo, C.; Dang, Q.; Xu, H.; Wang, L.; Lu, T.; Zhang, Y.; Sun, Z.; et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat. Commun. 2022, 13, 816. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Isella, C.; Terrasi, A.; Bellomo, S.E.; Petti, C.; Galatola, G.; Muratore, A.; Mellano, A.; Senetta, R.; Cassenti, A.; Sonetto, C.; et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015, 47, 312–319. [Google Scholar] [CrossRef]
- Wixted, J.T.; Mickes, L. ROC analysis measures objective discriminability for any eyewitness identification procedure. J. Appl. Res. Mem. Cogn. 2015, 4, 329–334. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.; Esteva, M.; Cabeza, E.; Llobera, J.; Ruiz, A. Lack of association between diagnostic and therapeutic delay and stage of colorectal cancer. Eur. J. Cancer 2008, 44, 510–521. [Google Scholar] [CrossRef]
- Gatalica, Z.; Torlakovic, E. Pathology of the hereditary colorectal carcinoma. Fam. Cancer 2008, 7, 15–26. [Google Scholar] [CrossRef]
- Bousis, D.; Verras, G.-I.; Bouchagier, K.; Antzoulas, A.; Panagiotopoulos, I.; Katinioti, A.; Kehagias, D.; Kaplanis, C.; Kotis, K.; Anagnostopoulos, C.-N. The role of deep learning in diagnosing colorectal cancer. Gastroenterol. Rev./Przegląd Gastroenterol. 2023, 18. [Google Scholar] [CrossRef]
- Khalili-Tanha, G.; Mohit, R.; Asadnia, A.; Khazaei, M.; Dashtiahangar, M.; Maftooh, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Kiani, M.A. Identification of ZMYND19 as a novel biomarker of colorectal cancer: RNA-sequencing and machine learning analysis. J. Cell Commun. Signal. 2023, 1–17. [Google Scholar] [CrossRef]
- Nazari, E.; Pourali, G.; Khazaei, M.; Asadnia, A.; Dashtiahangar, M.; Mohit, R.; Maftooh, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M. Identification of potential biomarkers in stomach adenocarcinoma using machine learning approaches. Curr. Bioinform. 2023, 18, 320–333. [Google Scholar] [CrossRef]
- Azari, H.; Nazari, E.; Mohit, R.; Asadnia, A.; Maftooh, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Shahidsales, S.; Khazaei, M. Machine learning algorithms reveal potential miRNAs biomarkers in gastric cancer. Sci. Rep. 2023, 13, 6147. [Google Scholar] [CrossRef]
- Wu, B.; Yang, J.; Qin, Z.; Yang, H.; Shao, J.; Shang, Y. Prognosis prediction of stage IV colorectal cancer patients by mRNA transcriptional profile. Cancer Med. 2022, 11, 4900–4912. [Google Scholar] [CrossRef]
- Chen, L.; Lu, D.; Sun, K.; Xu, Y.; Hu, P.; Li, X.; Xu, F. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene 2019, 692, 119–125. [Google Scholar] [CrossRef]
- Li, B.; Zhu, F.C.; Yu, S.X.; Liu, S.J.; Li, B.Y. Suppression of KIF22 Inhibits Cell Proliferation and Xenograft Tumor Growth in Colon Cancer. Cancer Biother. Radiopharm. 2020, 35, 50–57. [Google Scholar] [CrossRef]
- An, N.; Zhao, Y.; Lan, H.; Zhang, M.; Yin, Y.; Yi, C. SEZ6L2 knockdown impairs tumour growth by promoting caspase-dependent apoptosis in colorectal cancer. J. Cell Mol. Med. 2020, 24, 4223–4232. [Google Scholar] [CrossRef]
- Wang, L.; He, M.; Fu, L.; Jin, Y. Exosomal release of microRNA-454 by breast cancer cells sustains biological properties of cancer stem cells via the PRRT2/Wnt axis in ovarian cancer. Life Sci. 2020, 257, 118024. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.B.; Duan, C.Y.; Li, C.B.; Cui, L.; Ogino, S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: A systematic review and meta-analysis. Ann. Oncol. 2016, 27, 1836–1848. [Google Scholar] [CrossRef] [PubMed]
- Maffeis, V.; Nicolè, L.; Cappellesso, R. RAS, Cellular Plasticity, and Tumor Budding in Colorectal Cancer. Front. Oncol. 2019, 9, 1255. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, J.; Roiuk, M.; Neff, M.; Teleman, A.A. PRRC2 proteins impact translation initiation by promoting leaky scanning. Nucleic Acids Res. 2023, 51, 3391–3409. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Wang, Z.; Liu, L.; Zhang, G.; Li, J.; Ren, Z.; Dong, Z.; Yu, Z. PRRC2A Promotes Hepatocellular Carcinoma Progression and Associates with Immune Infiltration. J. Hepatocell. Carcinoma 2021, 8, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Nieters, A.; Conde, L.; Slager, S.L.; Brooks-Wilson, A.; Morton, L.; Skibola, D.R.; Novak, A.J.; Riby, J.; Ansell, S.M.; Halperin, E.; et al. PRRC2A and BCL2L11 gene variants influence risk of non-Hodgkin lymphoma: Results from the InterLymph consortium. Blood 2012, 120, 4645–4648. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hou, Q.; Wang, B.; Li, C.; Liu, L.; Gong, W.; Chai, J.; Guo, H. A novel mitochondria-related gene signature for controlling colon cancer cell mitochondrial respiration and proliferation. Hum. Cell 2022, 35, 1126–1139. [Google Scholar] [CrossRef]
- Chae, Y.S.; Lee, S.J.; Moon, J.H.; Kang, B.W.; Kim, J.G.; Sohn, S.K.; Jung, J.H.; Park, H.Y.; Park, J.Y.; Kim, H.J.; et al. VARS2 V552V variant as prognostic marker in patients with early breast cancer. Med. Oncol. 2011, 28, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef] [PubMed]
- Triner, D.; Castillo, C.; Hakim, J.B.; Xue, X.; Greenson, J.K.; Nuñez, G.; Chen, G.Y.; Colacino, J.A.; Shah, Y.M. Myc-Associated Zinc Finger Protein Regulates the Proinflammatory Response in Colitis and Colon Cancer via STAT3 Signaling. Mol. Cell Biol. 2018, 38, e00386-18. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Zhu, X.; Liu, W.; Ren, Y.; Bei, C.; Qin, L.; Miao, X.; Tang, F.; Tang, G.; Tan, S. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget 2016, 7, 86420–86432. [Google Scholar] [CrossRef]
- Ren, L.X.; Qi, J.C.; Zhao, A.N.; Shi, B.; Zhang, H.; Wang, D.D.; Yang, Z. Myc-associated zinc-finger protein promotes clear cell renal cell carcinoma progression through transcriptional activation of the MAP2K2-dependent ERK pathway. Cancer Cell Int. 2021, 21, 323. [Google Scholar] [CrossRef]
- Smits, M.; Wurdinger, T.; van het Hof, B.; Drexhage, J.A.; Geerts, D.; Wesseling, P.; Noske, D.P.; Vandertop, W.P.; de Vries, H.E.; Reijerkerk, A. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J. 2012, 26, 2639–2647. [Google Scholar] [CrossRef]
- He, J.; Wang, J.; Li, T.; Chen, K.; Li, S.; Zhang, S. SIPL1, Regulated by MAZ, Promotes Tumor Progression and Predicts Poor Survival in Human Triple-Negative Breast Cancer. Front. Oncol. 2021, 11, 766790. [Google Scholar] [CrossRef]
- Jiao, L.; Li, Y.; Shen, D.; Xu, C.; Wang, L.; Huang, G.; Chen, L.; Yang, Y.; Yang, C.; Yu, Y.; et al. The prostate cancer-up-regulated Myc-associated zinc-finger protein (MAZ) modulates proliferation and metastasis through reciprocal regulation of androgen receptor. Med. Oncol. 2013, 30, 570. [Google Scholar] [CrossRef]



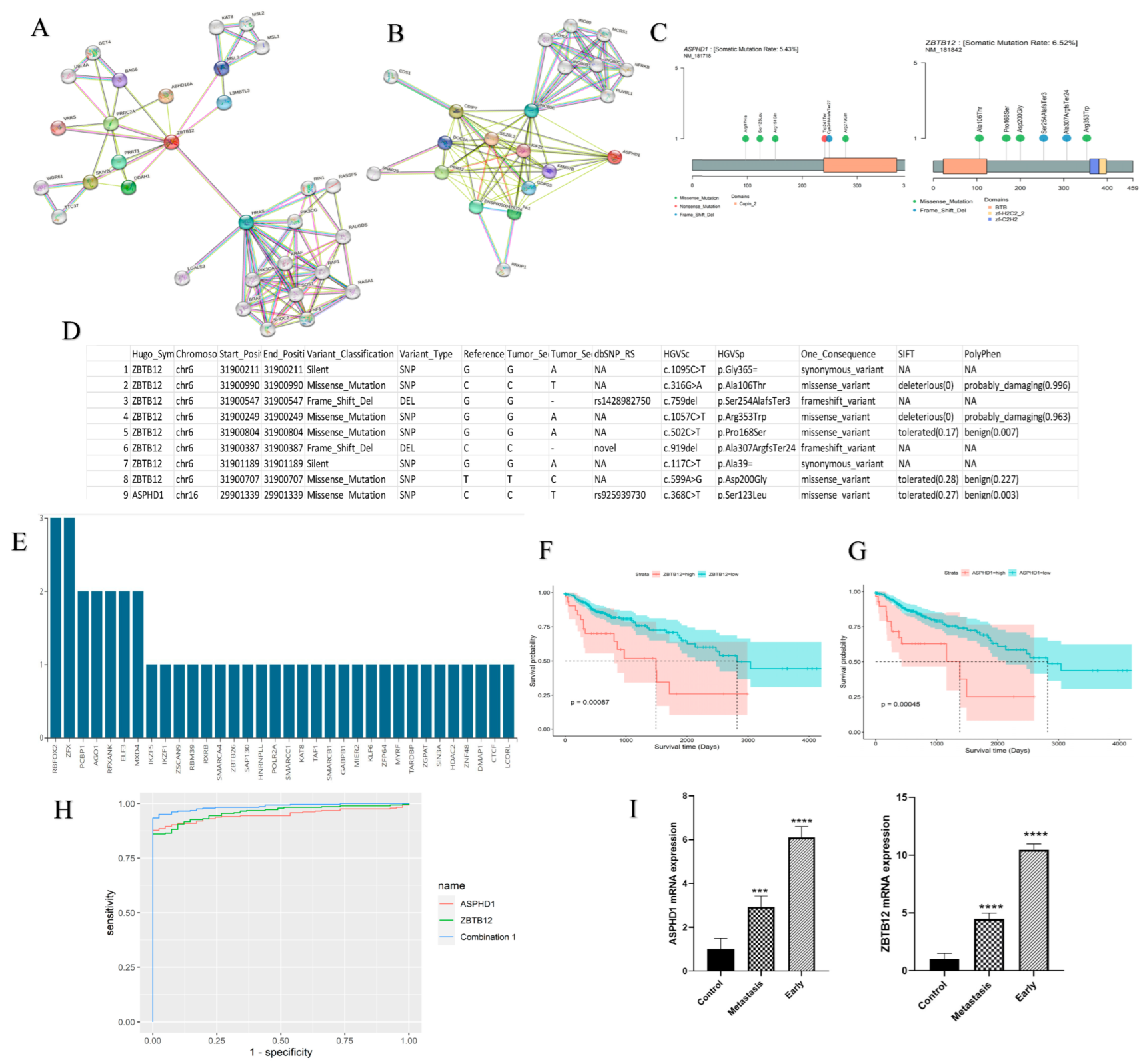
| Subgroups | R2 | AUC | MSE | RMSE | Accuracy | Prauc |
|---|---|---|---|---|---|---|
| MSI-H | 0.99 | 1.0 | 1.95 | 0.0044 | 97.14% | 1.0 |
| MSI-S | 0.99 | 1.0 | 0.0023 | 0.0489 | 97% | 1.0 |
| Receiving chemotherapy | 0.95 | 1.0 | 0.0076 | 0.0876 | 98% | 1.0 |
| Receiving targeted therapies | 0.64 | 0.88 | 0.0554 | 0.0235 | 92% | 0.95 |
| Biomarker | AUC | SE | SP | Cutoff | ACC | TN | TP | FN | FP | NPV | PPV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASPHD1 | 0.948 | 0.878 | 1 | 0.863 | 0.893 | 41 | 252 | 35 | 0 | 0.539 | 1 |
| ZBTB12 | 0.96 | 0.861 | 1 | 0.891 | 0.878 | 41 | 247 | 40 | . | 0.506 | 1 |
| Combination | 0.986 | 0.934 | 1 | 0.886 | 0.942 | 41 | 268 | 19 | . | 0.683 | 1 |
| Biomarker | Intercept | Coefficients | Degrees of Freedom | Null Deviance | Residual Deviance | AIC |
|---|---|---|---|---|---|---|
| ASPHD1 | −10.37 | Log (ASPHD1 + 1):3.032 | 327 | 247.2 | 136.3 | 140.3 |
| ZBTB12 | −22.345 | Log (ASPHD1 + 1):5.165 | 327 | 247.2 | 118.3 | 122.3 |
| Combination 1 | −36.814 | 5,6,2 | 327 | 247.2 | 63.99 | 69.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asadnia, A.; Nazari, E.; Goshayeshi, L.; Zafari, N.; Moetamani-Ahmadi, M.; Goshayeshi, L.; Azari, H.; Pourali, G.; Khalili-Tanha, G.; Abbaszadegan, M.R.; et al. The Prognostic Value of ASPHD1 and ZBTB12 in Colorectal Cancer: A Machine Learning-Based Integrated Bioinformatics Approach. Cancers 2023, 15, 4300. https://doi.org/10.3390/cancers15174300
Asadnia A, Nazari E, Goshayeshi L, Zafari N, Moetamani-Ahmadi M, Goshayeshi L, Azari H, Pourali G, Khalili-Tanha G, Abbaszadegan MR, et al. The Prognostic Value of ASPHD1 and ZBTB12 in Colorectal Cancer: A Machine Learning-Based Integrated Bioinformatics Approach. Cancers. 2023; 15(17):4300. https://doi.org/10.3390/cancers15174300
Chicago/Turabian StyleAsadnia, Alireza, Elham Nazari, Ladan Goshayeshi, Nima Zafari, Mehrdad Moetamani-Ahmadi, Lena Goshayeshi, Haneih Azari, Ghazaleh Pourali, Ghazaleh Khalili-Tanha, Mohammad Reza Abbaszadegan, and et al. 2023. "The Prognostic Value of ASPHD1 and ZBTB12 in Colorectal Cancer: A Machine Learning-Based Integrated Bioinformatics Approach" Cancers 15, no. 17: 4300. https://doi.org/10.3390/cancers15174300
APA StyleAsadnia, A., Nazari, E., Goshayeshi, L., Zafari, N., Moetamani-Ahmadi, M., Goshayeshi, L., Azari, H., Pourali, G., Khalili-Tanha, G., Abbaszadegan, M. R., Khojasteh-Leylakoohi, F., Bazyari, M., Kahaei, M. S., Ghorbani, E., Khazaei, M., Hassanian, S. M., Gataa, I. S., Kiani, M. A., Peters, G. J., ... Avan, A. (2023). The Prognostic Value of ASPHD1 and ZBTB12 in Colorectal Cancer: A Machine Learning-Based Integrated Bioinformatics Approach. Cancers, 15(17), 4300. https://doi.org/10.3390/cancers15174300










