Extracellular Vesicles—A Source of RNA Biomarkers for the Detection of Breast Cancer in Liquid Biopsies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. EV Isolation, Quantification, and Quality Control
2.3. Extraction of EV RNA and RNA Sequencing
2.4. RNA-Seq Data Processing and Analysis
2.5. Statistical Analysis
3. Results
3.1. Quality Control of EV Isolation
3.2. Composition of EV RNA Cargo in BC Patients and Cancer-Free Controls
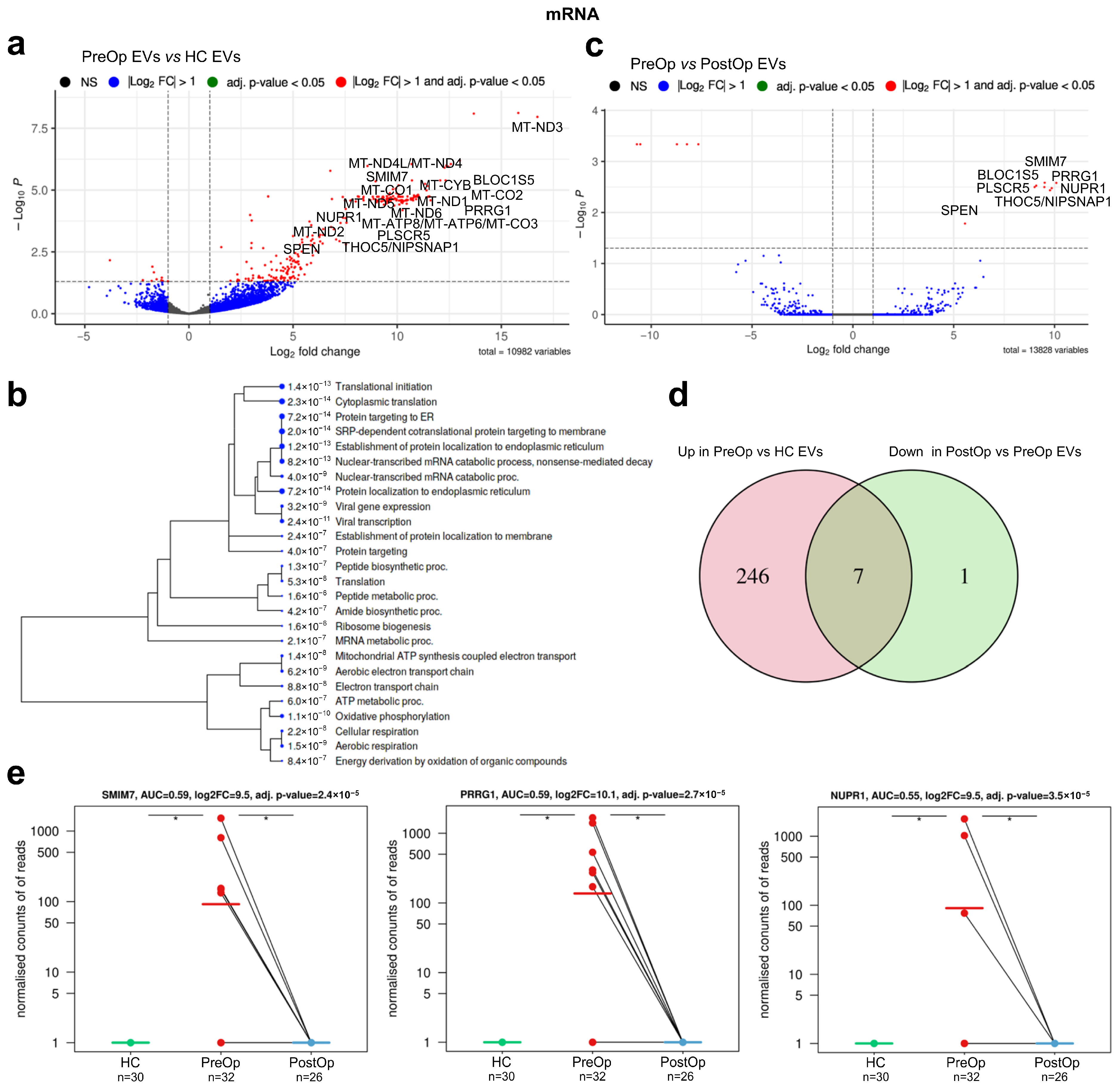
3.3. Identification of mRNA Biomarker Candidates
3.4. Identification of Biomarker Candidates in Non-Coding RNA Biotypes
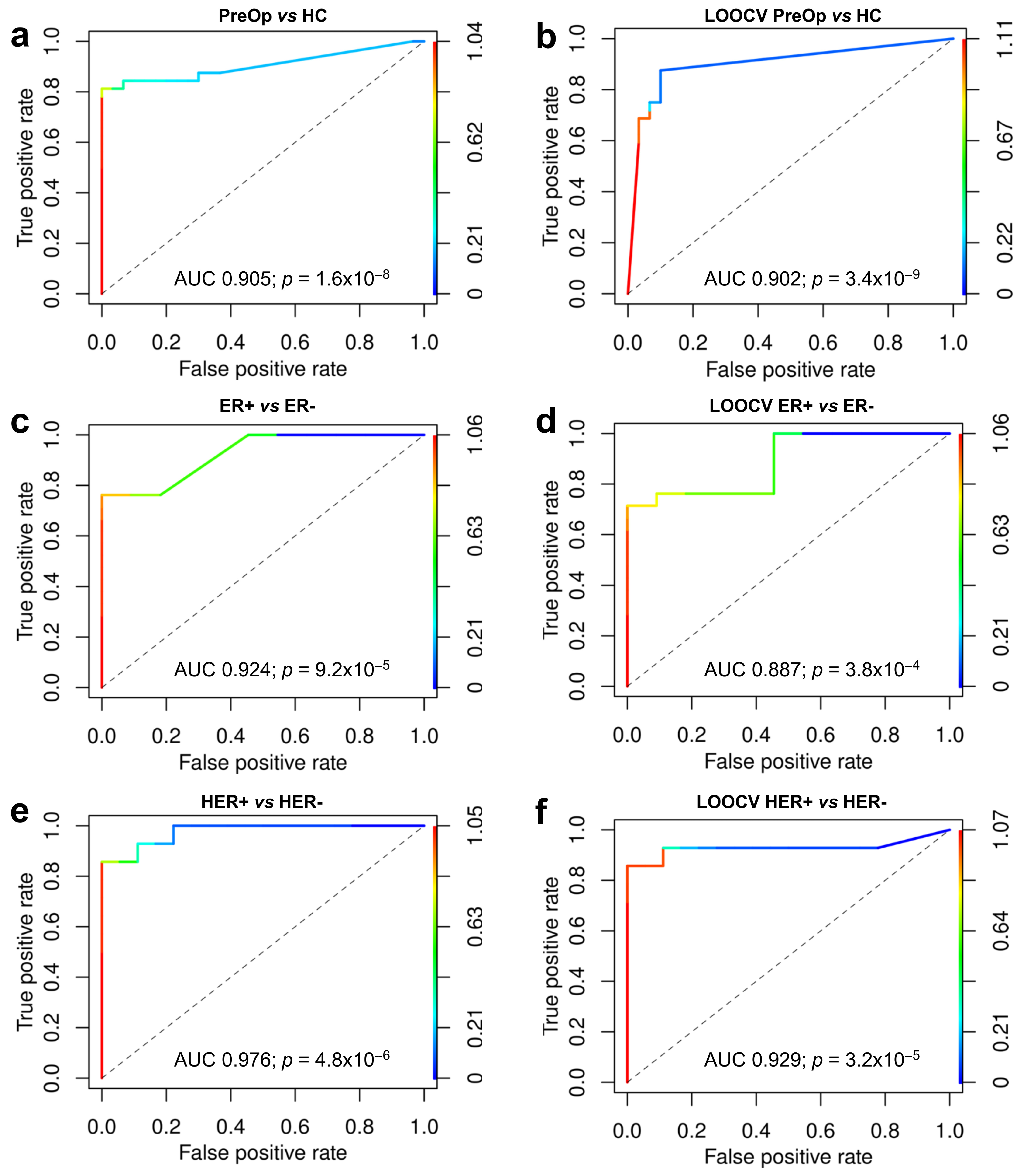
3.5. Construction of Biomarker Model
3.6. Association of RNA Biomarkers with Hormone Receptor Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.D.; Migliaccio, I.; Malorni, L.; Guarducci, C.; Biganzoli, L.; Di Leo, A. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 541–552. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24 (Suppl. S2), S26–S35. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef]
- Vasconcelos, M.H.; Caires, H.R.; Ābols, A.; Xavier, C.P.; Linē, A. Extracellular vesicles as a novel source of biomarkers in liquid biopsies for monitoring cancer progression and drug resistance. Drug Resist. Updat. 2019, 47, 100647. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Cayota, A.; Witwer, K. Exomeres and Supermeres: Monolithic or diverse? J. Extracell. Biol. 2022, 1, e45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, Á.; Yliperttula, M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef]
- García-Silva, S.; Benito-Martín, A.; Sánchez-Redondo, S.; Hernández-Barranco, A.; Ximénez-Embún, P.; Nogués, L.; Mazariegos, M.S.; Brinkmann, K.; López, A.A.; Meyer, L.; et al. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF V600E mutation. J. Exp. Med. 2019, 216, 1061–1070. [Google Scholar] [CrossRef]
- Broggi, M.A.; Maillat, L.; Clement, C.C.; Bordry, N.; Corthésy, P.; Auger, A.; Matter, M.; Hamelin, R.; Potin, L.; Demurtas, D.; et al. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J. Exp. Med. 2019, 216, 1091–1107. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.M.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; ’t Hoen, P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Lässer, C.; Shelke, G.V.; Yeri, A.; Kim, D.-K.; Crescitelli, R.; Raimondo, S.; Sjöstrand, M.; Gho, Y.S.; Van Keuren Jensen, K.; Lötvall, J. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017, 14, 58–72. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Yu, S.; Wang, Z.; He, X.; Su, Y.; Guo, T.; Sheng, H.; Chen, J.; Zheng, Q.; et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin. Chem. 2019, 65, 798–808. [Google Scholar] [CrossRef]
- Yi, Y.; Wu, M.; Zeng, H.; Hu, W.; Zhao, C.; Xiong, M.; Lv, W.; Deng, P.; Zhang, Q.; Wu, Y. Tumor-Derived Exosomal Non-Coding RNAs: The Emerging Mechanisms and Potential Clinical Applications in Breast Cancer. Front. Oncol. 2021, 11, 738945. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Curtaz, C.J.; Reifschläger, L.; Strähle, L.; Feldheim, J.; Feldheim, J.J.; Schmitt, C.; Kiesel, M.; Herbert, S.-L.; Wöckel, A.; Meybohm, P.; et al. Analysis of microRNAs in Exosomes of Breast Cancer Patients in Search of Molecular Prognostic Factors in Brain Metastases. Int. J. Mol. Sci. 2022, 23, 3683. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Q.; Zhong, H.; Li, L.; Zhang, Q.; Huang, Q.; Yu, Z. Differentially expressed microRNAs in exosomes of patients with breast cancer revealed by next-generation sequencing. Oncol. Rep. 2020, 43, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Li, Y.; Guo, R.; Zhao, J.; Chi, W.; Lai, H.; Wang, J.; Wang, Z.; Li, L.; Sang, Y.; et al. Plasma extracellular vesicle long RNA profiles in the diagnosis and prediction of treatment response for breast cancer. npj Breast Cancer 2021, 7, 154. [Google Scholar] [CrossRef]
- Cai, G.-X.; Lin, L.; Zhai, X.-M.; Guo, Z.-W.; Wu, Y.-S.; Ye, G.-L.; Liu, Q.; Chen, L.-S.; Xing, G.-Y.; Zhao, Q.-H.; et al. A plasma-derived extracellular vesicle mRNA classifier for the detection of breast cancer. Gland. Surg. 2021, 10, 2002–2009. [Google Scholar] [CrossRef]
- Sadovska, L.; Zayakin, P.; Eglītis, K.; Endzeliņš, E.; Radoviča-Spalviņa, I.; Avotiņa, E.; Auders, J.; Keiša, L.; Liepniece-Karele, I.; Leja, M.; et al. Comprehensive characterization of RNA cargo of extracellular vesicles in breast cancer patients undergoing neoadjuvant chemotherapy. Front. Oncol. 2022, 12, 1005812. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Axtell, M.J. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA 2013, 19, 740–751. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed]
- Endzelins, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Sobolevska, K.; Abols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef] [PubMed]
- Drula, R.; Pardini, B.; Fu, X.; De Los Santos, M.C.; Jurj, A.; Pang, L.; Daly, S.M.E.; Fabris, L.; Knutsen, E.; Dragomir, M.P.; et al. 17beta-estradiol promotes extracellular vesicle release and selective miRNA loading in ERalpha-positive breast cancer. Proc. Natl. Acad. Sci. USA 2023, 120, e2122053120. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Crescitelli, R.; Cvjetkovic, A.; Belgrano, V.; Bagge, R.O.; Sundfeldt, K.; Ochiya, T.; Kalluri, R.; Lötvall, J. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J. Extracell. Vesicles 2019, 8, 1635420. [Google Scholar] [CrossRef]
- D’acunzo, P.; Pérez-González, R.; Kim, Y.; Hargash, T.; Miller, C.; Alldred, M.J.; Erdjument-Bromage, H.; Penikalapati, S.C.; Pawlik, M.; Saito, M.; et al. Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci. Adv. 2021, 7, eabe5085. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Zhao, L.; Chen, X.; Lin, Z.; Zhang, L.; Li, Z. miR-224-5p Carried by Human Umbilical Cord Mesenchymal Stem Cells-Derived Exosomes Regulates Autophagy in Breast Cancer Cells via HOXA5. Front. Cell Dev. Biol. 2021, 9, 679185. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, Z.; Xie, J.; Wang, P.; Zhu, J.; Li, Y.; Wang, Y. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem. Biophys. Res. Commun. 2018, 506, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Duong, T.T.C.; Nguyen, T.H.N.; Nguyen, T.T.N.; Huynh, L.H.; Ngo, H.P.; Nguyen, H.T. Diagnostic and prognostic value of miR-200 family in breast cancer: A meta-analysis and systematic review. Cancer Epidemiol. 2022, 77, 102097. [Google Scholar] [CrossRef]
- Papadaki, C.; Stoupis, G.; Tsalikis, L.; Monastirioti, A.; Papadaki, M.; Maliotis, N.; Stratigos, M.; Mastrostamatis, G.; Mavroudis, D.; Agelaki, S. Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget 2019, 10, 966–981. [Google Scholar] [CrossRef]
- Fischer, C.; Deutsch, T.M.; Feisst, M.; Rippinger, N.; Riedel, F.; Hartkopf, A.D.; Brucker, S.Y.; Domschke, C.; Fremd, C.; Michel, L.; et al. Circulating miR-200 family as predictive markers during systemic therapy of metastatic breast cancer. Arch. Gynecol. Obstet. 2022, 306, 875–885. [Google Scholar] [CrossRef]
- Liu, X.; Cao, M.; Palomares, M.; Wu, X.; Li, A.; Yan, W.; Fong, M.Y.; Chan, W.-C.; Wang, S.E. Metastatic breast cancer cells overexpress and secrete miR-218 to regulate type I collagen deposition by osteoblasts. Breast Cancer Res. 2018, 20, 1–12. [Google Scholar] [CrossRef]
- Wang, M.; Cai, W.-R.; Meng, R.; Chi, J.-R.; Li, Y.-R.; Chen, A.-X.; Yu, Y.; Cao, X.-C. miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochem. Biophys. Res. Commun. 2018, 501, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, J.; Zheng, R.; Song, B.; Huang, L.; Liu, Y.; Hao, Y.; Bai, X. MiR-133 Targets YES1 and Inhibits the Growth of Triple-Negative Breast Cancer Cells. Technol. Cancer Res. Treat. 2020, 19, 1533033820927011. [Google Scholar] [CrossRef] [PubMed]
- van der Werf, J.; Chin, C.V.; Fleming, N.I. SnoRNA in Cancer Progression, Metastasis and Immunotherapy Response. Biology 2021, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Xia, S.; Xiao, F.; Peng, J.; Gao, Y.; Yu, F.; Wang, C.; Chen, X. The emerging role of snoRNAs in human disease. Genes Dis. 2023, 10, 2064–2081. [Google Scholar] [CrossRef]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNA s for detection of pancreatic cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef]
- Wang, K.; Song, X.; Li, X.; Zhang, Z.; Xie, L.; Song, X. Plasma SNORD83A as a potential biomarker for early diagnosis of non-small-cell lung cancer. Futur. Oncol. 2022, 18, 821–832. [Google Scholar] [CrossRef]
- Zhao, Y.; Yan, Y.; Ma, R.; Lv, X.; Zhang, L.; Wang, J.; Zhu, W.; Zhao, L.; Jiang, L.; Zhao, L.; et al. Expression signature of six-snoRNA serves as novel non-invasive biomarker for diagnosis and prognosis prediction of renal clear cell carcinoma. J. Cell. Mol. Med. 2020, 24, 2215–2228. [Google Scholar] [CrossRef]
- Dvinge, H.; Guenthoer, J.; Porter, P.L.; Bradley, R.K. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019, 29, 1591–1604. [Google Scholar] [CrossRef]
- Fu, M.; Gu, J.; Wang, M.; Zhang, J.; Chen, Y.; Jiang, P.; Zhu, T.; Zhang, X. Emerging roles of tRNA-derived fragments in cancer. Mol. Cancer 2023, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fu, Z.; Wang, S.; Li, J.; Li, Y.; Zhang, Y.; Yang, F.; Chu, J.; Wu, H.; Yin, Y.; et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018, 414, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Cayota, A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020, 17, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Koi, Y.; Tsutani, Y.; Nishiyama, Y.; Ueda, D.; Ibuki, Y.; Sasada, S.; Akita, T.; Masumoto, N.; Kadoya, T.; Tahara, H.; et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
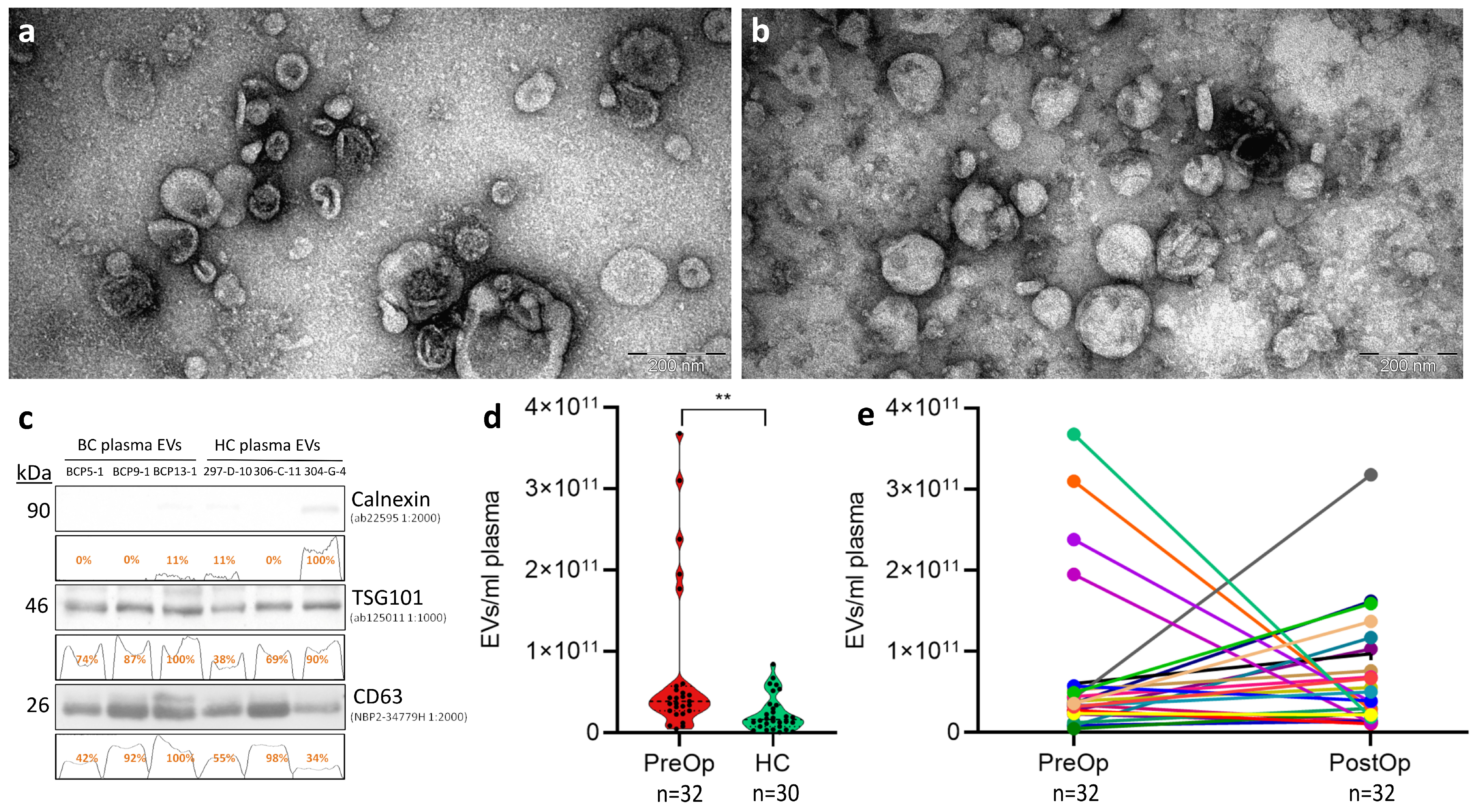
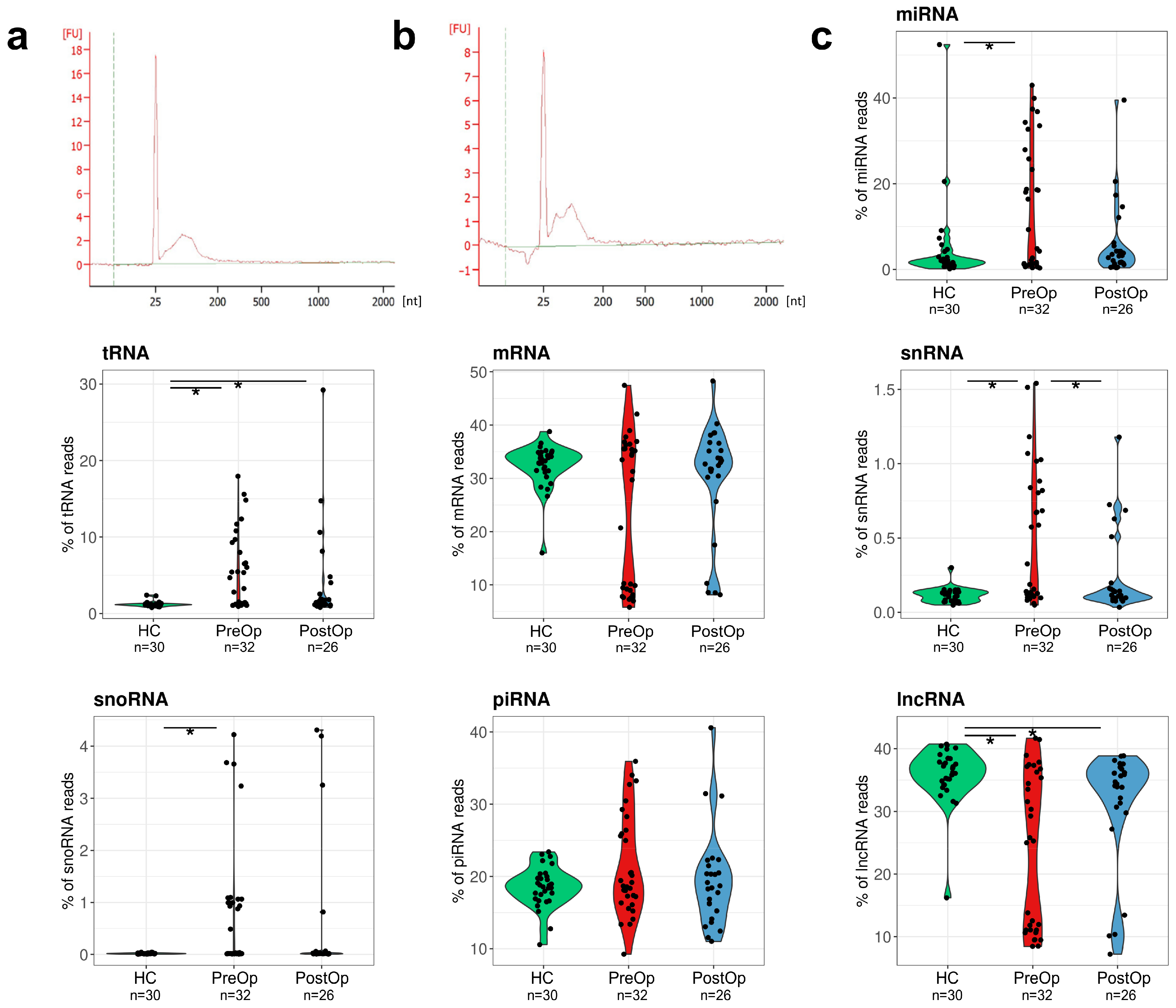

| Characteristic | BC Patients |
|---|---|
| Sample size (n) | 32 |
| Age mean, years | 50.9 |
| Age range, years | 34–77 |
| Tumor grade | |
| Grade 2 | 21 |
| Grade 3 | 11 |
| TNM stage | |
| T1 N1-3 M0 | 2 |
| T2 N1-3 M0 | 12 |
| T3 N1-3 M0 | 16 |
| T4 N1-3 M0 | 2 |
| Estrogen receptor | |
| Positive | 20 |
| Negative | 12 |
| Progesterone receptor | |
| Positive | 17 |
| Negative | 15 |
| HER2 overexpression | |
| 0 | 5 |
| 1 | 13 |
| 2 | 4 |
| 3 | 10 |
| TNBC | |
| Yes | 8 |
| No | 24 |
| E-cadherin | |
| Positive | 22 |
| Negative | 10 |
| Proliferation index (Ki-67) | |
| ≤14% | 5 |
| >14% | 27 |
| Gene | Full Name | Log2FC PreOp vs. HC | Adj. p Value | Log2FC PreOp vs. PostOp | Adj. p Value | Function * |
|---|---|---|---|---|---|---|
| SMIM7 | small integral membrane protein 7 | 9.508 | 2.35 × 10−5 | 9.508 | 0.0026 | Predicted to be integral component of membrane |
| PRRG1 | proline rich and Gla domain 1 | 10.086 | 2.66 × 10−5 | 10.086 | 0.0026 | Enables calcium ion and protein binding |
| BLOC1S5-TXNDC5 | biogenesis of lysosomal organelles complex 1 subunit 5 BLOC1S5-TXNDC5 readthrough (NMD candidate) | 9.090 | 2.71 × 10−5 | 9.090 | 0.0030 | BLOC1S5 enables protein binding, involved in the biogenesis of organelles. BLOC1S5-TXNDC5 is a naturally occurring read-through transcription between the neighboring MUTED and TXNDC5 genes on chromosome 6. A candidate for nonsense-mediated mRNA decay (NMD) and is unlikely to produce a protein product |
| PLSCR5 | phospholipid scramblase family member 5 | 9.007 | 3.11 × 10−5 | 9.007 | 0.0032 | Predicted to enable phospholipid scramblase activity and be involved in plasma membrane phospholipid scrambling |
| NUPR1 | nuclear protein 1, transcriptional regulator | 9.494 | 3.50 × 10−5 | 9.494 | 0.0032 | Enables DNA binding activity and transcription coactivator activity. Involved in regulation of cellular catabolic process; regulation of generation of precursor metabolites and energy; and regulation of programmed cell death. Acts upstream of or within negative regulation of cell cycle. |
| SPEN | spen family transcriptional repressor | 5.558 | 0.0002 | 5.558 | 0.0165 | Hormone inducible transcriptional repressor |
| THOC5/NIPSNAP1 (reads overlap both genes) | THO complex 5/nipsnap homolog 1 | 9.858 | 4.23 × 10−5 | 9.858 | 0.0033 | THOC5 is predicted to enable mRNA binding activity. Involved in monocyte differentiation, negative regulation of DNA damage checkpoint, and viral mRNA export from host cell nucleus. NIPSNAP1 may be involved in vesicular transport. A similar protein in mice inhibits the calcium channel TRPV6, and is also localized to the inner mitochondrial membrane where it may play a role in mitochondrial DNA maintenance. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zayakin, P.; Sadovska, L.; Eglītis, K.; Romanchikova, N.; Radoviča-Spalviņa, I.; Endzeliņš, E.; Liepniece-Karele, I.; Eglītis, J.; Linē, A. Extracellular Vesicles—A Source of RNA Biomarkers for the Detection of Breast Cancer in Liquid Biopsies. Cancers 2023, 15, 4329. https://doi.org/10.3390/cancers15174329
Zayakin P, Sadovska L, Eglītis K, Romanchikova N, Radoviča-Spalviņa I, Endzeliņš E, Liepniece-Karele I, Eglītis J, Linē A. Extracellular Vesicles—A Source of RNA Biomarkers for the Detection of Breast Cancer in Liquid Biopsies. Cancers. 2023; 15(17):4329. https://doi.org/10.3390/cancers15174329
Chicago/Turabian StyleZayakin, Pawel, Lilite Sadovska, Kristaps Eglītis, Nadezhda Romanchikova, Ilze Radoviča-Spalviņa, Edgars Endzeliņš, Inta Liepniece-Karele, Jānis Eglītis, and Aija Linē. 2023. "Extracellular Vesicles—A Source of RNA Biomarkers for the Detection of Breast Cancer in Liquid Biopsies" Cancers 15, no. 17: 4329. https://doi.org/10.3390/cancers15174329
APA StyleZayakin, P., Sadovska, L., Eglītis, K., Romanchikova, N., Radoviča-Spalviņa, I., Endzeliņš, E., Liepniece-Karele, I., Eglītis, J., & Linē, A. (2023). Extracellular Vesicles—A Source of RNA Biomarkers for the Detection of Breast Cancer in Liquid Biopsies. Cancers, 15(17), 4329. https://doi.org/10.3390/cancers15174329







