Artificial Intelligence in Lung Cancer Screening: The Future Is Now
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Screening Rationale
3. AI Terminology
4. AI Applications in Lung Cancer Screening
4.1. Personalized Screening Programs
4.2. Image Reconstruction
4.3. CAD System
4.3.1. Structure of the CAD Systems
4.3.2. Data Collection
4.3.3. Accuracy of CAD Systems
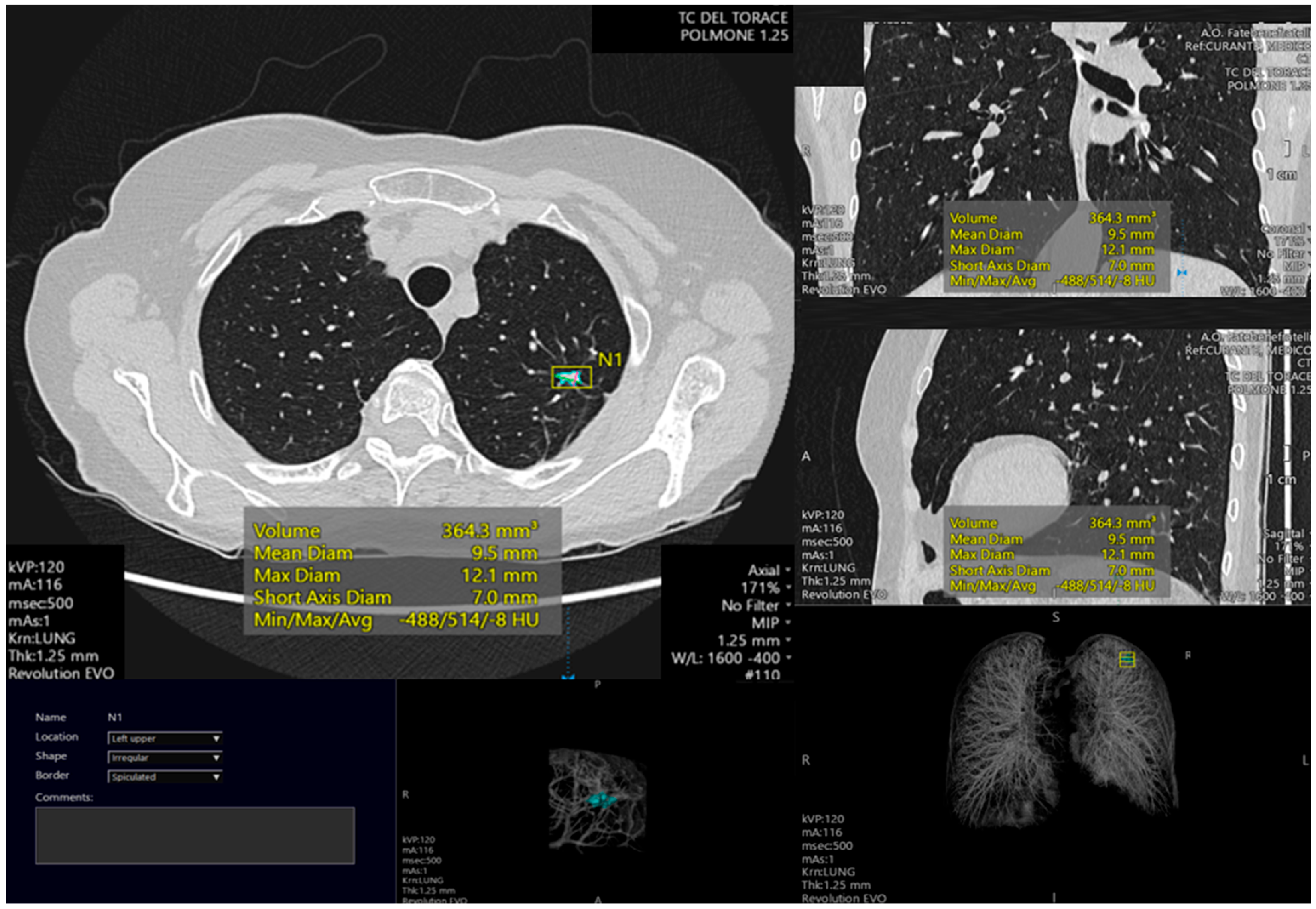
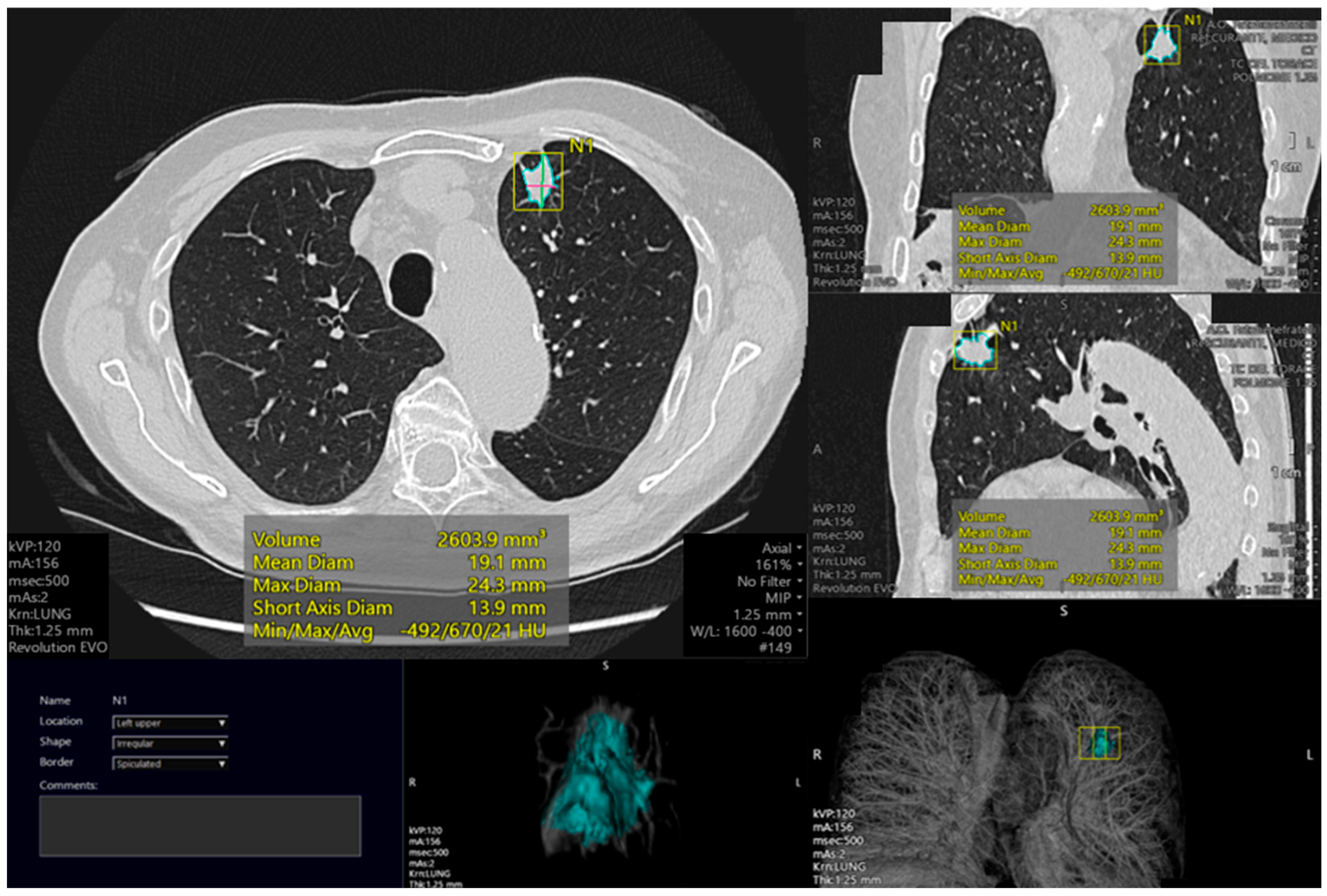
4.4. Nodule Segmentation
4.5. Nodule Characterization
Virtual Biopsy
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amisha, M.P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Family Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Khenkina, N.; Sinichich, P.; Cervelli, M.; Poggi, V.; Boemi, S.; Ierardi, A.M.; Carrafiello, G. Artificial Intelligence in the Era of Precision Oncological Imaging. Technol. Cancer Res. Treat. 2022, 21, 15330338221141793. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Caloro, E.; Bianchi, L.; Pellegrino, G.; D’Amico, N.; Papa, S.; Carrafiello, G. Artificial Intelligence in Emergency Radiology: Where Are We Going? Diagnostics 2022, 12, 3223. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Passiglia, F.; Calandri, M.; Guerrera, F.; Malapelle, U.; Mangone, L.; Ramella, S.; Trisolini, R.; Novello, S. Lung Cancer in Italy. J. Thorac. Oncol. 2019, 14, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Picozzi, G.; Sverzellati, N.; Anglesio, S.; Bartolucci, M.; Cavigli, E.; Deliperi, A.; Falchini, M.; Falaschi, F.; Ghio, D.; et al. Low-dose CT for lung cancer screening: Position paper from the Italian college of thoracic radiology. Radiol. Med. 2022, 127, 543–559. [Google Scholar] [CrossRef]
- Mao, Y.; Yang, D.; He, J.; Krasna, M.J. Epidemiology of Lung Cancer. Surg. Oncol. Clin. N. Am. 2016, 25, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Tyczynski, J.E.; Bray, F.; Maxwell Parkin, D. Lung cancer in Europe in 2000: Epidemiology, prevention, and early detection. Lancet Oncol. 2003, 4, 45–55. [Google Scholar] [CrossRef]
- Pastorino, U.; Boffi, R.; Marchianò, A.; Sestini, S.; Munarini, E.; Calareso, G.; Boeri, M.; Pelosi, G.; Sozzi, G.; Silva, M.; et al. Stopping Smoking Reduces Mortality in Low-Dose Computed Tomography Screening Participants. J. Thorac. Oncol. 2016, 11, 693–699. [Google Scholar] [CrossRef]
- Cruickshank, A.; Stieler, G.; Ameer, F. Evaluation of the solitary pulmonary nodule. Intern. Med. J. 2019, 49, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Skalski, M.; Gaillard, F. The utilisation of convolutional neural networks in detecting pulmonary nodules: A review. Br. J. Radiol. 2018, 91, 20180028. [Google Scholar] [CrossRef] [PubMed]
- Wender, R.; Fontham, E.T.; Barrera, E., Jr.; Colditz, G.A.; Church, T.R.; Ettinger, D.S.; Etzioni, R.; Flowers, C.R.; Gazelle, G.S.; Kelsey, D.K.; et al. American Cancer Society lung cancer screening guidelines. CA Cancer J. Clin. 2013, 63, 107–117. [Google Scholar] [CrossRef] [PubMed]
- De Koning, H.J.; Van der Aalst, C.M.; De Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Paci, E.; Puliti, D.; Lopes Pegna, A.; Carrozzi, L.; Picozzi, G.; Falaschi, F.; Pistelli, F.; Aquilini, F.; Ocello, C.; Zappa, M.; et al. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax 2017, 72, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, U.; Silva, M.; Sestini, S.; Sabia, F.; Boeri, M.; Cantarutti, A.; Sverzellati, N.; Sozzi, G.; Corrao, G.; Marchianò, A. Prolonged Lung Cancer Screening Reduced 10-year Mortality in the MILD Trial. Ann. Oncol. 2019, 30, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Motsch, E.; Trotter, A.; Heussel, C.P.; Dienemann, H.; Schnabel, P.A.; Kauczor, H.U.; Maldonado, S.G.; Miller, A.B.; Kaaks, R.; et al. Lung cancer mortality reduction by LDCT screening-Results from the randomized German LUSI trial. Int. J. Cancer 2020, 146, 1503–1513. [Google Scholar] [CrossRef]
- Passiglia, F.; Cinquini, M.; Bertolaccini, L.; Del Re, M.; Facchinetti, F.; Ferrara, R.; Franchina, T.; Larici, A.R.; Malapelle, U.; Menis, J.; et al. Benefits and Harms of Lung Cancer Screening by Chest Computed Tomography: A Systematic Review and Meta-Analysis. J. Clin. Oncol. 2021, 39, 2574–2585. [Google Scholar] [CrossRef]
- Puliti, D.; Picozzi, G.; Gorini, G.; Carrozzi, L.; Mascalchi, M. Gender effect in the ITALUNG screening trial. A comparison with UKLS and other trials. Lancet Reg. Health Eur. 2022, 13, 100300. [Google Scholar] [CrossRef]
- Joy Mathew, C.; David, A.M.; Joy Mathew, C.M. Artificial Intelligence and its future potential in lung cancer screening. EXCLI J. 2020, 19, 1552–1562. [Google Scholar] [CrossRef]
- International Early Lung Cancer Action Program Investigators; Henschke, C.I.; Yankelevitz, D.F.; Libby, D.M.; Pasmantier, M.W.; Smith, J.P.; Miettinen, O.S. Survival of patients with stage I lung cancer detected on CT screening. N. Engl. J. Med. 2006, 355, 1763–1771. [Google Scholar] [CrossRef]
- Field, J.K.; Smith, R.A.; Aberle, D.R.; Oudkerk, M.; Baldwin, D.R.; Yankelevitz, D.; Pedersen, J.H.; Swanson, S.J.; Travis, W.D.; Wisbuba, I.I.; et al. International Association for the Study of Lung Cancer Computed Tomography Screening Workshop 2011 report. J. Thorac. Oncol. 2012, 7, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.D. Lung nodule and cancer detection in computed tomography screening. J. Thorac. Imaging 2015, 30, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, G.; Maisonneuve, P.; Rampinelli, C.; Bertolotti, R.; Petrella, F.; Spaggiari, L.; Bellomi, M. Computed tomography screening for lung cancer: Results of ten years of annual screening and validation of cosmos prediction model. Lung Cancer 2013, 82, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, U.; Boeri, M.; Sestini, S.; Sabia, F.; Milanese, G.; Silva, M.; Suatoni, P.; Verri, C.; Cantarutti, A.; Sverzellati, N.; et al. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann. Oncol. 2022, 33, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Oudkerk, M.; Devaraj, A.; Vliegenthart, R.; Henzler, T.; Prosch, H.; Heussel, C.P.; Bastarrika, G.; Sverzellati, N.; Mascalchi, M.; Delorme, S.; et al. European position statement on lung cancer screening. Lancet Oncol. 2017, 18, e754–e766. [Google Scholar] [CrossRef] [PubMed]
- Kauczor, H.U.; Baird, A.M.; Blum, T.G.; Bonomo, L.; Bostantzoglou, C.; Burghuber, O.; Čepická, B.; Comanescu, A.; Couraud, S.; Devaraj, A.; et al. European Society of Radiology (ESR) and the European Respiratory Society (ERS). ESR/ERS statement paper on lung cancer screening. Eur. Radiol. 2020, 30, 3277–3294. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, G.; Baldwin, D.R.; Henschke, C.I.; Ghislandi, S.; Iavicoli, S.; Oudkerk, M.; De Koning, H.J.; Shemesh, J.; Field, J.K.; Zulueta, J.J.; et al. Recommendations for Implementing Lung Cancer Screening with Low-Dose Computed Tomography in Europe. Cancers 2020, 12, 1672. [Google Scholar] [CrossRef]
- US Preventive Services Task Force; Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962–970. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644. [Google Scholar] [CrossRef]
- Matsoukas, S.; Scaggiante, J.; Schuldt, B.R.; Smith, C.J.; Chennareddy, S.; Kalagara, R.; Majidi, S.; Bederson, J.B.; Fifi, J.T.; Mocco, J.; et al. Accuracy of artificial intelligence for the detection of intracranial hemorrhage and chronic cerebral microbleeds: A systematic review and pooled analysis. Radiol. Med. 2022, 127, 1106–1123. [Google Scholar] [CrossRef]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Zerunian, M.; Pucciarelli, F.; Caruso, D.; Polici, M.; Masci, B.; Guido, G.; De Santis, D.; Polverari, D.; Principessa, D.; Benvenga, A.; et al. Artificial intelligence based image quality enhancement in liver MRI: A quantitative and qualitative evaluation. Radiol. Med. 2022, 127, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Cipollari, S.; Pecoraro, M.; Forookhi, A.; Laschena, L.; Bicchetti, M.; Messina, E.; Lucciola, S.; Catalano, C.; Panebianco, V. Biparametric prostate MRI: Impact of a deep learning-based software and of quantitative ADC values on the inter-reader agreement of experienced and inexperienced readers. Radiol. Med. 2022, 127, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Bian, S.; Zhu, D.; Yuan, Y.; Pan, K.; Pan, Z.; Feng, X.; Tang, K.; Yang, Y. Machine learning-based radiomics for multiple primary prostate cancer biological characteristics prediction with 18F-PSMA-1007 PET: Comparison among different volume segmentation thresholds. Radiol. Med. 2022, 127, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Cardobi, N.; Benetti, G.; Cardano, G.; Arena, C.; Micheletto, C.; Cavedon, C.; Montemezzi, S. CT radiomic models to distinguish COVID-19 pneumonia from other interstitial pneumonias. Radiol. Med. 2021, 126, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Kohli, M.; Prevedello, L.M.; Filice, R.W.; Geis, J.R. Implementing Machine Learning in Radiology Practice and Research. AJR Am. J. Roentgenol. 2017, 208, 754–760. [Google Scholar] [CrossRef]
- Abdullah, S.S.; Rajasekaran, M.P. Automatic detection and classification of knee osteoarthritis using deep learning approach. Radiol. Med. 2022, 127, 398–406. [Google Scholar] [CrossRef]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Jaklitsch, M.T.; Jacobson, F.L.; Austin, J.H.; Field, J.K.; Jett, J.R.; Keshavjee, S.; MacMahon, H.; Mulshine, J.L.; Munden, R.F.; Salgia, R.; et al. The American Association for Thoracic Surgery guidelines for lung cancer screening using low-dose computed tomography scans for lung cancer survivors and other high-risk groups. J. Thorac. Cardiovasc. Surg. 2012, 144, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Kazerooni, E.A.; Baum, S.L.; Eapen, G.A.; Ettinger, D.S.; Hou, L.; Jackman, D.M.; Klippenstein, D.; Kumar, R.; Lackner, R.P.; et al. Lung Cancer Screening, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 412–441. [Google Scholar] [CrossRef]
- Cheung, L.C.; Berg, C.D.; Castle, P.E.; Katki, H.A.; Chaturvedi, A.K. Life-Gained-Based Versus Risk-Based Selection of Smokers for Lung Cancer Screening. Ann. Intern. Med. 2019, 171, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Cohen, J.T.; van Klaveren, D.; Soeteman, D.I.; Wong, J.B.; Neumann, P.J.; Kent, D.M. Risk-Targeted Lung Cancer Screening: A Cost-Effectiveness Analysis. Ann. Intern. Med. 2018, 168, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ten Haaf, K.; Bastani, M.; Cao, P.; Jeon, J.; Toumazis, I.; Han, S.S.; Plevritis, S.K.; Blom, E.F.; Kong, C.Y.; Tammemägi, M.C.; et al. A Comparative Modeling Analysis of Risk-Based Lung Cancer Screening Strategies. J. Natl. Cancer Inst. 2020, 112, 466–479. [Google Scholar] [CrossRef]
- Ali, N.; Lifford, K.J.; Carter, B.; McRonald, F.; Yadegarfar, G.; Baldwin, D.R.; Weller, D.; Hansell, D.M.; Duffy, S.W.; Field, J.K.; et al. Barriers to uptake among high-risk individuals declining participation in lung cancer screening: A mixed methods analysis of the UK Lung Cancer Screening (UKLS) trial. BMJ Open 2015, 5, e008254. [Google Scholar] [CrossRef]
- Huo, J.; Shen, C.; Volk, R.J.; Shih, Y.T. Use of CT and Chest Radiography for Lung Cancer Screening Before and After Publication of Screening Guidelines: Intended and Unintended Uptake. JAMA Intern. Med. 2017, 177, 439–441. [Google Scholar] [CrossRef]
- Van der Aalst, C.M.; Ten Haaf, K.; De Koning, H.J. Lung cancer screening: Latest developments and unanswered questions. Lancet Respir. Med. 2016, 4, 749–761. [Google Scholar] [CrossRef]
- Ten Haaf, K.; Van der Aalst, C.M.; De Koning, H.J.; Kaaks, R.; Tammemägi, M.C. Personalising lung cancer screening: An overview of risk-stratification opportunities and challenges. Int. J. Cancer 2021, 149, 250–263. [Google Scholar] [CrossRef]
- Bach, P.B.; Kattan, M.W.; Thornquist, M.D.; Kris, M.G.; Tate, R.C.; Barnett, M.J.; Hsieh, L.J.; Begg, C.B. Variations in lung cancer risk among smokers. J. Natl. Cancer Inst. 2003, 95, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Katki, H.A.; Kovalchik, S.A.; Berg, C.D.; Cheung, L.C.; Chaturvedi, A.K. Development and Validation of Risk Models to Select Ever-Smokers for CT Lung Cancer Screening. JAMA 2016, 315, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.; Myles, J.P.; Van Tongeren, M.; Page, R.D.; Liloglou, T.; Duffy, S.W.; Field, J.K. The LLP risk model: An individual risk prediction model for lung cancer. Br. J. Cancer 2008, 98, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Field, J.K.; Vulkan, D.; Davies, M.P.A.; Duffy, S.W.; Gabe, R. Liverpool Lung Project lung cancer risk stratification model: Calibration and prospective validation. Thorax 2021, 76, 161–168. [Google Scholar] [CrossRef]
- Tammemägi, M.C.; Katki, H.A.; Hocking, W.G.; Church, T.R.; Caporaso, N.; Kvale, P.A.; Chaturvedi, A.K.; Silvestri, G.A.; Riley, T.L.; Commins, J.; et al. Selection criteria for lung-cancer screening. N. Engl. J. Med. 2013, 368, 728–736. [Google Scholar] [CrossRef]
- McRonald, F.E.; Yadegarfar, G.; Baldwin, D.R.; Devaraj, A.; Brain, K.E.; Eisen, T.; Holemans, J.A.; Ledson, M.; Screaton, N.; Rintoul, R.C.; et al. The UK Lung Screen (UKLS): Demographic profile of first 88,897 approaches provides recommendations for population screening. Cancer Prev. Res. 2014, 7, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Schütte, S.; Dietrich, D.; Montet, X.; Flahault, A. Participation in lung cancer screening programs: Are there gender and social differences? A systematic review. Public Health Rev. 2018, 39, 23. [Google Scholar] [CrossRef] [PubMed]
- Yousaf-Khan, U.; Horeweg, N.; van der Aalst, C.; Ten Haaf, K.; Oudkerk, M.; de Koning, H. Baseline Characteristics and Mortality Outcomes of Control Group Participants and Eligible Non-Responders in the NELSON Lung Cancer Screening Study. J. Thorac. Oncol. 2015, 10, 747–753. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Clapp, J.D.; Clingan, K.L.; Gareen, I.F.; Lynch, D.A.; Marcus, P.M.; Pinsky, P.F. Baseline characteristics of participants in the randomized national lung screening trial. J. Natl. Cancer Inst. 2010, 102, 1771–1779. [Google Scholar] [CrossRef]
- Patz, E.F., Jr.; Greco, E.; Gatsonis, C.; Pinsky, P.; Kramer, B.S.; Aberle, D.R. Lung cancer incidence and mortality in National Lung Screening Trial participants who underwent low-dose CT prevalence screening: A retrospective cohort analysis of a randomised, multicentre, diagnostic screening trial. Lancet Oncol. 2016, 17, 590–599. [Google Scholar] [CrossRef]
- Horeweg, N.; van der Aalst, C.M.; Vliegenthart, R.; Zhao, Y.; Xie, X.; Scholten, E.T.; Mali, W.; Thunnissen, E.; Weenink, C.; Groen, H.J.; et al. Volumetric computed tomography screening for lung cancer: Three rounds of the NELSON trial. Eur. Respir. J. 2013, 42, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Horeweg, N.; van Rosmalen, J.; Heuvelmans, M.A.; van der Aalst, C.M.; Vliegenthart, R.; Scholten, E.T.; ten Haaf, K.; Nackaerts, K.; Lammers, J.W.; Weenink, C.; et al. Lung cancer probability in patients with CT-detected pulmonary nodules: A prespecified analysis of data from the NELSON trial of low-dose CT screening. Lancet Oncol. 2014, 15, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- McCaffery, K.J.; Smith, S.K.; Wolf, M. The challenge of shared decision making among patients with lower literacy: A framework for research and development. Med. Decis. Mak. 2010, 30, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Ten Haaf, K.; Tammemägi, M.C.; Bondy, S.J.; van der Aalst, C.M.; Gu, S.; McGregor, S.E.; Nicholas, G.; de Koning, H.J.; Paszat, L.F. Performance and Cost-Effectiveness of Computed Tomography Lung Cancer Screening Scenarios in a Population-Based Setting: A Microsimulation Modeling Analysis in Ontario, Canada. PLoS Med. 2017, 14, e1002225. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, Y.; Ten Haaf, K.; Frauenfelder, T.; Kohler, M.; Kouyos, R.D.; Shilaih, M.; Lorez, M.; de Koning, H.J.; Schwenkglenks, M.; Puhan, M.A. Cost-effectiveness of low-dose CT screening for lung cancer in a European country with high prevalence of smoking-A modelling study. Lung Cancer 2018, 121, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Criss, S.D.; Cao, P.; Bastani, M.; Ten Haaf, K.; Chen, Y.; Sheehan, D.F.; Blom, E.F.; Toumazis, I.; Jeon, J.; de Koning, H.J.; et al. Cost-Effectiveness Analysis of Lung Cancer Screening in the United States: A Comparative Modeling Study. Ann. Intern. Med. 2019, 171, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, F.H.; Hughes, D.R.; Wang, J.M.; Mullins, M.E.; Hawkins, C.M.; Duszak, R., Jr. Diagnostic Radiology Resident and Fellow Workloads: A 12-Year Longitudinal Trend Analysis Using National Medicare Aggregate Claims Data. J. Am. Coll. Radiol. 2015, 12, 664–669. [Google Scholar] [CrossRef]
- Reicher, J.; Currie, S.; Birchall, D. Safety of working patterns among UK neuroradiologists: What can we learn from the aviation industry and cognitive science? Br. J. Radiol. 2018, 91, 20170284. [Google Scholar] [CrossRef]
- Nishie, A.; Kakihara, D.; Nojo, T.; Nakamura, K.; Kuribayashi, S.; Kadoya, M.; Ohtomo, K.; Sugimura, K.; Honda, H. Current radiologist workload and the shortages in Japan: How many full-time radiologists are required? Jpn. J. Radiol. 2015, 33, 266–272. [Google Scholar] [CrossRef]
- Schreuder, A.; Schaefer-Prokop, C.M.; Scholten, E.T.; Jacobs, C.; Prokop, M.; van Ginneken, B. Lung cancer risk to personalise annual and biennial follow-up computed tomography screening. Thorax 2018, 73, 626–633. [Google Scholar] [CrossRef]
- Robbins, H.A.; Berg, C.D.; Cheung, L.C.; Chaturvedi, A.K.; Katki, H.A. Identification of Candidates for Longer Lung Cancer Screening Intervals Following a Negative Low-Dose Computed Tomography Result. J. Natl. Cancer Inst. 2019, 111, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Tammemägi, M.C.; Ten Haaf, K.; Toumazis, I.; Kong, C.Y.; Han, S.S.; Jeon, J.; Commins, J.; Riley, T.; Meza, R. Development and Validation of a Multivariable Lung Cancer Risk Prediction Model That Includes Low-Dose Computed Tomography Screening Results: A Secondary Analysis of Data From the National Lung Screening Trial. JAMA Netw. Open 2019, 2, e190204. [Google Scholar] [CrossRef] [PubMed]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M.; Ostrin, E.J.; Fahrmann, J.F. Blood based biomarkers beyond genomics for lung cancer screening. Transl. Lung Cancer Res. 2018, 7, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.H.; Zheng, H.L.; Li, J.T.; Li, P.; Zheng, C.H.; Chen, Q.Y.; Huang, C.M.; Xie, J.W. Prediction of recurrence-free survival and adjuvant therapy benefit in patients with gastrointestinal stromal tumors based on radiomics features. Radiol. Med. 2022, 127, 1085–1097. [Google Scholar] [CrossRef]
- Fehlmann, T.; Kahraman, M.; Ludwig, N.; Backes, C.; Galata, V.; Keller, V.; Geffers, L.; Mercaldo, N.; Hornung, D.; Weis, T.; et al. Evaluating the Use of Circulating MicroRNA Profiles for Lung Cancer Detection in Symptomatic Patients. JAMA Oncol. 2020, 6, 714–723. [Google Scholar] [CrossRef]
- Hung, R.J.; Warkentin, M.T.; Brhane, Y.; Chatterjee, N.; Christiani, D.C.; Landi, M.T.; Caporaso, N.E.; Liu, G.; Johansson, M.; Albanes, D.; et al. Assessing Lung Cancer Absolute Risk Trajectory Based on a Polygenic Risk Model. Cancer Res. 2021, 81, 1607–1615. [Google Scholar] [CrossRef]
- Horst, C.; Dickson, J.L.; Tisi, S.; Ruparel, M.; Nair, A.; Devaraj, A.; Janes, S.M. Delivering low-dose CT screening for lung cancer: A pragmatic approach. Thorax 2020, 75, 831–832. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Triphuridet, N.; Zhang, S.S.; Nagasaka, M.; Gao, Y.; Zhao, J.J.; Syn, N.L.; Hanaoka, T.; Ou, S.I.; Shum, E. Low-Dose Computed Tomography (LDCT) Lung Cancer Screening in Asian Female Never-Smokers Is as Efficacious in Detecting Lung Cancer as in Asian Male Ever-Smokers: A Systematic Review and Meta-Analysis. J. Thorac. Oncol. 2023, 18, 698–717. [Google Scholar] [CrossRef]
- Agostini, A.; Floridi, C.; Borgheresi, A.; Badaloni, M.; Esposto Pirani, P.; Terilli, F.; Ottaviani, L.; Giovagnoni, A. Proposal of a low-dose, long-pitch, dual-source chest CT protocol on third-generation dual-source CT using a tin filter for spectral shaping at 100 kVp for CoronaVirus Disease 2019 (COVID-19) patients: A feasibility study. Radiol. Med. 2020, 125, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Tagliati, C.; Lanza, C.; Pieroni, G.; Amici, L.; Carotti, M.; Giuseppetti, G.M.; Giovagnoni, A. Ultra-low-dose chest CT in adult patients with cystic fibrosis using a third-generation dual-source CT scanner. Radiol. Med. 2021, 126, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Yaguchi, A.; Okazaki, T.; Aoyagi, K.; Yamagata, H.; Sugihara, N.; Koyama, H.; Yoshikawa, T.; Sugimura, K. Comparative evaluation of newly developed model-based and commercially available hybrid-type iterative reconstruction methods and filter back projection method in terms of accuracy of computer-aided volumetry (cadv) for low-dose ct protocols in phantom study. Eur. J. Radiol. 2016, 85, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Fusco, R.; Setola, S.V.; Raiano, N.; Granata, V.; Cerciello, V.; Pecori, B.; Petrillo, A. Analysis of a monocentric computed tomography dosimetric database using a radiation dose index monitoring software: Dose levels and alerts before and after the implementation of the adaptive statistical iterative reconstruction on CT images. Radiol. Med. 2022, 127, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Agostini, A.; Borgheresi, A.; Carotti, M.; Ottaviani, L.; Badaloni, M.; Floridi, C.; Giovagnoni, A. Third-generation iterative reconstruction on a dual-source, high-pitch, low-dose chest CT protocol with tin filter for spectral shaping at 100 kV: A study on a small series of COVID-19 patients. Radiol. Med. 2021, 126, 388–398. [Google Scholar] [CrossRef] [PubMed]
- DSun, J.; Li, H.; Gao, J.; Li, J.; Li, M.; Zhou, Z.; Peng, Y. Performance evaluation of a deep learning image reconstruction (DLIR) algorithm in “double low” chest CTA in children: A feasibility study. Radiol. Med. 2021, 126, 1181–1188. [Google Scholar] [CrossRef]
- Kang, E.; Min, J.; Ye, J.C. A deep convolutional neural network using directional wavelets for low-dose X-ray ct reconstruction. Med. Phys. 2017, 44, e360–e375. [Google Scholar] [CrossRef] [PubMed]
- Higaki, T.; Nakamura, Y.; Zhou, J.; Yu, Z.; Nemoto, T.; Tatsugami, F.; Awai, K. Deep learning reconstruction at ct: Phantom study of the image characteristics. Acad. Radiol. 2020, 27, 82–87. [Google Scholar] [CrossRef]
- Zhang, D.; Mu, C.; Zhang, X.; Yan, J.; Xu, M.; Wang, Y.; Wang, Y.; Xue, H.; Chen, Y.; Jin, Z. Image quality comparison of lower extremity CTA between CT routine reconstruction algorithms and deep learning reconstruction. BMC Med. Imaging 2023, 23, 33. [Google Scholar] [CrossRef]
- Mikayama, R.; Shirasaka, T.; Kojima, T.; Sakai, Y.; Yabuuchi, H.; Kondo, M.; Kato, T. Deep-learning reconstruction for ultra-low-dose lung CT: Volumetric measurement accuracy and reproducibility of artificial ground-glass nodules in a phantom study. Br. J. Radiol. 2022, 95, 20210915. [Google Scholar] [CrossRef]
- Borghesi, A.; Sverzellati, N.; Polverosi, R.; Balbi, M.; Baratella, E.; Busso, M.; Calandriello, L.; Cortese, G.; Farchione, A.; Iezzi, R.; et al. Impact of the COVID-19 pandemic on the selection of chest imaging modalities and reporting systems: A survey of Italian radiologists. Radiol. Med. 2021, 126, 1258–1272. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.G.; Ahn, C.; Choi, H.; Hong, W.; Park, J.; Kim, J.H.; Goo, J.M. Image quality of ultralow-dose chest CT using deep learning techniques: Potential superiority of vendor-agnostic post-processing over vendor-specific techniques. Eur. Radiol. 2021, 31, 5139–5147. [Google Scholar] [CrossRef] [PubMed]
- Ziyad, S.R.; Radha, V.; Vayyapuri, T. Overview of Computer Aided Detection and Computer Aided Diagnosis Systems for Lung Nodule Detection in Computed Tomography. Curr. Med. Imaging Rev. 2020, 16, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Castellino, R.A. Computer aided detection (CAD): An overview. Cancer Imaging 2005, 5, 17–19. [Google Scholar] [CrossRef]
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep Learning Applications in Computed Tomography Images for Pulmonary Nodule Detection and Diagnosis: A Review. Diagnostics 2022, 12, 298. [Google Scholar] [CrossRef]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef]
- Mansoor, A.; Bagci, U.; Foster, B.; Xu, Z.; Papadakis, G.Z.; Folio, L.R.; Udupa, J.K.; Mollura, D.J. Segmentation and Image Analysis of Abnormal Lungs at CT: Current Approaches, Challenges, and Future Trends. Radiographics 2015, 35, 1056–1076. [Google Scholar] [CrossRef]
- Armato, S.G., 3rd; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- McNitt-Gray, M.F.; Armato, S.G., 3rd; Meyer, C.R.; Reeves, A.P.; McLennan, G.; Pais, R.C.; Freymann, J.; Brown, M.S.; Engelmann, R.M.; Bland, P.H.; et al. The Lung Image Database Consortium (LIDC) data collection process for nodule detection and annotation. Acad. Radiol. 2007, 14, 1464–1474. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; Bogaard, C.V.D.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Armato, S.G., 3rd; de Hoop, B.; van Amelsvoort-van de Vorst, S.; Duindam, T.; Niemeijer, M.; Murphy, K.; Schilham, A.; Retico, A.; Fantacci, M.E.; et al. Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study. Med. Image Anal. 2010, 14, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Grasso, R.F.; Bernetti, C.; Pacella, G.; Altomare, C.; Castiello, G.; Andresciani, F.; Sarli, M.; Zobel, B.B.; Faiella, E. A comparative analysis of thermal ablation techniques in the treatment of primary and secondary lung tumors: A single-center experience. Radiol. Med. 2022, 127, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.; Zhang, S.; Yu, X.; Wu, C.; Jiang, Y. A Novel Pulmonary Nodule Detection Model Based on Multi-Step Cascaded Networks. Sensors 2020, 20, 4301. [Google Scholar] [CrossRef] [PubMed]
- Khosravan, N.; Bagci, U. S4ND: Single-Shot Single-Scale Lung Nodule Detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 794–802. [Google Scholar] [CrossRef]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated Lung Nodule Detection and Classification Using Deep Learning Combined with Multiple Strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.A.; Malik, H.A.M.; Muhammad, A.; Alourani, A.; Butt, Z.A. Deep learning ensemble 2D CNN approach towards the detection of lung cancer. Sci. Rep. 2023, 13, 2987. [Google Scholar] [CrossRef]
- Cai, L.; Long, T.; Dai, Y.; Huang, Y. Mask R-CNN-Based Detection and Segmentation for Pulmonary Nodule 3D Visualization Diagnosis. IEEE Access 2020, 8, 44400–44409. [Google Scholar] [CrossRef]
- Manickavasagam, R.; Selvan, S.; Selvan, M. CAD system for lung nodule detection using deep learning with CNN. Med. Biol. Eng. Comput. 2022, 60, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, D.; Kandil, H.; Khelifi, A.; Yaghi, M.; Ghazal, M.; Sharafeldeen, A.; Mahmoud, A.; El-Baz, A. How AI Can Help in the Diagnostic Dilemma of Pulmonary Nodules. Cancers 2022, 14, 1840. [Google Scholar] [CrossRef]
- Wu, P.; Sun, X.; Zhao, Z.; Wang, H.; Pan, S.; Schuller, B. Classification of Lung Nodules Based on Deep Residual Networks and Migration Learning. Comput. Intell. Neurosci. 2020, 2020, 8975078. [Google Scholar] [CrossRef]
- Mastouri, R.; Khlifa, N.; Neji, H.; Hantous-Zannad, S. A Bilinear Convolutional Neural Network for Lung Nodules Classification on CT Images. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 91–101. [Google Scholar] [CrossRef]
- Zhang, G.; Lin, L.; Wang, J. Lung Nodule Classification in CT Images Using 3D DenseNet. J. Phys. Conf. Ser. 2021, 1827, 012155. [Google Scholar] [CrossRef]
- Al-Shabi, M.; Lee, H.K.; Tan, M. Gated-Dilated Networks for Lung Nodule Classification in CT Scans. IEEE Access 2019, 7, 178827–178838. [Google Scholar] [CrossRef]
- Liu, H.; Cao, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Liu, C.; Hung, C.-C. Multi-Model Ensemble Learning Architecture Based on 3D CNN for Lung Nodule Malignancy Suspiciousness Classification. J. Digit. Imaging 2020, 33, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sun, W.; Tseng, T.B.; Li, C.; Qian, W. Fast and fully-automated detection and segmentation of pulmonary nodules in thoracic CT scans using deep convolutional neural networks. Comput. Med. Imaging Graph. 2019, 74, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Lee, B.-D.; Byon, S.-S.; Kim, S.-H.; Lee, B.-I.; Shin, Y.-G. Volumetric lung nodule segmentation using adaptive ROI with multi-view residual learning. Sci. Rep. 2020, 10, 12839. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Han, J.; Jia, Y.; Gou, F. Lung nodule detection via 3D U-Net and contextual convolutional neural network. In Proceedings of the 2018 International Conference on Networking and Network Applications (NaNA), Xi’an, China, 12–15 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 356–361. [Google Scholar] [CrossRef]
- Kumar, S.; Raman, S. Lung Nodule Segmentation Using 3-Dimensional Convolutional Neural Networks. In Soft Computing for Problem Solving; Springer: Singapore, 2020; pp. 585–596. [Google Scholar] [CrossRef]
- Keetha, N.V.; Annavarapu, C.S.R. U-Det: A Modified U-Net architecture with bidirectional feature network for lung nodule segmentation. arXiv 2020, arXiv:2003.09293. [Google Scholar]
- Zhang, J.; Xia, Y.; Zeng, H.; Zhang, Y. NODULe: Combining constrained multi-scale LoG filters with densely dilated 3D deep convolutional neural network for pulmonary nodule detection. Neurocomputing 2018, 317, 159–167. [Google Scholar] [CrossRef]
- Cao, H.; Liu, H.; Song, E.; Hung, C.-C.; Ma, G.; Xu, X.; Jin, R.; Lu, J. Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 2019, 86, 105934. [Google Scholar] [CrossRef]
- Wu, B.; Zhou, Z.; Wang, J.; Wang, Y. Joint learning for pulmonary nodule segmentation, attributes and malignancy prediction. arXiv 2018, 1109–1113. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 2017, 40, 172–183. [Google Scholar] [CrossRef]
- Pezzano, G.; Ripoll, V.R.; Radeva, P. CoLe-CNN: Context-learning convolutional neural network with adaptive loss function for lung nodule segmentation. Comput. Methods Programs Biomed. 2021, 198, 105792. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Xu, S.; Liu, Y.; Wang, A.; Saripan, M.I.; Li, L.; Zhang, X.; Lu, L. Multi-view secondary input collaborative deep learning for lung nodule 3D segmentation. Cancer Imaging 2020, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabi, M.; Lan, B.L.; Chan, W.Y.; Ng, K.H.; Tan, M. Lung nodule classification using deep local–global networks. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1815–1819. [Google Scholar] [CrossRef] [PubMed]
- Ledda, R.E.; Silva, M.; McMichael, N.; Sartorio, C.; Branchi, C.; Milanese, G.; Nayak, S.M.; Sverzellati, N. The diagnostic value of grey-scale inversion technique in chest radiography. Radiol. Med. 2022, 127, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, U.; Sverzellati, N.; Sestini, S.; Silva, M.; Sabia, F.; Boeri, M.; Cantarutti, A.; Sozzi, G.; Corrao, G.; Marchianò, A. Ten-year results of the Multicentric Italian Lung Detection trial demonstrate the safety and efficacy of biennial lung cancer screening. Eur. J. Cancer 2019, 118, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.; Pucciarelli, F.; Zerunian, M.; Ganeshan, B.; De Santis, D.; Polici, M.; Rucci, C.; Polidori, T.; Guido, G.; Bracci, B.; et al. Chest CT texture-based radiomics analysis in differentiating COVID-19 from other interstitial pneumonia. Radiol. Med. 2021, 126, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Schreuder, A.; Scholten, E.T.; van Ginneken, B.; Jacobs, C. Artificial intelligence for detection and characterization of pulmonary nodules in lung cancer CT screening: Ready for practice? Transl. Lung Cancer Res. 2021, 10, 2378–2388. [Google Scholar] [CrossRef]
- Cozzi, D.; Bicci, E.; Cavigli, E.; Danti, G.; Bettarini, S.; Tortoli, P.; Mazzoni, L.N.; Busoni, S.; Pradella, S.; Miele, V. Radiomics in pulmonary neuroendocrine tumours (NETs). Radiol. Med. 2022, 127, 609–615. [Google Scholar] [CrossRef]
- Cellina, M.; Pirovano, M.; Ciocca, M.; Gibelli, D.; Floridi, C.; Oliva, G. Radiomic analysis of the optic nerve at the first episode of acute optic neuritis: An indicator of optic nerve pathology and a predictor of visual recovery? Radiol. Med. 2021, 126, 698–706. [Google Scholar] [CrossRef]
- McCague, C.; Ramlee, S.; Reinius, M.; Selby, I.; Hulse, D.; Piyatissa, P.; Bura, V.; Crispin-Ortuzar, M.; Sala, E.; Woitek, R. Introduction to radiomics for a clinical audience. Clin. Radiol. 2023, 78, 83–98. [Google Scholar] [CrossRef]
- Caruso, D.; Polici, M.; Rinzivillo, M.; Zerunian, M.; Nacci, I.; Marasco, M.; Magi, L.; Tarallo, M.; Gargiulo, S.; Iannicelli, E.; et al. CT-based radiomics for prediction of therapeutic response to Everolimus in metastatic neuroendocrine tumors. Radiol. Med. 2022, 127, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Morales, J.; Tunali, I.; Stringfield, O.; Eschrich, S.A.; Balagurunathan, Y.; Gillies, R.J.; Schabath, M.B. Peritumoral and intratumoral radiomic features predict survival outcomes among patients diagnosed in lung cancer screening. Sci. Rep. 2020, 10, 10528. [Google Scholar] [CrossRef] [PubMed]
- Mega, S.; Fiore, M.; Carpenito, M.; Novembre, M.L.; Miele, M.; Trodella, L.E.; Grigioni, F.; Ippolito, E.; Ramella, S. Early GLS changes detection after chemoradiation in locally advanced non-small cell lung cancer (NSCLC). Radiol. Med. 2022, 127, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.J.; Wu, F.Z.; Yang, S.C.; Tang, E.K.; Liang, C.H. Radiomics in Early Lung Cancer Diagnosis: From Diagnosis to Clinical Decision Support and Education. Diagnostics 2022, 12, 1064. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhao, Z.; Wang, X.; Ai, H.; Yang, C.; Luo, Y.; Jiang, X. Radiomics for prediction of response to EGFR-TKI based on metastasis/brain parenchyma (M/BP)-interface. Radiol. Med. 2022, 127, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Binczyk, F.; Prazuch, W.; Bozek, P.; Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res. 2021, 10, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Gitto, S.; Bologna, M.; Corino, V.D.A.; Emili, I.; Albano, D.; Messina, C.; Armiraglio, E.; Parafioriti, A.; Luzzati, A.; Mainardi, L.; et al. Diffusion-weighted MRI radiomics of spine bone tumors: Feature stability and machine learning-based classification performance. Radiol. Med. 2022, 127, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Coroller, T.P.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother. Oncol. 2016, 119, 480–486. [Google Scholar] [CrossRef]
- Satake, H.; Ishigaki, S.; Ito, R.; Naganawa, S. Radiomics in breast MRI: Current progress toward clinical application in the era of artificial intelligence. Radiol. Med. 2022, 127, 39–56. [Google Scholar] [CrossRef]
- Lee, G.; Park, H.; Bak, S.H.; Lee, H.Y. Radiomics in Lung Cancer from Basic to Advanced: Current Status and Future Directions. Korean J. Radiol. 2020, 21, 159–171. [Google Scholar] [CrossRef]
- Tu, S.J.; Wang, C.W.; Pan, K.T.; Wu, Y.C.; Wu, C.T. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys. Med. Biol. 2018, 63, 065005. [Google Scholar] [CrossRef] [PubMed]
- Gibelli, D.; Cellina, M.; Gibelli, S.; Cappella, A.; Oliva, A.G.; Termine, G.; Dolci, C.; Sforza, C. Relationship between sphenoid sinus volume and protrusion of internal carotid artery and optic nerve: A 3D segmentation study on maxillofacial CT-scans. Surg. Radiol. Anat. 2019, 41, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Chiti, G.; Grazzini, G.; Flammia, F.; Matteuzzi, B.; Tortoli, P.; Bettarini, S.; Pasqualini, E.; Granata, V.; Busoni, S.; Messserini, L.; et al. Gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs): A radiomic model to predict tumor grade. Radiol. Med. 2022, 127, 928–938. [Google Scholar] [CrossRef]
- Baeßler, B.; Weiss, K.; Pinto Dos Santos, D. Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging: A Phantom Study. Investig. Radiol. 2019, 54, 221–228. [Google Scholar] [CrossRef]
- Autorino, R.; Gui, B.; Panza, G.; Boldrini, L.; Cusumano, D.; Russo, L.; Nardangeli, A.; Persiani, S.; Campitelli, M.; Ferrandina, G.; et al. Radiomics-based prediction of two-year clinical outcome in locally advanced cervical cancer patients undergoing neoadjuvant chemoradiotherapy. Radiol. Med. 2022, 127, 498–506. [Google Scholar] [CrossRef] [PubMed]
- MacMahon, H.; Naidich, D.P.; Goo, J.M.; Lee, K.S.; Leung, A.N.C.; Mayo, J.R.; Mehta, A.C.; Ohno, Y.; Powell, C.A.; Prokop, M.; et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017, 284, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Bueno, J.; Landeras, L.; Chung, J. Updated Fleischner Society Guidelines for Managing Incidental Pulmonary Nodules: Common Questions and Challenging Scenarios. Radiographics 2018, 38, 1337–1350. [Google Scholar] [CrossRef]
- Gould, M.K.; Donington, J.; Lynch, W.R.; Mazzone, P.J.; Midthun, D.E.; Naidich, D.P.; Wiener, R.S. Evaluation of individuals with pulmonary nodules: When is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143, e93S–e120S. [Google Scholar] [CrossRef]
- Callister, M.E.; Baldwin, D.R.; Akram, A.R.; Barnard, S.; Cane, P.; Draffan, J.; Franks, K.; Gleeson, F.; Graham, R.; Malhotra, P.; et al. British Thoracic Society Pulmonary Nodule Guideline Development Group; British Thoracic Society Standards of Care Committee. British Thoracic Society guidelines for the investigation and management of pulmonary nodules. Thorax 2015, 70 (Suppl. 2), ii1–ii54. [Google Scholar] [CrossRef]
- Graham, R.N.; Baldwin, D.R.; Callister, M.E.; Gleeson, F.V. Return of the pulmonary nodule: The radiologist’s key role in implementing the 2015 BTS guidelines on the investigation and management of pulmonary nodules. Br. J. Radiol. 2016, 89, 20150776. [Google Scholar] [CrossRef]
- Chassagnon, G.; De Margerie-Mellon, C.; Vakalopoulou, M.; Marini, R.; Hoang-Thi, T.N.; Revel, M.P.; Soyer, P. Artificial intelligence in lung cancer: Current applications and perspectives. Jpn. J. Radiol. 2023, 41, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Chen, Y.; Li, X.; Li, W.; Zhang, X.; He, T.; Yu, Y.; Dou, Y.; Duan, H.; Yu, N. Development and validation of a 3D-convolutional neural network model based on chest CT for differentiating active pulmonary tuberculosis from community-acquired pneumonia. Radiol. Med. 2023, 128, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Devaraj, A. Radiomics of pulmonary nodules and lung cancer. Transl. Lung Cancer Res. 2017, 6, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Q.; Ren, Y.; Hu, H.; Zhao, J. Automatic lung nodule classification with radiomics approach. In Medical Imaging 2016, Proceedings of the PACS and Imaging Informatics: Next Generation and Innovations 2016, San Diego, CA, USA, 5 April 2016; SPIE: Bellingham, WA, USA, 2016; Volume 9879. [Google Scholar]
- Chae, H.D.; Park, C.M.; Park, S.J.; Lee, S.M.; Kim, K.G.; Goo, J.M. Computerized texture analysis of persistent part-solid ground-glass nodules: Differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 2014, 273, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, W.; Xie, D.; Yu, Y.; Cao, H.; Liao, G.; Wu, S.; Yao, L.; Wang, Z.; Wang, M.; et al. Development and validation of a preoperative CT-based radiomic nomogram to predict pathology invasiveness in patients with a solitary pulmonary nodule: A machine learning approach, multicenter, diagnostic study. Eur. Radiol. 2022, 32, 1983–1996. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Tang, C.; Hobbs, B.P.; Li, X.; Koay, E.J.; Wistuba, I.I.; Sepesi, B.; Behrens, C.; Rodriguez Canales, J.; Parra Cuentas, E.R.; et al. Development and Validation of a Predictive Radiomics Model for Clinical Outcomes in Stage I Non-small Cell Lung Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Cousin, F.; Louis, T.; Dheur, S.; Aboubakar, F.; Ghaye, B.; Occhipinti, M.; Vos, W.; Bottari, F.; Paulus, A.; Sibille, A.; et al. Radiomics and Delta-Radiomics Signatures to Predict Response and Survival in Patients with Non-Small-Cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Cancers 2023, 15, 1968. [Google Scholar] [CrossRef]
- Tunali, I.; Gillies, R.J.; Schabath, M.B. Application of Radiomics and Artificial Intelligence for Lung Cancer Precision Medicine. Cold Spring Harb. Perspect. Med. 2021, 11, a039537. [Google Scholar] [CrossRef]
- Danti, G.; Flammia, F.; Matteuzzi, B.; Cozzi, D.; Berti, V.; Grazzini, G.; Pradella, S.; Recchia, L.; Brunese, L.; Miele, V. Gastrointestinal neuroendocrine neoplasms (GI-NENs): Hot topics in morphological, functional, and prognostic imaging. Radiol. Med. 2021, 126, 1497–1507. [Google Scholar] [CrossRef]
- Hou, K.Y.; Chen, J.R.; Wang, Y.C.; Chiu, M.H.; Lin, S.P.; Mo, Y.H.; Peng, S.C.; Lu, C.F. Radiomics-Based Deep Learning Prediction of Overall Survival in Non-Small-Cell Lung Cancer Using Contrast-Enhanced Computed Tomography. Cancers 2022, 14, 3798. [Google Scholar] [CrossRef]
- Mitra, S.; Shankar, B.U. Integrating Radio Imaging With Gene Expressions Toward a Personalized Management of Cancer. IEEE Trans. Hum.-Mach. Syst. 2014, 44, 664–677. [Google Scholar] [CrossRef]
- Vicini, S.; Bortolotto, C.; Rengo, M.; Ballerini, D.; Bellini, D.; Carbone, I.; Preda, L.; Laghi, A.; Coppola, F.; Faggioni, L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: Focus on the three most common cancers. Radiol. Med. 2022, 127, 819–836. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, J.; Yang, X.; Xu, F.; Wang, L.; He, C.; Lin, L.; Qing, H.; Ren, J.; Zhou, P. Radiomic and quantitative-semantic models of low-dose computed tomography for predicting the poorly differentiated invasive non-mucinous pulmonary adenocarcinoma. Radiol. Med. 2023, 128, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. Radiol. Med. 2021, 126, 1296–1311. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- De Robertis, R.; Geraci, L.; Tomaiuolo, L.; Bortoli, L.; Beleù, A.; Malleo, G.; D’Onofrio, M. Liver metastases in pancreatic ductal adenocarcinoma: A predictive model based on CT texture analysis. Radiol. Med. 2022, 127, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Bracci, S.; Dolciami, M.; Trobiani, C.; Izzo, A.; Pernazza, A.; D’Amati, G.; Manganaro, L.; Ricci, P. Quantitative CT texture analysis in predicting PD-L1 expression in locally advanced or metastatic NSCLC patients. Radiol. Med. 2021, 126, 1425–1433. [Google Scholar] [CrossRef]
- Chen, K.; Wang, M.; Song, Z. Multi-task learning-based histologic subtype classification of non-small cell lung cancer. Radiol. Med. 2023, 128, 537–543. [Google Scholar] [CrossRef]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’Ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I.; et al. Delta radiomics: A systematic review. Radiol. Med. 2021, 126, 1571–1583. [Google Scholar] [CrossRef]
- Gregucci, F.; Fiorentino, A.; Mazzola, R.; Ricchetti, F.; Bonaparte, I.; Surgo, A.; Figlia, V.; Carbonara, R.; Caliandro, M.; Ciliberti, M.P.; et al. Radiomic analysis to predict local response in locally advanced pancreatic cancer treated with stereotactic body radiation therapy. Radiol. Med. 2022, 127, 100–107. [Google Scholar] [CrossRef]
- Anagnostopoulos, A.K.; Gaitanis, A.; Gkiozos, I.; Athanasiadis, E.I.; Chatziioannou, S.N.; Syrigos, K.N.; Thanos, D.; Chatziioannou, A.N.; Papanikolaou, N. Radiomics/Radiogenomics in Lung Cancer: Basic Principles and Initial Clinical Results. Cancers 2022, 14, 1657. [Google Scholar] [CrossRef]
- Lee, J.; Bartholmai, B.; Peikert, T.; Chun, J.; Kim, H.; Kim, J.S.; Park, S.Y. Evaluation of Computer-Aided Nodule Assessment and Risk Yield (CANARY) in Korean patients for prediction of invasiveness of ground-glass opacity nodule. PLoS ONE 2021, 16, e0253204. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.K.R.; Saeed, U.A.; McDougall, C.C.; Sabri, A.; Kovacina, B.; Raidu, B.V.S.; Khokhar, R.A.; Probst, S.; Hirsh, V.; Chankowsky, J.; et al. Radiogenomic Models Using Machine Learning Techniques to Predict EGFR Mutations in Non-Small Cell Lung Cancer. Can. Assoc. Radiol. J. 2021, 72, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Lafata, K.J.; Corradetti, M.N.; Gao, J.; Jacobs, C.D.; Weng, J.; Chang, Y.; Wang, C.; Hatch, A.; Xanthopoulos, E.; Jones, G.; et al. Radiogenomic Analysis of Locally Advanced Lung Cancer Based on CT Imaging and Intratreatment Changes in Cell-Free DNA. Radiol. Imaging Cancer 2021, 3, e200157. [Google Scholar] [CrossRef] [PubMed]
- Xue, K.; Liu, L.; Liu, Y.; Guo, Y.; Zhu, Y.; Zhang, M. Radiomics model based on multi-sequence MR images for predicting preoperative immunoscore in rectal cancer. Radiol. Med. 2022, 127, 702–713. [Google Scholar] [CrossRef]
- Singh, R.; Kalra, M.K.; Homayounieh, F.; Nitiwarangkul, C.; McDermott, S.; Little, B.P.; Lennes, I.T.; Shepard, J.O.; Digumarthy, S.R. Artificial intelligence-based vessel suppression for detection of sub-solid nodules in lung cancer screening computed tomography. Quant. Imaging Med. Surg. 2021, 11, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Sardanelli, F.; Colarieti, A. Open issues for education in radiological research: Data integrity, study reproducibility, peer-review, levels of evidence, and cross-fertilization with data scientists. Radiol. Med. 2023, 128, 133–135. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cellina, M.; Cacioppa, L.M.; Cè, M.; Chiarpenello, V.; Costa, M.; Vincenzo, Z.; Pais, D.; Bausano, M.V.; Rossini, N.; Bruno, A.; et al. Artificial Intelligence in Lung Cancer Screening: The Future Is Now. Cancers 2023, 15, 4344. https://doi.org/10.3390/cancers15174344
Cellina M, Cacioppa LM, Cè M, Chiarpenello V, Costa M, Vincenzo Z, Pais D, Bausano MV, Rossini N, Bruno A, et al. Artificial Intelligence in Lung Cancer Screening: The Future Is Now. Cancers. 2023; 15(17):4344. https://doi.org/10.3390/cancers15174344
Chicago/Turabian StyleCellina, Michaela, Laura Maria Cacioppa, Maurizio Cè, Vittoria Chiarpenello, Marco Costa, Zakaria Vincenzo, Daniele Pais, Maria Vittoria Bausano, Nicolò Rossini, Alessandra Bruno, and et al. 2023. "Artificial Intelligence in Lung Cancer Screening: The Future Is Now" Cancers 15, no. 17: 4344. https://doi.org/10.3390/cancers15174344
APA StyleCellina, M., Cacioppa, L. M., Cè, M., Chiarpenello, V., Costa, M., Vincenzo, Z., Pais, D., Bausano, M. V., Rossini, N., Bruno, A., & Floridi, C. (2023). Artificial Intelligence in Lung Cancer Screening: The Future Is Now. Cancers, 15(17), 4344. https://doi.org/10.3390/cancers15174344






