Automatic Segmentation with Deep Learning in Radiotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Related Work
3. Data Collection
- Title: “segmenting” or “*segmentation”;
- Title, abstract, or keywords: “CT”, “MRI”, “PET”, “DWI”, or “*medical image*”;
- Title, abstract, or keywords: “deep learning” or “artificial neural network*”;
- Title, abstract, or keywords: “radiotherapy”;
- Document type: article or conference proceeding;
- Language: English.
- Is the paper a review article? (Yes/No).
- How many patients were used in the study? Only answer with a number.
- What type of images were used in the study? Choose from MRI/PET/CT/DWI/Ultrasound/Other.
- Did the paper use images from multiple sources or multiple organs? (Yes/No).
- Is the paper about organ or tumor segmentation? Choose from Organ/Tumor/Both.
- Did the paper propose a novel segmentation method or deep learning architecture? (Yes/No).
- Was the code made public? (Yes/No).
- Was the data made public? (Yes/No).
- What is the key takeaway of the paper? Answer in one sentence.
“You will be asked to read a scientific paper and then answer questions about it. The paper is about automatic segmentation with deep learning in radiotherapy. Please prioritize correctness in your answers. If you don’t know the answer, respond with ‘I don’t know.’”
4. Analysis
4.1. Statistics
4.2. Region-Specific Analysis
4.3. Subjective Analysis
- What is missing from the current corpus?
- What should researchers think about when starting a segmentation study?
- How can research practices in medical image segmentation be improved?
- Do authors agree on conclusions, and is it possible to spot trends in the employed methods?
5. Results
5.1. Statistics
5.2. Region-Specific Analysis
5.2.1. Head and Neck
5.2.2. Brain
5.2.3. Prostate
5.2.4. Lung
5.2.5. Liver
5.2.6. Heart
5.2.7. Colon
5.2.8. Kidney
5.2.9. Breast
5.2.10. Pancreas
5.2.11. Cervix
5.2.12. Spleen
5.2.13. Skin
5.3. Subjective Analysis
5.3.1. What Is Missing from the Current Corpus?
- Benchmark datasets: At present, direct comparisons of results remain unfeasible since very few studies use the same data. Specifically, only 92 studies employed data sourced from at least one of the following datasets: UK Biobank, BraTS, KiTS, HECKTOR, OASIS, or PROMISE. Moreover, there is a very strong preference for novelty over replication in the current corpus. This prevents researchers from knowing how their model fares against other models without having to implement them themselves (often from scratch), which in itself is often not possible due to insufficient details having been given in the publications of other groups. Efforts to publish high-quality benchmarks, analogous to the CIFAR10 and ImageNet datasets in general computer vision, are much more likely to yield high-impact results, especially since the literature is dominated by presentations of novel “state of the art” models. It is possible that this overabundance is a direct consequence of the lack of benchmarks, possibly convincing researchers that minor improvements on very small datasets result in a state-of-the-art model (while, in reality, it may just be the result of statistical variations).
- Open sourcing and making code available: Out of 807 papers, only three made code of their models available; an abysmal number compared to numbers in related research disciplines. Open-sourcing code is a great way to promote research and enable other researchers to benchmark their models and implement strong baselines, ultimately driving the field forward. If there are privacy objections to open-sourcing, a good compromise is to publish the code without the model weights.
- More research on, e.g., spleen and pancreas: As seen clearly in Figure 3, relatively few papers have been published on the spleen and pancreas compared to, e.g., H and N or lung cancer. It might be worthwhile for institutions to put more effort into collecting data in these areas so models and methods can be tested in a wider variety of settings.
- Evaluation of alternative training techniques: Computer vision is a rapidly evolving field with successful novel paradigms being introduced relatively frequently. Some notable training strategies appear to be less studied in the field of medical image segmentation. In particular, only 48 papers made use of transfer learning or pretraining, only 21 papers studied self-supervised, semi-supervised, or contrastive learning, and only 11 papers explicitly studied transformer-based architectures. It is possible that segmenting medical images could be more amenable to these methods than what is currently being realized.
5.3.2. What Should Researchers Consider When Starting a Segmentation Study?
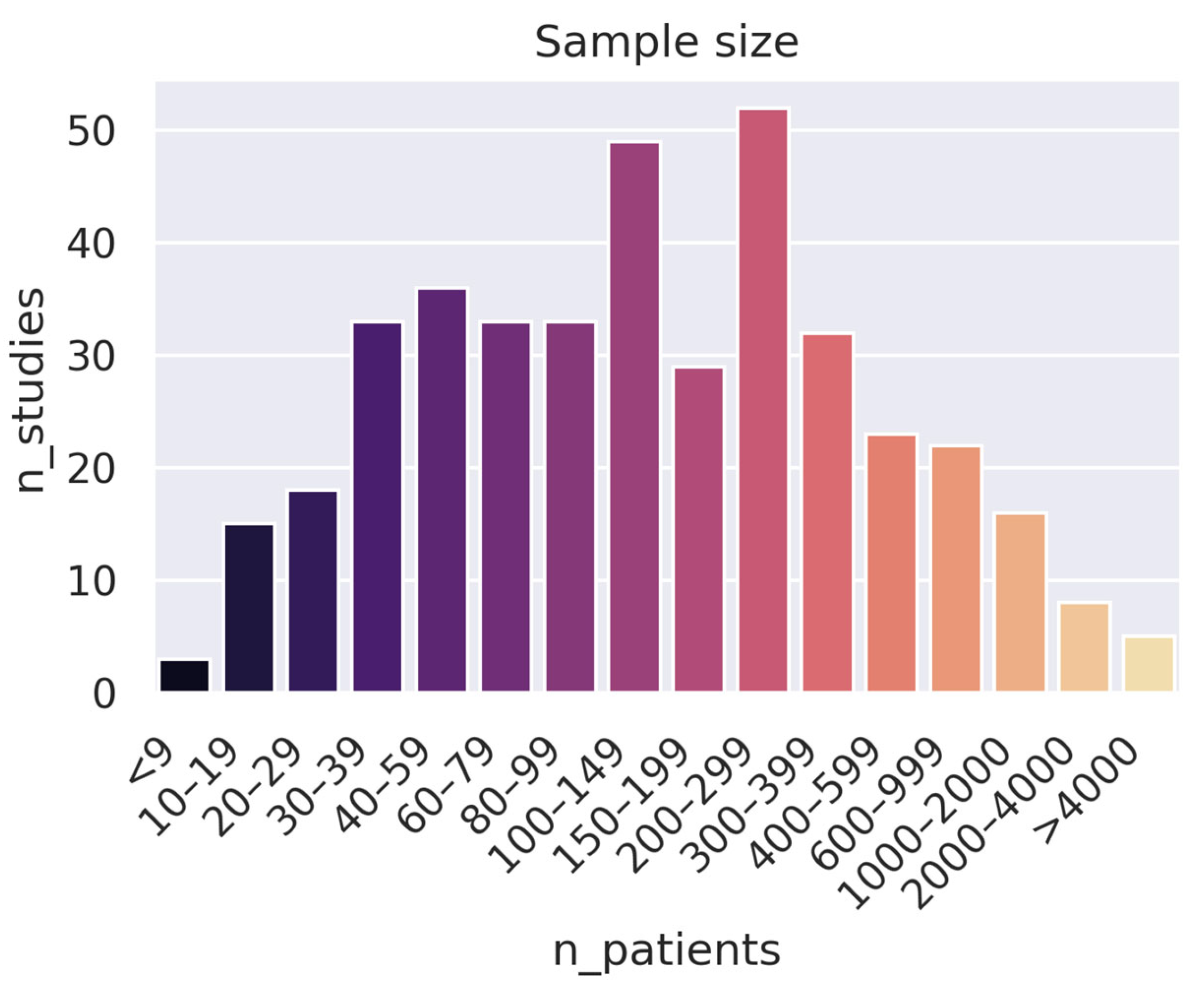
- Collect more (high-quality) data: An alarming number of studies use very small datasets: 163 studies used fewer than 100 patients. While this can be good as a proof of concept, it is hard to draw conclusions from such a narrow scope. Furthermore, increasing the number of data samples can often be more effective than developing a stronger training pipeline (e.g., a better model) when it comes to both performance and generalizability.
- Test models on multiple datasets and cancer sites: Diversifying a model by training it on qualitatively different images is a great way to demonstrate the capacity of a model, even if it is intended for a very specific purpose. Moreover, models trained on multiple modalities (e.g., both images and patient data) often show superior performance compared to single-modality training [50,51,52,53,54]. One way to do this is to include images from open sources like the UK Biobank, the Cancer Imaging Archive (TCIA), the BraTS (brain tumor segmentation) dataset, the MM-WHS (Multi-modality whole heart segmentation) dataset, and even The Cancer Genome Atlas (TCGA).
- Evaluate existing models and training techniques over developing new ones: There is a relative overabundance of papers proposing novel deep learning architectures/modules and claiming state-of-the-art performance on rather narrow datasets. It seems exceedingly unlikely that a model trained on just about 50 patients can really be considered state-of-the-art within a certain field. As such, the scientific community should focus more on evaluating existing models and training techniques on larger and more representative datasets and benchmarking them against established baselines.
- Focus on clinical viability over minor improvements in metrics: A limited number of publications assess the practical feasibility of their models, such as subjecting them to downstream applications, investigating their generalizability across different patient populations and clinical scenarios, or evaluating the contours they produce from a medical standpoint. It seems likely that the difficult legislative environment and distance to market limits the number of groups willing to dedicate time and energy to rendering their models suitable for clinics. The regulatory landscape for healthcare is complex and can be a significant obstacle to the implementation of new technologies, which is why research on these issues is important.
5.3.3. How Can Research Practices in Medical Image Segmentation Be Improved?
- Follow good reporting/documentation guidelines: We found a surprising number of studies lacking critical information needed to reproduce or even implement the methods given in the study. In particular, multiple studies (and abstracts) fail to report details about the patients (e.g., the class balance), the training procedure (e.g., learning rate, data augmentation parameters, and dropout rate), or the validation procedure (e.g., whether the train-test split was random or not). A good place to start is to follow proposed guidelines such as [55] or [56].
- Do cross-validation: Despite being a standard practice in machine learning, a surprising number of studies do not cross-validate their models and instead opt to use a single train-test split. This severely limits the validity of the conclusions, particularly for smaller datasets, and is strongly discouraged.
- Implement more robust baselines: Implementing a strong baseline is critical in order to adequately assess a new model (particularly due to the lack of open benchmarks). Yet, a number of studies appear to use dated or suboptimally implemented references such as a naïve U-net. A good place to start is models like SegFormer [57] or nn-Unet [58], which are open-source and can be copied directly from their respective repositories.
5.3.4. Do Authors Agree on Conclusions, and Is It Possible to Spot Trends in the Employed Methods?
6. Limitations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brouwer, C.L.; Steenbakkers, R.J.; van den Heuvel, E.; Duppen, J.C.; Navran, A.; Bijl, H.P.; Chouvalova, O.; Burlage, F.R.; Meertens, H.; Langendijk, J.A.; et al. 3D Variation in Delineation of Head and Neck Organs at Risk. Radiat. Oncol. 2012, 7, 32. [Google Scholar] [CrossRef]
- Veiga-Canuto, D.; Cerdà-Alberich, L.; Sangüesa Nebot, C.; Martínez de las Heras, B.; Pötschger, U.; Gabelloni, M.; Carot Sierra, J.M.; Taschner-Mandl, S.; Düster, V.; Cañete, A.; et al. Comparative Multicentric Evaluation of Inter-Observer Variability in Manual and Automatic Segmentation of Neuroblastic Tumors in Magnetic Resonance Images. Cancers 2022, 14, 3648. [Google Scholar] [CrossRef]
- Van de Steene, J.; Linthout, N.; de Mey, J.; Vinh-Hung, V.; Claassens, C.; Noppen, M.; Bel, A.; Storme, G. Definition of Gross Tumor Volume in Lung Cancer: Inter-Observer Variability. Radiother. Oncol. 2002, 62, 37–49. [Google Scholar] [CrossRef]
- Senan, S.; van Sörnsen de Koste, J.; Samson, M.; Tankink, H.; Jansen, P.; Nowak, P.J.C.M.; Krol, A.D.G.; Schmitz, P.; Lagerwaard, F.J. Evaluation of a Target Contouring Protocol for 3D Conformal Radiotherapy in Non-Small Cell Lung Cancer. Radiother. Oncol. 1999, 53, 247–255. [Google Scholar] [CrossRef]
- Covert, E.C.; Fitzpatrick, K.; Mikell, J.; Kaza, R.K.; Millet, J.D.; Barkmeier, D.; Gemmete, J.; Christensen, J.; Schipper, M.J.; Dewaraja, Y.K. Intra- and Inter-Operator Variability in MRI-Based Manual Segmentation of HCC Lesions and Its Impact on Dosimetry. EJNMMI Phys. 2022, 9, 90. [Google Scholar] [CrossRef]
- Rasch, C.; Eisbruch, A.; Remeijer, P.; Bos, L.; Hoogeman, M.; van Herk, M.; Lebesque, J.V. Irradiation of Paranasal Sinus Tumors, a Delineation and Dose Comparison Study. Int. J. Radiat. Oncol. *Biol. *Phys. 2002, 52, 120–127. [Google Scholar] [CrossRef]
- Becker, A.S.; Chaitanya, K.; Schawkat, K.; Muehlematter, U.J.; Hötker, A.M.; Konukoglu, E.; Donati, O.F. Variability of Manual Segmentation of the Prostate in Axial T2-Weighted MRI: A Multi-Reader Study. Eur. J. Radiol. 2019, 121, 108716. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes. arXiv 2019, arXiv:1904.00445. [Google Scholar]
- Ji, Y.; Bai, H.; Ge, C.; Yang, J.; Zhu, Y.; Zhang, R.; Li, Z.; Zhanng, L.; Ma, W.; Wan, X.; et al. AMOS: A Large-Scale Abdominal Multi-Organ Benchmark for Versatile Medical Image Segmentation. Adv. Neural Inf. Process. Syst. 2022, 35, 36722–36732. [Google Scholar]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.; et al. Evaluation of Prostate Segmentation Algorithms for MRI: The PROMISE12 Challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef]
- Li, Z.; Kamnitsas, K.; Glocker, B. Overfitting of Neural Nets Under Class Imbalance: Analysis and Improvements for Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 402–410. [Google Scholar]
- Gu, J.; Zhao, H.; Tresp, V.; Torr, P. Adversarial Examples on Segmentation Models Can Be Easy to Transfer. Available online: https://arxiv.org/abs/2111.11368v1 (accessed on 25 June 2023).
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A Survey of Transfer Learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Sagi, O.; Rokach, L. Ensemble Learning: A Survey. WIREs Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Yang, C.; Qin, L.; Xie, Y.; Liao, J. Deep Learning in CT Image Segmentation of Cervical Cancer: A Systematic Review and Meta-Analysis. Radiat. Oncol. 2022, 17, 175. [Google Scholar] [CrossRef]
- Kalantar, R.; Lin, G.; Winfield, J.M.; Messiou, C.; Lalondrelle, S.; Blackledge, M.D.; Koh, D.-M. Automatic Segmentation of Pelvic Cancers Using Deep Learning: State-of-the-Art Approaches and Challenges. Diagnostics 2021, 11, 1964. [Google Scholar] [CrossRef]
- Liu, X.; Li, K.-W.; Yang, R.; Geng, L.-S. Review of Deep Learning Based Automatic Segmentation for Lung Cancer Radiotherapy. Front. Oncol. 2021, 11, 717039. [Google Scholar] [CrossRef]
- Fernando, K.R.M.; Tsokos, C.P. Deep and Statistical Learning in Biomedical Imaging: State of the Art in 3D MRI Brain Tumor Segmentation. Inf. Fusion 2023, 92, 450–465. [Google Scholar] [CrossRef]
- Altini, N.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; et al. Liver, Kidney and Spleen Segmentation from CT Scans and MRI with Deep Learning: A Survey. Neurocomputing 2022, 490, 30–53. [Google Scholar] [CrossRef]
- Badrigilan, S.; Nabavi, S.; Abin, A.A.; Rostampour, N.; Abedi, I.; Shirvani, A.; Ebrahimi Moghaddam, M. Deep Learning Approaches for Automated Classification and Segmentation of Head and Neck Cancers and Brain Tumors in Magnetic Resonance Images: A Meta-Analysis Study. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 529–542. [Google Scholar] [CrossRef]
- Zöllner, F.G.; Kociński, M.; Hansen, L.; Golla, A.-K.; Trbalić, A.Š.; Lundervold, A.; Materka, A.; Rogelj, P. Kidney Segmentation in Renal Magnetic Resonance Imaging—Current Status and Prospects. IEEE Access 2021, 9, 71577–71605. [Google Scholar] [CrossRef]
- Ali, H.M.; Kaiser, M.S.; Mahmud, M. Application of Convolutional Neural Network in Segmenting Brain Regions from MRI Data. In Proceedings of the Brain Informatics; Liang, P., Goel, V., Shan, C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 136–146. [Google Scholar]
- Ranjbarzadeh, R.; Dorosti, S.; Jafarzadeh Ghoushchi, S.; Caputo, A.; Tirkolaee, E.B.; Ali, S.S.; Arshadi, Z.; Bendechache, M. Breast Tumor Localization and Segmentation Using Machine Learning Techniques: Overview of Datasets, Findings, and Methods. Comput. Biol. Med. 2023, 152, 106443. [Google Scholar] [CrossRef]
- Habijan, M.; Babin, D.; Galić, I.; Leventić, H.; Romić, K.; Velicki, L.; Pižurica, A. Overview of the Whole Heart and Heart Chamber Segmentation Methods. Cardiovasc. Eng. Technol. 2020, 11, 725–747. [Google Scholar] [CrossRef]
- Islam, M.Z.; Naqvi, R.A.; Haider, A.; Kim, H.S. Deep Learning for Automatic Tumor Lesions Delineation and Prognostic Assessment in Multi-Modality PET/CT: A Prospective Survey. Eng. Appl. Artif. Intell. 2023, 123, 106276. [Google Scholar] [CrossRef]
- Mohammed, Y.M.A.; El Garouani, S.; Jellouli, I. A Survey of Methods for Brain Tumor Segmentation-Based MRI Images. J. Comput. Des. Eng. 2023, 10, 266–293. [Google Scholar] [CrossRef]
- Punn, N.S.; Agarwal, S. Modality Specific U-Net Variants for Biomedical Image Segmentation: A Survey. Artif. Intell. Rev. 2022, 55, 5845–5889. [Google Scholar] [CrossRef]
- El Jurdi, R.; Petitjean, C.; Honeine, P.; Cheplygina, V.; Abdallah, F. High-Level Prior-Based Loss Functions for Medical Image Segmentation: A Survey. Comput. Vis. Image Underst. 2021, 210, 103248. [Google Scholar] [CrossRef]
- Sharma, N.; Aggarwal, L.M. Automated Medical Image Segmentation Techniques. J. Med. Phys. 2010, 35, 3–14. [Google Scholar] [CrossRef]
- Lin, X.; Li, X. Image Based Brain Segmentation: From Multi-Atlas Fusion to Deep Learning. Curr. Med. Imaging Rev. 2019, 15, 443–452. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Latif, G.; Alghazo, J.; Sibai, F.N.; Iskandar, D.N.F.A.; Khan, A.H. Recent Advancements in Fuzzy C-Means Based Techniques for Brain MRI Segmentation. Curr. Med. Imaging 2021, 17, 917–930. [Google Scholar] [CrossRef]
- Trimpl, M.J.; Primakov, S.; Lambin, P.; Stride, E.P.J.; Vallis, K.A.; Gooding, M.J. Beyond Automatic Medical Image Segmentation-the Spectrum between Fully Manual and Fully Automatic Delineation. Phys. Med. Biol. 2022, 67, 12TR01. [Google Scholar] [CrossRef]
- Sakshi; Kukreja, V. Image Segmentation Techniques: Statistical, Comprehensive, Semi-Automated Analysis and an Application Perspective Analysis of Mathematical Expressions. Arch. Comput. Methods Eng. 2023, 30, 457–495. [Google Scholar] [CrossRef]
- Shao, J.; Chen, S.; Zhou, J.; Zhu, H.; Wang, Z.; Brown, M. Application of U-Net and Optimized Clustering in Medical Image Segmentation: A Review. CMES 2023, 136, 2173–2219. [Google Scholar] [CrossRef]
- Qureshi, I.; Yan, J.; Abbas, Q.; Shaheed, K.; Riaz, A.B.; Wahid, A.; Khan, M.W.J.; Szczuko, P. Medical Image Segmentation Using Deep Semantic-Based Methods: A Review of Techniques, Applications and Emerging Trends. Inf. Fusion 2023, 90, 316–352. [Google Scholar] [CrossRef]
- Huang, L.; Ruan, S.; Denœux, T. Application of Belief Functions to Medical Image Segmentation: A Review. Inf. Fusion 2023, 91, 737–756. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. A Review of Deep Learning Based Methods for Medical Image Multi-Organ Segmentation. Phys. Medica 2021, 85, 107–122. [Google Scholar] [CrossRef]
- OpenAI Introducing ChatGPT. Available online: https://openai.com/blog/chatgpt (accessed on 25 June 2023).
- Koss, J.E.; Newman, F.D.; Johnson, T.K.; Kirch, D.L. Abdominal Organ Segmentation Using Texture Transforms and a Hopfield Neural Network. IEEE Trans. Med. Imaging 1999, 18, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Popple, R.A.; Griffith, H.R.; Sawrie, S.M.; Fiveash, J.B.; Brezovich, I.A. Implementation of Talairach Atlas Based Automated Brain Segmentation for Radiation Therapy Dosimetry. Technol. Cancer Res. Treat. 2006, 5, 15–21. [Google Scholar] [CrossRef]
- Sharma, N.; Ray, A.K.; Sharma, S.; Shukla, K.K.; Pradhan, S.; Aggarwal, L.M. Segmentation and Classification of Medical Images Using Texture-Primitive Features: Application of BAM-Type Artificial Neural Network. J. Med. Phys. 2008, 33, 119–126. [Google Scholar] [CrossRef]
- Sharif, M.S.; Abbod, M.; Amira, A.; Zaidi, H. Novel Hybrid Approach Combining ANN and MRA for PET Volume Segmentation. In Proceedings of the 2010 IEEE Asia Pacific Conference on Circuits and Systems, Kuala Lumpur, Malaysia, 6–9 December 2010; pp. 596–599. [Google Scholar]
- Sharif, M.S.; Abbod, M.; Amira, A.; Zaidi, H. Artificial Neural Network-Based System for PET Volume Segmentation. J. Biomed. Imaging 2010, 2010, 105610. [Google Scholar] [CrossRef]
- Zeleznik, R.; Weiss, J.; Taron, J.; Guthier, C.; Bitterman, D.S.; Hancox, C.; Kann, B.H.; Kim, D.W.; Punglia, R.S.; Bredfeldt, J.; et al. Deep-Learning System to Improve the Quality and Efficiency of Volumetric Heart Segmentation for Breast Cancer. NPJ Digit. Med. 2021, 4, 43. [Google Scholar] [CrossRef]
- Deng, Y.; Hou, D.; Li, B.; Lv, X.; Ke, L.; Qiang, M.; Li, T.; Jing, B.; Li, C. A Novel Fully Automated MRI-Based Deep-Learning Method for Segmentation of Nasopharyngeal Carcinoma Lymph Nodes. J. Med. Biol. Eng. 2022, 42, 604–612. [Google Scholar] [CrossRef]
- Graffy, P.M.; Liu, J.; O’Connor, S.; Summers, R.M.; Pickhardt, P.J. Automated Segmentation and Quantification of Aortic Calcification at Abdominal CT: Application of a Deep Learning-Based Algorithm to a Longitudinal Screening Cohort. Abdom. Radiol. 2019, 44, 2921–2928. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.J.; Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Lipkova, J.; Noor, Z.; Shaban, M.; Shady, M.; Williams, M.; Joo, B.; et al. Pan-Cancer Integrative Histology-Genomic Analysis via Multimodal Deep Learning. Cancer Cell 2022, 40, 865–878.e6. [Google Scholar] [CrossRef] [PubMed]
- Hager, P.; Menten, M.J.; Rueckert, D. Best of Both Worlds: Multimodal Contrastive Learning with Tabular and Imaging Data. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 23924–23935. [Google Scholar]
- Hong, D.; Gao, L.; Yokoya, N.; Yao, J.; Chanussot, J.; Du, Q.; Zhang, B. More Diverse Means Better: Multimodal Deep Learning Meets Remote-Sensing Imagery Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 4340–4354. [Google Scholar] [CrossRef]
- Vale-Silva, L.A.; Rohr, K. Long-Term Cancer Survival Prediction Using Multimodal Deep Learning. Sci. Rep. 2021, 11, 13505. [Google Scholar] [CrossRef]
- Venugopalan, J.; Tong, L.; Hassanzadeh, H.R.; Wang, M.D. Multimodal Deep Learning Models for Early Detection of Alzheimer’s Disease Stage. Sci. Rep. 2021, 11, 3254. [Google Scholar] [CrossRef]
- Luo, W.; Phung, D.; Tran, T.; Gupta, S.; Rana, S.; Karmakar, C.; Shilton, A.; Yearwood, J.; Dimitrova, N.; Ho, T.B.; et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J. Med. Internet Res. 2016, 18, e323. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. In Proceedings of the Advances in Neural Information Processing Systems, Virtual, 6–14 December 2021; Curran Associates, Inc.: Red Hook, NY, USA, 2021; Volume 34, pp. 12077–12090. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. NnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isaksson, L.J.; Summers, P.; Mastroleo, F.; Marvaso, G.; Corrao, G.; Vincini, M.G.; Zaffaroni, M.; Ceci, F.; Petralia, G.; Orecchia, R.; et al. Automatic Segmentation with Deep Learning in Radiotherapy. Cancers 2023, 15, 4389. https://doi.org/10.3390/cancers15174389
Isaksson LJ, Summers P, Mastroleo F, Marvaso G, Corrao G, Vincini MG, Zaffaroni M, Ceci F, Petralia G, Orecchia R, et al. Automatic Segmentation with Deep Learning in Radiotherapy. Cancers. 2023; 15(17):4389. https://doi.org/10.3390/cancers15174389
Chicago/Turabian StyleIsaksson, Lars Johannes, Paul Summers, Federico Mastroleo, Giulia Marvaso, Giulia Corrao, Maria Giulia Vincini, Mattia Zaffaroni, Francesco Ceci, Giuseppe Petralia, Roberto Orecchia, and et al. 2023. "Automatic Segmentation with Deep Learning in Radiotherapy" Cancers 15, no. 17: 4389. https://doi.org/10.3390/cancers15174389
APA StyleIsaksson, L. J., Summers, P., Mastroleo, F., Marvaso, G., Corrao, G., Vincini, M. G., Zaffaroni, M., Ceci, F., Petralia, G., Orecchia, R., & Jereczek-Fossa, B. A. (2023). Automatic Segmentation with Deep Learning in Radiotherapy. Cancers, 15(17), 4389. https://doi.org/10.3390/cancers15174389







