Modulation of Notch Signaling by Small-Molecular Compounds and Its Potential in Anticancer Studies
Simple Summary
Abstract
1. Canonical and Non-Canonical Notch Pathway
2. Two Sides of the Same Coin—Notch Gain- vs. Loss-of-Function
3. Compounds Interfering with Endoplasmic Reticulum-to-Golgi and Membrane Trafficking of NOTCH Receptors
4. NOTCH–Ligand Interaction, Γ-Secretase, and ADAM Proteases Inhibition
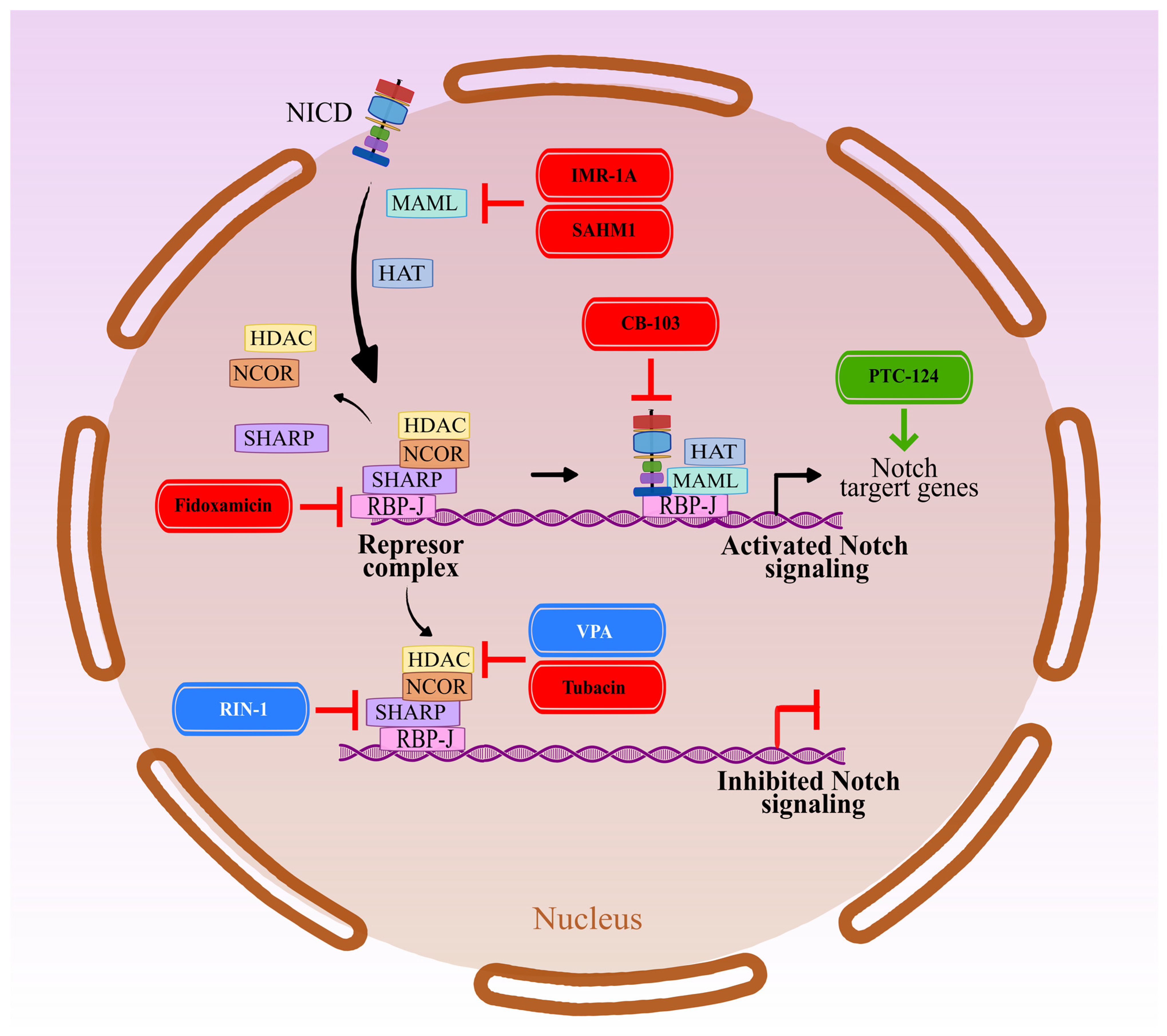
5. NICD-Dependent Transcription in the Nucleus
6. Small Molecular Weight Compounds with Pleiotropic Functions in Relation to Notch Signaling
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sherry, K.P.; Das, R.K.; Pappu, R.V.; Barrick, D. Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proc. Natl. Acad. Sci. USA 2017, 114, E9243–E9252. [Google Scholar] [CrossRef]
- Steinbuck, M.P.; Winandy, S. A Review of Notch Processing with New Insights Into Ligand-Independent Notch Signaling in T-Cells. Front. Immunol. 2018, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Aster, J.C. Notch signaling in cancer: Complexity and challenges on the path to clinical translation. Semin. Cancer Biol. 2022, 85, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Apodaca, G.; Brown, W.J. Membrane traffic research: Challenges for the next decade. Front. Cell Dev. Biol. 2014, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Antfolk, D.; Antila, C.; Kemppainen, K.; Landor, S.K.-J.; Sahlgren, C. Decoding the PTM-switchboard of Notch. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118507. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fischer, M.; Satkunarajah, M.; Zhou, D.; Withers, S.G.; Rini, J.M. Structural basis of Notch O-glucosylation and O–xylosylation by mammalian protein–O-glucosyltransferase 1 (POGLUT1). Nat. Commun. 2017, 8, 185. [Google Scholar] [CrossRef]
- Mugisha, S.; Di, X.; Disoma, C.; Jiang, H.; Zhang, S. Fringe family genes and their modulation of Notch signaling in cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188746. [Google Scholar] [CrossRef]
- Majumder, S.; Crabtree, J.S.; Golde, T.E.; Minter, L.M.; Osborne, B.A.; Miele, L. Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 2021, 20, 125–144. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Gordon, W.R.; Zimmerman, B.; He, L.; Miles, L.J.; Huang, J.; Tiyanont, K.; McArthur, D.G.; Aster, J.C.; Perrimon, N.; Loparo, J.J.; et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell 2015, 33, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. 2017, 12, 245–275. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Sliz, P.; Pear, W.S.; Aster, J.C.; Blacklow, S.C. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 2103–2108. [Google Scholar] [CrossRef] [PubMed]
- Kovall, R.A.; Blacklow, S.C. Mechanistic insights into notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 2010, 92, 31–71. [Google Scholar] [PubMed]
- Orzechowska, M.; Anusewicz, D.; Bednarek, A.K. Functional Gene Expression Differentiation of the Notch Signaling Pathway in Female Reproductive Tract Tissues—A Comprehensive Review with Analysis. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Castel, D.; Mourikis, P.; Bartels, S.J.J.; Brinkman, A.B.; Tajbakhsh, S.; Stunnenberg, H.G. Dynamic binding of RBPJ is determined by notch signaling status. Genes Dev. 2013, 27, 592616. [Google Scholar] [CrossRef]
- Misiorek, J.O.; Przybyszewska-Podstawka, A.; Kałafut, J.; Paziewska, B.; Rolle, K.; Rivero-Müller, A.; Nees, M. Context matters: Notch signatures and pathway in cancer progression and metastasis. Cells 2021, 10, 94. [Google Scholar] [CrossRef]
- Dongre, A.; Surampudi, L.; Lawlor, R.G.; Fauq, A.H.; Miele, L.; Golde, T.E.; Minter, L.M.; Osborne, B.A. Non-canonical Notch signaling drives activation and differentiation of peripheral CD4+ T cells. Front. Immunol. 2014, 5, 54. [Google Scholar] [CrossRef]
- Charbonnier, L.M.; Wang, S.; Georgiev, P.; Sefik, E.; Chatila, T.A. Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat. Immunol. 2015, 16, 1162–1173. [Google Scholar] [CrossRef]
- Ayaz, F.; Osborne, B.A. Non-canonical Notch signaling in cancer and immunity. Front. Oncol. 2014, 4, 345. [Google Scholar] [CrossRef]
- Jin, S.; Mutvei, A.P.; Chivukula, I.V.; Andersson, E.R.; Ramsköld, D.; Sandberg, R.; Lee, K.L.; Kronqvist, P.; Mamaeva, V.; Östling, P.; et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene 2013, 32, 4892–4902. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Sorrentino, C.; Ucar, D.A.; Peng, Y.; Matossian, M.; Wyczechowska, D.; Crabtree, J.; Zabaleta, J.; Morello, S.; Del Valle, L.; et al. Notch signaling regulates mitochondrial metabolism and NF-κB activity in triple-negative breast cancer cells via IKKα-dependent non-canonical pathways. Front. Oncol. 2018, 8, 575. [Google Scholar] [CrossRef] [PubMed]
- Lakhan, R.; Rathinam, C.V. Deficiency of Rbpj Leads to Defective Stress-Induced Hematopoietic Stem Cell Functions and Hif Mediated Activation of Non-canonical Notch Signaling Pathways. Front. Cell Dev. Biol. 2021, 8, 622190. [Google Scholar] [CrossRef] [PubMed]
- Conner, S.D. Regulation of Notch Signaling Through Intracellular Transport. Int. Rev. Cell Mol. Biol. 2016, 323, 107–127. [Google Scholar] [PubMed]
- Johnson, F.M.; Janku, F.; Gouda, M.A.; Tran, H.T.; Kawedia, J.D.; Schmitz, D.; Streefkerk, H.; Lee, J.J.; Andersen, C.R.; Deng, D.; et al. Inhibition of the Phosphatidylinositol-3 Kinase Pathway Using Bimiralisib in Loss-of-Function NOTCH1-Mutant Head and Neck Cancer. Oncologist 2022, 27, 1004-e926. [Google Scholar] [CrossRef]
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Front. Cell Dev. Biol. 2021, 9, 857. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Reichrath, J.; Reichrath, S. Notch Signaling and Embryonic Development: An Ancient Friend, Revisited. Adv. Exp. Med. Biol. 2020, 1218, 9–37. [Google Scholar]
- Kiyokawa, H.; Morimoto, M. Notch signaling in the mammalian respiratory system, specifically the trachea and lungs, in development, homeostasis, regeneration, and disease. Dev. Growth Differ. 2020, 62, 67–79. [Google Scholar] [CrossRef]
- Vanderbeck, A.; Maillard, I. Notch signaling at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 2021, 109, 535–548. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Hu, H.; Huang, Q.; Chen, Y.; Wang, G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int. J. Biol. Macromol. 2018, 117, 445–453. [Google Scholar] [CrossRef]
- Kukcinaviciute, E.; Jonusiene, V.; Sasnauskiene, A.; Dabkeviciene, D.; Eidenaite, E.; Laurinavicius, A. Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116. J. Cell. Biochem. 2018, 119, 5913–5920. [Google Scholar] [CrossRef] [PubMed]
- Janghorban, M.; Xin, L.; Rosen, J.M.; Zhang, X.H.F. Notch signaling as a regulator of the tumor immune response: To target or not to target? Front. Immunol. 2018, 9, 1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Huang, C.; Long, J.; Zhao, Z.B.; Ma, H.Q.; Yao, X.Q.; Li, L.; Lian, Z.X. Notch signaling mutations increase intra-tumor chemokine expression and predict response to immunotherapy in colorectal cancer. BMC Cancer 2022, 22, 933. [Google Scholar] [CrossRef]
- Nandi, A.; Debnath, R.; Nayak, A.; To, T.K.J.; Thacker, G.; Reilly, M.; Gumber, S.; Karagounis, I.; Li, N.; Lengner, C.J.; et al. Dll1-Mediated Notch Signaling Drives Tumor Cell Cross-talk with Cancer-Associated Fibroblasts to Promote Radioresistance in Breast Cancer. Cancer Res. 2022, 82, 3718–3733. [Google Scholar] [CrossRef]
- Xie, Q.; Guo, H.; He, P.; Deng, H.; Gao, Y.; Dong, N.; Niu, W.; Liu, T.; Li, M.; Wang, S.; et al. Tspan5 promotes epithelial–mesenchymal transition and tumour metastasis of hepatocellular carcinoma by activating Notch signalling. Mol. Oncol. 2021, 15, 3184. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yu, J.; Park, A.; Dubon, M.J.; Do, J.; Kim, Y.; Nam, D.; Noh, J.; Park, K.S. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci. Rep. 2019, 9, 11724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sha, J.; Yang, G.; Huang, X.; Bo, J.; Huang, Y. Activation of Notch pathway is linked with epithelial-mesenchymal transition in prostate cancer cells. Cell Cycle 2017, 16, 999–1007. [Google Scholar] [CrossRef]
- Xie, Q.; Cheng, Z.; Chen, X.; Lobe, C.G.; Liu, J. The role of Notch signalling in ovarian angiogenesis. J. Ovarian Res. 2017, 10, 13. [Google Scholar] [CrossRef]
- Xiao, W.; Gao, Z.; Duan, Y.; Yuan, W.; Ke, Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 41. [Google Scholar] [CrossRef]
- Forghanifard, M.M.; Kasebi, P.; Abbaszadegan, M.R. SOX2/SALL4 stemness axis modulates Notch signaling genes to maintain self-renewal capacity of esophageal squamous cell carcinoma. Mol. Cell. Biochem. 2021, 476, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The Role of Notch3 Signaling in Cancer Stemness and Chemoresistance: Molecular Mechanisms and Targeting Strategies. Front. Mol. Biosci. 2021, 8, 694141. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Seki, M.; Yoshida, K.; Shiraishi, Y.; Akiyama, M.; Koh, K.; Imamura, T.; Manabe, A.; Hayashi, Y.; Kobayashi, M.; et al. NOTCH1 pathway activating mutations and clonal evolution in pediatric T-cell acute lymphoblastic leukemia. Cancer Sci. 2019, 110, 784–794. [Google Scholar] [CrossRef]
- Takam Kamga, P.; Collo, G.D.; Resci, F.; Bazzoni, R.; Mercuri, A.; Quaglia, F.M.; Tanasi, I.; Delfino, P.; Visco, C.; Bonifacio, M.; et al. Notch Signaling Molecules as Prognostic Biomarkers for Acute Myeloid Leukemia. Cancers 2019, 11, 1958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Berndt-Paetz, M.; Neuhaus, J. A comprehensive bioinformatics analysis of notch pathways in bladder cancer. Cancers 2021, 13, 3089. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.-Q.; Sun, J.-L.; Wang, F.; Zhang, H.-Z.; Zhou, G.; Xi, Q. Current understanding of adenoid cystic carcinoma in the gene expression and targeted therapy. Holist. Integr. Oncol. 2023, 2, 7. [Google Scholar] [CrossRef]
- Sorrentino, C.; Cuneo, A.; Roti, G. Therapeutic targeting of notch signaling pathway in hematological malignancies. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019037. [Google Scholar]
- Choi, S.H.; Severson, E.; Pear, W.S.; Liu, X.S.; Aster, J.C.; Blacklow, S.C. The common oncogenomic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0185762. [Google Scholar] [CrossRef]
- Toribio, M.L.; González-García, S. Notch Partners in the Long Journey of T-ALL Pathogenesis. Int. J. Mol. Sci. 2023, 24, 1383. [Google Scholar] [CrossRef]
- Chimento, A.; D’amico, M.; Pezzi, V.; De Amicis, F. Notch Signaling in Breast Tumor Microenvironment as Mediator of Drug Resistance. Int. J. Mol. Sci. 2022, 23, 6296. [Google Scholar] [CrossRef]
- Nandi, A.; Chakrabarti, R. The many facets of Notch signaling in breast cancer: Toward overcoming therapeutic resistance. Genes Dev. 2020, 34, 1422–1438. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Brennan, K. Notch Signalling in Breast Development and Cancer. Front. Cell Dev. Biol. 2021, 9, 692173. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, H.; Bahramy, A.; Zafari, N.; Delavar, M.R.; Nguyen, K.; Haghi, A.; Kandelouei, T.; Vittori, C.; Jazireian, P.; Maleki, S.; et al. Notch signaling pathway: A comprehensive prognostic and gene expression profile analysis in breast cancer. BMC Cancer 2022, 22, 1282. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Zhang, M.; Singh, R.; Peng, S.; Mazumdar, T.; Sambandam, V.; Shen, L.; Tong, P.; Li, L.; Kalu, N.N.; Pickering, C.R.; et al. Mutations of the LIM protein AJUBA mediate sensitivity of head and neck squamous cell carcinoma to treatment with cell-cycle inhibitors. Cancer Lett. 2017, 392, 71–82. [Google Scholar] [CrossRef]
- Yokobori, T.; Mimori, K.; Iwatsuki, M.; Ishii, H.; Tanaka, F.; Sato, T.; Toh, H.; Sudo, T.; Iwaya, T.; Tanaka, Y.; et al. Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma. Int. J. Oncol. 2012, 41, 253–259. [Google Scholar]
- Schleicher, K.; Schramek, D. AJUBA: A regulator of epidermal homeostasis and cancer. Exp. Dermatol. 2021, 30, 546–559. [Google Scholar] [CrossRef]
- Luo, Z.; Mu, L.; Zheng, Y.; Shen, W.; Li, J.; Xu, L.; Zhong, B.; Liu, Y.; Zhou, Y. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J. Mol. Cell Biol. 2020, 12, 345–358. [Google Scholar] [CrossRef]
- Kar, R.; Jha, S.K.; Ojha, S.; Sharma, A.; Dholpuria, S.; Raju, V.S.R.; Prasher, P.; Chellappan, D.K.; Gupta, G.; Kumar Singh, S.; et al. The FBXW7-NOTCH interactome: A ubiquitin proteasomal system-induced crosstalk modulating oncogenic transformation in human tissues. Cancer Rep. 2021, 4, e1369. [Google Scholar] [CrossRef] [PubMed]
- Grilli, G.; Hermida-Prado, F.; Álvarez-Fernández, M.; Allonca, E.; Álvarez-González, M.; Astudillo, A.; Moreno-Bueno, G.; Cano, A.; García-Pedrero, J.M.; Rodrigo, J.P. Impact of notch signaling on the prognosis of patients with head and neck squamous cell carcinoma. Oral Oncol. 2020, 110, 105003. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, L.; Pellegrini, C.; Di Stefani, A.; Del Regno, L.; Sollena, P.; Piccerillo, A.; Longo, C.; Garbe, C.; Fargnoli, M.C.; Peris, K. Molecular genetics of cutaneous squamous cell carcinoma: Perspective for treatment strategies. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 932–941. [Google Scholar] [CrossRef]
- Wang, N.J.; Sanborn, Z.; Arnett, K.L.; Bayston, L.J.; Liao, W.; Proby, C.M.; Leigh, I.M.; Collisson, E.A.; Gordon, P.B.; Jakkula, L.; et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2011, 108, 17761–17766. [Google Scholar] [CrossRef]
- Schwaederle, M.; Elkin, S.K.; Tomson, B.N.; Carter, J.L.; Kurzrock, R. Squamousness: Next-generation sequencing reveals shared molecular features across squamous tumor types. Cell Cycle 2015, 14, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Nyman, P.E.; Buehler, D.; Lambert, P.F. Loss of function of canonical Notch signaling drives head and neck carcinogenesis. Clin. Cancer Res. 2018, 24, 6308–6318. [Google Scholar] [CrossRef]
- Kałafut, J.; Czerwonka, A.; Anameriç, A.; Przybyszewska-Podstawka, A.; Misiorek, J.O.; Rivero-Müller, A.; Nees, M. Shooting at Moving and Hidden Targets-Tumour Cell Plasticity and the Notch Signalling Pathway in Head and Neck Squamous Cell Carcinomas. Cancers 2021, 13, 6219. [Google Scholar] [CrossRef]
- Shah, P.A.; Huang, C.; Li, Q.; Kazi, S.A.; Byers, L.A.; Wang, J.; Johnson, F.M.; Frederick, M.J. NOTCH1 Signaling in Head and Neck Squamous Cell Carcinoma. Cells 2020, 9, 2677. [Google Scholar] [CrossRef]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef]
- Zhang, M.; Biswas, S.; Qin, X.; Gong, W.; Deng, W.; Yu, H. Does Notch play a tumor suppressor role across diverse squamous cell carcinomas? Cancer Med. 2016, 5, 2048–2060. [Google Scholar] [CrossRef]
- Doody, R.S.; Raman, R.; Farlow, M.; Iwatsubo, T.; Vellas, B.; Joffe, S.; Kieburtz, K.; He, F.; Sun, X.; Thomas, R.G.; et al. A Phase 3 Trial of Semagacestat for Treatment of Alzheimer’s Disease. N. Engl. J. Med. 2013, 369, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, L.; Savage, S.R.; Eguez, R.V.; Dou, Y.; Li, Y.; da Veiga Leprevost, F.; Jaehnig, E.J.; Lei, J.T.; Wen, B.; et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell 2021, 39, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.R.; Zhang, J.; Yoo, S.Y.; Bengtsson, L.; Moorthy, S.; Neskey, D.M.; Zhao, M.; Ortega Alves, M.V.; Chang, K.; Drummond, J.; et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013, 3, 770–781. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef]
- Yap, L.F.; Lee, D.; Khairuddin, A.; Pairan, M.F.; Puspita, B.; Siar, C.H.; Paterson, I.C. The opposing roles of NOTCH signalling in head and neck cancer: A mini review. Oral Dis. 2015, 21, 850–857. [Google Scholar] [CrossRef]
- Schmidl, B.; Siegl, M.; Boxberg, M.; Stögbauer, F.; Jira, D.; Winter, C.; Stark, L.; Pickhard, A.; Wollenberg, B.; Wirth, M. NOTCH1 Intracellular Domain and the Tumor Microenvironment as Prognostic Markers in HNSCC. Cancers 2022, 14, 1080. [Google Scholar] [CrossRef]
- Sun, W.; Gaykalova, D.A.; Ochs, M.F.; Mambo, E.; Arnaoutakis, D.; Liu, Y.; Loyo, M.; Agrawal, N.; Howard, J.; Li, R.; et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014, 74, 1091–1104. [Google Scholar] [CrossRef]
- Porcheri, C.; Mitsiadis, T.A. Notch in Head and Neck Cancer. Adv. Exp. Med. Biol. 2021, 1287, 81–103. [Google Scholar]
- Porcheri, C.; Meisel, C.T.; Mitsiadis, T. Multifactorial Contribution of Notch Signaling in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 1520. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.N.; Benjamin Burch, M.; Cooper, T.S.; Manna, D.L.D.; Wei, S.; Ojesina, A.I.; Rosenthal, E.L.; Yang, E.S. Notch signaling activation is associated with patient mortality and increased FGF1-mediated invasion in squamous cell carcinoma of the oral cavity. Mol. Cancer Res. 2016, 14, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, S.K.; Schleicher, K.; Malik, A.; Quevedo, R.; Langille, E.; Teng, K.; Oh, R.H.; Rathod, B.; Tsai, R.; Samavarchi-Tehrani, P.; et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science 2020, 367, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, T.; Takahashi, K.; Takimoto, H.; Tomizuka, K.; Hayasaka, M.; Tabira, T.; Hanaoka, K. Functional redundancy of the Notch gene family during mouse embryogenesis: Analysis of Notch gene expression in Notch3-deficient mice. Biochem. Biophys. Res. Commun. 2005, 331, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cain-Hom, C.; Choy, L.; Hagenbeek, T.J.; De Leon, G.P.; Chen, Y.; Finkle, D.; Venook, R.; Wu, X.; Ridgway, J.; et al. Therapeutic antibody targeting of individual Notch receptors. Nature 2010, 464, 1052–1057. [Google Scholar] [CrossRef]
- Riccio, O.; van Gijn, M.E.; Bezdek, A.C.; Pellegrinet, L.; van Es, J.H.; Zimber-Strobl, U.; Strobl, L.J.; Honjo, T.; Clevers, H.; Radtke, F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008, 9, 377–383. [Google Scholar] [CrossRef]
- Hatano, K.; Saigo, C.; Kito, Y.; Shibata, T.; Takeuchi, T. Overexpression of JAG2 is related to poor outcomes in oral squamous cell carcinoma. Clin. Exp. Dent. Res. 2020, 6, 174–180. [Google Scholar] [CrossRef]
- Lin, J.T.; Chen, M.K.; Yeh, K.T.; Chang, C.S.; Chang, T.H.; Lin, C.Y.; Wu, Y.C.; Su, B.W.; Lee, K.D.; Chang, P.J. Association of high levels of Jagged-1 and Notch-1 expression with poor prognosis in head and neck cancer. Ann. Surg. Oncol. 2010, 17, 2976–2983. [Google Scholar] [CrossRef]
- Zhang, T.; Liang, L.; Liu, X.; Wu, J.N.; Chen, J.; Su, K.; Zheng, Q.; Huang, H.; Liao, G. qing TGFβ1-Smad3-Jagged1-Notch1-Slug signaling pathway takes part in tumorigenesis and progress of tongue squamous cell carcinoma. J. Oral Pathol. Med. 2016, 45, 486–493. [Google Scholar] [CrossRef]
- Parmigiani, E.; Taylor, V.; Giachino, C. Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma. Cells 2020, 9, 2304. [Google Scholar] [CrossRef]
- Marignol, L. Notch signalling: The true driver of small cell lung cancer? Transl. Cancer Res. 2017, 6, 1191–1196. [Google Scholar] [CrossRef]
- Leonetti, A.; Facchinetti, F.; Minari, R.; Cortellini, A.; Rolfo, C.D.; Giovannetti, E.; Tiseo, M. Notch pathway in small-cell lung cancer: From preclinical evidence to therapeutic challenges. Cell. Oncol. 2019, 42, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.A.; Sambandam, V.; Fernandez, A.M.; Zhao, H.; Mazumdar, T.; Shen, L.; Wang, Q.; Ahmed, K.M.; Ghosh, S.; Frederick, M.J.; et al. Sustained Aurora Kinase B Expression Confers Resistance to PI3K Inhibition in Head and Neck Squamous Cell Carcinoma. Cancer Res. 2022, 82, 4444–4456. [Google Scholar] [CrossRef] [PubMed]
- Nolin, E.; Gans, S.; Llamas, L.; Bandyopadhyay, S.; Brittain, S.M.; Bernasconi-Elias, P.; Carter, K.P.; Loureiro, J.J.; Thomas, J.R.; Schirle, M.; et al. Discovery of a ZIP7 inhibitor from a Notch pathway screen. Nat. Chem. Biol. 2019, 15, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Roti, G.; Carlton, A.; Ross, K.N.; Markstein, M.; Pajcini, K.; Su, A.H.; Perrimon, N.; Pear, W.S.; Kung, A.L.; Blacklow, S.C.; et al. Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer. Cancer Cell 2013, 23, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, J.T.; Brennen, W.N.; Christensen, S.B.; Denmeade, S.R. Mipsagargin: The beginning—Not the end—Of thapsigargin prodrug-based cancer therapeutics. Molecules 2021, 26, 7469. [Google Scholar] [CrossRef]
- De Ford, C.; Heidersdorf, B.; Haun, F.; Murillo, R.; Friedrich, T.; Borner, C.; Merfort, I. The clerodane diterpene casearin J induces apoptosis of T-all cells through SERCA inhibition, oxidative stress, and interference with notch1 signaling. Cell Death Dis. 2016, 7, e2070. [Google Scholar] [CrossRef]
- Marchesini, M.; Gherli, A.; Montanaro, A.; Patrizi, L.; Sorrentino, C.; Pagliaro, L.; Rompietti, C.; Kitara, S.; Heit, S.; Olesen, C.E.; et al. Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia. Cell Chem. Biol. 2020, 27, 678–697. [Google Scholar] [CrossRef]
- Pagliaro, L.; Marchesini, M.; Roti, G. Targeting oncogenic Notch signaling with SERCA inhibitors. J. Hematol. Oncol. 2021, 14, 8. [Google Scholar] [CrossRef]
- Pagliaro, L.; Sorrentino, C.; Roti, G. Targeting Notch Trafficking and Processing in Cancers. Cells 2020, 9, 2212. [Google Scholar] [CrossRef]
- Baldoni, S.; Del Papa, B.; Dorillo, E.; Aureli, P.; De Falco, F.; Rompietti, C.; Sorcini, D.; Varasano, E.; Cecchini, D.; Zei, T.; et al. Bepridil exhibits anti-leukemic activity associated with NOTCH1 pathway inhibition in chronic lymphocytic leukemia. Int. J. Cancer 2018, 143, 958–970. [Google Scholar] [CrossRef]
- Krämer, A.; Mentrup, T.; Kleizen, B.; Rivera-Milla, E.; Reichenbach, D.; Enzensperger, C.; Nohl, R.; Täuscher, E.; Görls, H.; Ploubidou, A.; et al. Small molecules intercept Notch signaling and the early secretory pathway. Nat. Chem. Biol. 2013, 9, 731–738. [Google Scholar] [CrossRef]
- Zhang, M.; Han, Y.; Zheng, Y.; Zhang, Y.; Zhao, X.; Gao, Z.; Liu, X. ZEB1-activated LINC01123 accelerates the malignancy in lung adenocarcinoma through NOTCH signaling pathway. Cell Death Dis. 2020, 11, 981. [Google Scholar] [CrossRef]
- Lu, Z.; Ren, Y.; Zhang, M.; Fan, T.; Wang, Y.; Zhao, Q.; Liu, H.M.; Zhao, W.; Hou, G. FLI-06 suppresses proliferation, induces apoptosis and cell cycle arrest by targeting LSD1 and Notch pathway in esophageal squamous cell carcinoma cells. Biomed. Pharmacother. 2018, 107, 1370–1376. [Google Scholar] [CrossRef]
- Gan, R.H.; Lin, L.S.; Xie, J.; Huang, L.; Ding, L.C.; Su, B.H.; Peng, X.E.; Zheng, D.L.; Lu, Y.G. FLI-06 Intercepts Notch Signaling and Suppresses the Proliferation and Self-renewal of Tongue Cancer Cells. OncoTargets Ther. 2019, 12, 7663–7674. [Google Scholar] [CrossRef] [PubMed]
- Hounjet, J.; Habets, R.; Schaaf, M.B.; Hendrickx, T.C.; Barbeau, L.M.O.; Yahyanejad, S.; Rouschop, K.M.; Groot, A.J.; Vooijs, M. The anti-malarial drug chloroquine sensitizes oncogenic NOTCH1 driven human T-ALL to γ-secretase inhibition. Oncogene 2019, 38, 5457–5468. [Google Scholar] [CrossRef] [PubMed]
- Kobia, F.; Duchi, S.; Deflorian, G.; Vaccari, T. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol. Oncol. 2014, 8, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Kumar, V.; Nordstrøm, L.U.; Feng, L.; Takeuchi, H.; Hao, H.; Luca, V.C.; Garcia, K.C.; Stanley, P.; Wu, P.; et al. Inhibition of Delta-induced Notch signaling using fucose analogs. Nat. Chem. Biol. 2018, 14, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. Structure and Function of the γ-Secretase Complex. Biochemistry 2019, 58, 2953–2966. [Google Scholar] [CrossRef]
- Bai, X.C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H.W. Sampling the conformational space of the catalytic subunit of human g-secretase. Elife 2015, 4, e11182. [Google Scholar] [CrossRef]
- Hur, J.Y. γ-Secretase in Alzheimer’s disease. Exp. Mol. Med. 2022, 54, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.E.; Li, Y.M. Turning the tide on Alzheimer’s disease: Modulation of γ-secretase. Cell Biosci. 2022, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Deangelo, D.J.; Stone, R.M.; Silverman, L.B.; Stock, W.; Attar, E.C.; Fearen, I.; Dallob, A.; Matthews, C.; Stone, J.; Freedman, S.J.; et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J. Clin. Oncol. 2006, 24, 6585. [Google Scholar] [CrossRef]
- López-Nieva, P.; González-Sánchez, L.; Cobos-Fernández, M.Á.; Córdoba, R.; Santos, J.; Fernández-Piqueras, J. More Insights on the Use of γ-Secretase Inhibitors in Cancer Treatment. Oncologist 2021, 26, e298–e305. [Google Scholar] [CrossRef]
- Pine, S.R. Rethinking Gamma-secretase Inhibitors for Treatment of Non–small-Cell Lung Cancer: Is notch the target? Clin. Cancer Res. 2018, 24, 6136–6141. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Hossain, F.; Pannuti, A.; Lessard, C.B.; Ladd, G.Z.; Jung, J.I.; Minter, L.M.; Osborne, B.A.; Miele, L.; Golde, T.E. γ-Secretase inhibitors in cancer clinical trials are pharmacologically and functionally distinct. EMBO Mol. Med. 2017, 9, 950–966. [Google Scholar] [CrossRef]
- Samon, J.B.; Castillo-Martin, M.; Hadler, M.; Ambesi-Impiobato, A.; Paietta, E.; Racevskis, J.; Wiernik, P.H.; Rowe, J.M.; Jakubczak, J.; Randolph, S.; et al. Preclinical analysis of the γ-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol. Cancer Ther. 2012, 11, 1565–1575. [Google Scholar] [CrossRef]
- Collins, M.; Michot, J.M.; Bellanger, C.; Mussini, C.; Benhadji, K.; Massard, C.; Carbonnel, F. Notch inhibitors induce diarrhea, hypercrinia and secretory cell Metaplasia in the human colon. EXCLI J. 2021, 20, 819–827. [Google Scholar]
- Sancho, R.; Cremona, C.A.; Behrens, A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 2015, 16, 571–581. [Google Scholar] [CrossRef]
- Chan, D.; Kaplan, J.; Gordon, G.; Desai, J. Activity of the gamma secretase inhibitor al101 in desmoid tumors: A case report of 2 adult cases. Curr. Oncol. 2021, 28, 3659–3667. [Google Scholar] [CrossRef]
- Federman, N. Molecular pathogenesis of desmoid tumor and the role of γ-secretase inhibition. NPJ Precis. Oncol. 2022, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Gounder, M.; Ratan, R.; Alcindor, T.; Schöffski, P.; van der Graaf, W.T.; Wilky, B.A.; Riedel, R.F.; Lim, A.; Smith, L.M.; Moody, S.; et al. Nirogacestat, a γ-Secretase Inhibitor for Desmoid Tumors. N. Engl. J. Med. 2023, 388, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Even, C.; Lassen, U.; Merchan, J.; Le Tourneau, C.; Soria, J.C.; Ferte, C.; Ricci, F.; Diener, J.T.; Yuen, E.; Smith, C.; et al. Safety and clinical activity of the Notch inhibitor, crenigacestat (LY3039478), in an open-label phase I trial expansion cohort of advanced or metastatic adenoid cystic carcinoma. Investig. New Drugs 2020, 38, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Cassier, P.A.; Azaro, A.; Anderson, B.; Yuen, E.; Yu, D.; Oakley, G.; Benhadji, K.A.; Pant, S. A phase 1b study of crenigacestat (LY3039478) in combination with gemcitabine and cisplatin or gemcitabine and carboplatin in patients with advanced or metastatic solid tumors. Cancer Chemother. Pharmacol. 2022, 90, 335–344. [Google Scholar] [CrossRef]
- Doi, T.; Tajimi, M.; Mori, J.; Asou, H.; Inoue, K.; Benhadji, K.A.; Naito, Y. A phase 1 study of crenigacestat (LY3039478), the Notch inhibitor, in Japanese patients with advanced solid tumors. Investig. New Drugs 2021, 39, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, G.; Martinelli, G.; Raffoux, E.; Chevallier, P.; Chromik, J.; Lithio, A.; Smith, C.L.; Yuen, E.; Oakley, G.J.; Benhadji, K.A.; et al. Phase 1 study to evaluate Crenigacestat (LY3039478) in combination with dexamethasone in patients with T-cell acute lymphoblastic leukemia and lymphoma. Cancer 2021, 127, 372–380. [Google Scholar] [CrossRef]
- McCaw, T.R.; Inga, E.; Chen, H.; Jaskula-Sztul, R.; Dudeja, V.; Bibb, J.A.; Ren, B.; Rose, J.B. Gamma Secretase Inhibitors in Cancer: A Current Perspective on Clinical Performance. Oncologist 2021, 26, e608–e621. [Google Scholar] [CrossRef]
- Tagami, S.; Yanagida, K.; Kodama, T.S.; Takami, M.; Mizuta, N.; Oyama, H.; Nishitomi, K.; Chiu, Y.W.; Okamoto, T.; Ikeuchi, T.; et al. Semagacestat Is a Pseudo-Inhibitor of γ-Secretase. Cell Rep. 2017, 21, 259–273. [Google Scholar] [CrossRef]
- Gu, K.; Li, Q.; Lin, H.; Zhu, J.; Mo, J.; He, S.; Lu, X.; Jiang, X.; Sun, H. Gamma secretase inhibitors: A patent review (2013–2015). Expert Opin. Ther. Pat. 2017, 27, 851–866. [Google Scholar] [CrossRef]
- Zhu, R.R.; Chen, Q.; Liu, Z.B.; Ruan, H.G.; Wu, Q.C.; Zhou, X. liang Inhibition of the Notch1 pathway induces peripartum cardiomyopathy. J. Cell. Mol. Med. 2020, 24, 7907–7914. [Google Scholar] [CrossRef]
- Sajadimajd, S.; Bahrami, G.; Mohammadi, B.; Madani, S.H. Notch signaling-induced cyclin d1 in diabetes ameliorating effects of the isolated polysaccharide from Rosa canina: In vitro and in vivo studies. Cell Biochem. Funct. 2022, 40, 935–945. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, D.W.; Kim, E.; Jeong, I.; Kim, S.; Kim, B.J.; Park, H.C. Notch signaling controls oligodendrocyte regeneration in the injured telencephalon of adult zebrafish. Exp. Neurobiol. 2020, 29, 417–424. [Google Scholar] [CrossRef]
- Ohuchi, K.; Funato, M.; Yoshino, Y.; Ando, S.; Inagaki, S.; Sato, A.; Kawase, C.; Seki, J.; Saito, T.; Nishio, H.; et al. Notch Signaling Mediates Astrocyte Abnormality in Spinal Muscular Atrophy Model Systems. Sci. Rep. 2019, 9, 3701. [Google Scholar] [CrossRef] [PubMed]
- Aguayo-Ortiz, R.; Guzmán-Ocampo, D.C.; Dominguez, L. Toward the Characterization of DAPT Interactions with γ-Secretase. ChemMedChem 2019, 14, 1005–1010. [Google Scholar] [CrossRef]
- Dai, G.; Deng, S.; Guo, W.; Yu, L.; Yang, J.; Zhou, S.; Gao, T. Notch pathway inhibition using DAPT, a γ-secretase inhibitor (GSI), enhances the antitumor effect of cisplatin in resistant osteosarcoma. Mol. Carcinog. 2019, 58, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Pindiprolu, S.K.S.S.; Krishnamurthy, P.T.; Dev, C.; Chintamaneni, P.K. DR5 antibody conjugated lipid-based nanocarriers of gamma-secretase inhibitor for the treatment of triple negative breast cancer. Chem. Phys. Lipids 2021, 235, 105033. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari-Movahed, M.; Shiri Varnamkhasti, B.; Shourian, M. Inhibiting Notch activity in breast cancer stem cells by functionalized gold nanoparticles with gamma-secretase inhibitor DAPT and vitamin C. Chem. Pap. 2022, 76, 1157–1170. [Google Scholar] [CrossRef]
- Wan, X.; Liu, C.; Lin, Y.; Fu, J.; Lu, G.; Lu, Z. pH sensitive peptide functionalized nanoparticles for co-delivery of erlotinib and DAPT to restrict the progress of triple negative breast cancer. Drug Deliv. 2019, 26, 470–480. [Google Scholar] [CrossRef]
- Lee, C.; Kim, M.; Park, C.; Jo, W.; Seo, J.K.; Kim, S.; Oh, J.; Kim, C.S.; Ryu, H.S.; Lee, K.H.; et al. Epigenetic regulation of Neuregulin 1 promotes breast cancer progression associated to hyperglycemia. Nat. Commun. 2023, 14, 439. [Google Scholar] [CrossRef]
- Bocchicchio, S.; Tesone, M.; Irusta, G. Convergence of Wnt and Notch signaling controls ovarian cancer cell survival. J. Cell. Physiol. 2019, 234, 22130–22143. [Google Scholar] [CrossRef]
- He, G.; Mu, T.; Yuan, Y.; Yang, W.; Zhang, Y.; Chen, Q.; Bian, M.; Pan, Y.; Xiang, Q.; Chen, Z.; et al. Effects of notch signaling pathway in cervical cancer by curcumin mediated photodynamic therapy and its possible mechanisms in vitro and in vivo. J. Cancer 2019, 10, 4114–4122. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, D.; Wang, Z.; Zhao, B.; Li, Y.; Sun, X.; Liu, J.; Wang, X.; Sheng, J. Inhibition of the notch signaling pathway overcomes resistance of cervical cancer cells to paclitaxel through retardation of the epithelial–mesenchymal transition process. Environ. Toxicol. 2021, 36, 1758–1764. [Google Scholar] [CrossRef]
- Rice, M.A.; Hsu, E.C.; Aslan, M.; Ghoochani, A.; Su, A.; Stoyanova, T. Loss of Notch1 activity inhibits prostate cancer growth and metastasis and sensitizes prostate cancer cells to antiandrogen therapies. Mol. Cancer Ther. 2019, 18, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhou, J.; Li, B.; Zhang, T.; Zuo, Y.; Gu, X. Notch1 and PI3K/Akt signaling blockers DAPT and LY294002 coordinately inhibit metastasis of gastric cancer through mutual enhancement. Cancer Chemother. Pharmacol. 2020, 85, 309–320. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; He, P.; Lu, S.; Dong, W. KIFC3 Regulates the progression and metastasis of gastric cancer via Notch1 pathway. Dig. Liver Dis. 2023, 55, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.G.; Bertrand, F.E.; Sigounas, G. A potential requirement for Smad3 phosphorylation in Notch-mediated EMT in colon cancer. Adv. Biol. Regul. 2023, 88, 100957. [Google Scholar] [CrossRef]
- Liu, F.; Chu, H.X.; Han, J.S.; Sun, X.; Chen, J.; Qiu, X.L.; Zheng, X.H.; Jia, B.; Zhao, J.J. Inhibitory effect of the Notch pathway-inhibitor DAPT on invasion and metastasis of tongue cancer via lncRNA-KAT14 regulation. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 189–199. [Google Scholar]
- Liu, S.; Dou, L.; Miao, M.; Man, X.; Wei, B.; Jiang, Z.; Ouyang, Y.; Ozaki, T.; Yu, M.; Zhu, Y. HES1-mediated down-regulation of miR-138 sustains NOTCH1 activation and promotes proliferation and invasion in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2023, 42, 72. [Google Scholar] [CrossRef]
- Lou, W.; Gao, K.; Xu, C.; Li, Q. Bromodomain-containing protein 9 is a prognostic biomarker associated with immune infiltrates and promotes tumor malignancy through activating notch signaling pathway in negative HIF-2α clear cell renal cell carcinoma. IUBMB Life 2021, 73, 1334–1347. [Google Scholar] [CrossRef]
- Wu, Q.; Guo, J.; Liu, Y.; Zheng, Q.; Li, X.; Wu, C.; Fang, D.; Chen, X.; Ma, L.; Xu, P.; et al. YAP drives fate conversion and chemoresistance of small cell lung cancer. Sci. Adv. 2021, 7, eabg1850. [Google Scholar] [CrossRef]
- Mancarella, S.; Serino, G.; Coletta, S.; Armentano, R.; Dituri, F.; Ardito, F.; Ruzzenente, A.; Fabregat, I.; Giannelli, G. The Tumor Microenvironment Drives Intrahepatic Cholangiocarcinoma Progression. Int. J. Mol. Sci. 2022, 23, 4187. [Google Scholar] [CrossRef] [PubMed]
- Mancarella, S.; Serino, G.; Dituri, F.; Cigliano, A.; Ribback, S.; Wang, J.; Chen, X.; Calvisi, D.F.; Giannelli, G. Crenigacestat, a selective NOTCH1 inhibitor, reduces intrahepatic cholangiocarcinoma progression by blocking VEGFA/DLL4/MMP13 axis. Cell Death Differ. 2020, 27, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Danielpour, D.; Corum, S.; Leahy, P.; Bangalore, A. Jagged-1 is induced by mTOR inhibitors in renal cancer cells through an Akt/ALK5/Smad4-dependent mechanism. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100117. [Google Scholar] [CrossRef] [PubMed]
- Low, H.Y.; Lee, Y.C.; Lee, Y.J.; Wang, H.L.; Chen, Y.I.; Chien, P.J.; Li, S.T.; Chang, W.W. Reciprocal regulation between indoleamine 2,3-dioxigenase 1 and notch1 involved in radiation response of cervical cancer stem cells. Cancers 2020, 12, 1547. [Google Scholar] [CrossRef] [PubMed]
- Giordano, F.; Montalto, F.I.; Panno, M.L.; Andò, S.; De Amicis, F. A Notch inhibitor plus Resveratrol induced blockade of autophagy drives glioblastoma cell death by promoting a switch to apoptosis. Am. J. Cancer Res. 2021, 11, 5933–5950. [Google Scholar]
- Bill, M.; Pathmanathan, A.; Karunasiri, M.; Shen, C.; Burke, M.H.; Ranganathan, P.; Papaioannou, D.; Zitzer, N.C.; Snyder, K.; Larocco, A.; et al. EGFL7 antagonizes notch signaling and represents a novel therapeutic target in acute myeloid leukemia. Clin. Cancer Res. 2020, 26, 669–678. [Google Scholar] [CrossRef]
- Li, T.; Xu, X.H.; Guo, X.; Yuan, T.; Tang, Z.H.; Jiang, X.M.; Xu, Y.L.; Zhang, L.L.; Chen, X.; Zhu, H.; et al. Activation of notch 3/c-MYC/CHOP axis regulates apoptosis and promotes sensitivity of lung cancer cells to mTOR inhibitor everolimus. Biochem. Pharmacol. 2020, 175, 113921. [Google Scholar] [CrossRef]
- Ghanbari-Movahed, M.; Ghanbari-Movahed, Z.; Momtaz, S.; Kilpatrick, K.L.; Farzaei, M.H.; Bishayee, A. Unlocking the secrets of cancer stem cells with -Secretase inhibitors: A novel anticancer strategy. Molecules 2021, 26, 972. [Google Scholar] [CrossRef]
- Pathak, Y.; Camps, I.; Mishra, A.; Tripathi, V. Targeting notch signaling pathway in breast cancer stem cells through drug repurposing approach. Mol. Divers. 2022. [Google Scholar] [CrossRef]
- Singh, A.K.; Prajapati, K.S.; Kumar, S. Hesperidin potentially interacts with the catalytic site of gamma-secretase and modifies notch sensitive genes and cancer stemness marker expression in colon cancer cells and colonosphere. J. Biomol. Struct. Dyn. 2022, 41, 8432–8444. [Google Scholar] [CrossRef]
- Subramaniam, D.; Ponnurangam, S.; Ramamoorthy, P.; Standing, D.; Battafarano, R.J.; Anant, S.; Sharma, P. Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS ONE 2012, 7, e30590. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Huang, Y.; Wang, Y.; Qi, X.; Su, T.; Lu, L. Evodiamine suppresses Notch3 signaling in lung tumorigenesis via direct binding to γ-secretases. Phytomedicine 2020, 68, 153176. [Google Scholar] [CrossRef]
- Nie, P.; Kalidindi, T.; Nagle, V.L.; Wu, X.; Li, T.; Liao, G.P.; Frost, G.; Henry, K.E.; Punzalan, B.; Carter, L.M.; et al. Imaging of Cancer γ-Secretase activity using an Inhibitor-Based PET Probe. Clin. Cancer Res. 2021, 27, 6145–6155. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, R.; Guo, X.; Yan, C.; Lei, J.; Shi, Y. Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell 2021, 184, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yu, T.; Lin, L.; Xing, L.; Cho, S.F.; Wen, K.; Aardalen, K.; Oka, A.; Lam, J.; Daley, M.; et al. γ-secretase inhibitors augment efficacy of BCMA-targeting bispecific antibodies against multiple myeloma cells without impairing T-cell activation and differentiation. Blood Cancer J. 2022, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Ambrogio, C.; Gómez-López, G.; Falcone, M.; Vidal, A.; Nadal, E.; Crosetto, N.; Blasco, R.B.; Fernández-Marcos, P.J.; Sánchez-Céspedes, M.; Ren, X.; et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat. Med. 2016, 22, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Mancarella, S.; Gigante, I.; Serino, G.; Pizzuto, E.; Dituri, F.; Valentini, M.F.; Wang, J.; Chen, X.; Armentano, R.; Calvisi, D.F.; et al. Crenigacestat blocking notch pathway reduces liver fibrosis in the surrounding ecosystem of intrahepatic CCA viaTGF-β inhibition. J. Exp. Clin. Cancer Res. 2022, 41, 331. [Google Scholar] [CrossRef]
- Abd El-Rhman, R.H.; El-Naga, R.N.; Gad, A.M.; Tadros, M.G.; Hassaneen, S.K. Dibenzazepine Attenuates Against Cisplatin-Induced Nephrotoxicity in Rats: Involvement of NOTCH Pathway. Front. Pharmacol. 2020, 11, 567852. [Google Scholar] [CrossRef]
- Alsemeh, A.; Abd El- Fatah, S.; Abdel Hamid, R.; Abbas, N.; Abdullah, D. Notch γ-Secretase Inhibitor Dibenzazepine Attenuates Cisplatin-induced Spleen Toxicity in Rats: Role of Notch Signaling Pathway. Zagazig Univ. Med. J. 2022, 28, 1242–1253. [Google Scholar] [CrossRef]
- Ahmed, L.A.; Abd El-Rhman, R.H.; Gad, A.M.; Hassaneen, S.K.; El-Yamany, M.F. Dibenzazepine combats acute liver injury in rats via amendments of Notch signaling and activation of autophagy. Naunyn-Schmiedebergs Arch. Pharmacol. 2021, 394, 337–348. [Google Scholar] [CrossRef]
- Wu, J.; Dong, X.; Li, W.; Zhao, L.; Zhou, L.; Sun, S.; Li, H. Dibenzazepine promotes cochlear supporting cell proliferation and hair cell regeneration in neonatal mice. Cell Prolif. 2020, 53, e12872. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Hesari, A.R.; Azizian, M.; Sheikhi, A.; Nesaei, A.; Sanaei, S.; Mahinparvar, N.; Derakhshani, M.; Hedayt, P.; Ghasemi, F.; Mirzaei, H. Chemopreventive and therapeutic potential of curcumin in esophageal cancer: Current and future status. Int. J. Cancer 2019, 144, 1215–1226. [Google Scholar] [CrossRef]
- Liu, S.; Cao, Y.; Qu, M.; Zhang, Z.; Feng, L.; Ye, Z.; Xiao, M.; Hou, S.T.; Zheng, R.; Han, Z. Curcumin protects against stroke and increases levels of Notch intracellular domain. Neurol. Res. 2016, 38, 553–559. [Google Scholar] [CrossRef]
- Tandon, A.; Singh, S.J.; Gupta, M.; Singh, N.; Shankar, J.; Arjaria, N.; Goyal, S.; Chaturvedi, R.K. Notch pathway up-regulation via curcumin mitigates bisphenol-A (BPA) induced alterations in hippocampal oligodendrogenesis. J. Hazard. Mater. 2020, 392, 122052. [Google Scholar] [CrossRef] [PubMed]
- Zhdanovskaya, N.; Lazzari, S.; Caprioglio, D.; Firrincieli, M.; Maioli, C.; Pace, E.; Imperio, D.; Talora, C.; Bellavia, D.; Checquolo, S.; et al. Identification of a Novel Curcumin Derivative Influencing Notch Pathway and DNA Damage as a Potential Therapeutic Agent in T-ALL. Cancers 2022, 14, 5772. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhang, J.; Zhang, K.; Zhao, Y. Curcumin inhibits cell viability, migration, and invasion of thymic carcinoma cells via downregulation of microRNA-27a. Phyther. Res. 2020, 34, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Q.; Chai, K.Q.; Zhu, X.M.; Jiang, H.; Wang, X.; Xue, Q.; Zheng, A.H.; Zhou, H.Y.; Chen, Y.; Chen, X.C.; et al. Anti-cancer effects of curcumin on lung cancer through the inhibition of EZH2 and NOTCH1. Oncotarget 2016, 7, 26535–26550. [Google Scholar] [CrossRef]
- Sha, J.; Li, J.; Wang, W.; Pan, L.; Cheng, J.; Li, L.; Zhao, H.; Lin, W. Curcumin induces G0/G1 arrest and apoptosis in hormone independent prostate cancer DU-145 cells by down regulating Notch signaling. Biomed. Pharmacother. 2016, 84, 177–184. [Google Scholar] [CrossRef]
- Tang, Y.; Cao, Y. Curcumin Inhibits the Growth and Metastasis of Melanoma via miR-222-3p/SOX10/Notch Axis. Dis. Markers 2022, 2022, 3129781. [Google Scholar] [CrossRef]
- Naujokat, C.; McKee, D.L. The “Big Five” Phytochemicals Targeting Cancer Stem Cells: Curcumin, EGCG, Sulforaphane, Resveratrol and Genistein. Curr. Med. Chem. 2020, 28, 4321–4342. [Google Scholar] [CrossRef] [PubMed]
- Avila-Carrasco, L.; Majano, P.; Sánchez-Toméro, J.A.; Selgas, R.; López-Cabrera, M.; Aguilera, A.; González Mateo, G. Natural Plants Compounds as Modulators of Epithelial-to-Mesenchymal Transition. Front. Pharmacol. 2019, 10, 715. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T. Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett. 2014, 346, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Desmoulin, S.; Banerjee, S.; Kong, D.; Li, Y.; Deraniyagala, R.L.; Abbruzzese, J.; Sarkar, F.H. Synergistic effects of multiple natural products in pancreatic cancer cells. Life Sci. 2008, 83, 293–300. [Google Scholar] [CrossRef]
- Allen, J.L.; Hames, R.A.; Mastroianni, N.M.; Greenstein, A.E.; Weed, S.A. Evaluation of the matrix metalloproteinase 9 (MMP9) inhibitor Andecaliximab as an Anti-invasive therapeutic in Head and neck squamous cell carcinoma. Oral Oncol. 2022, 132, 106008. [Google Scholar] [CrossRef]
- Guo, Z.; Jin, X.; Jia, H. Inhibition of ADAM-17 more effectively down-regulates the Notch pathway than that of γ-secretase in renal carcinoma. J. Exp. Clin. Cancer Res. 2013, 32, 26. [Google Scholar] [CrossRef]
- Lu, H.Y.; Zu, Y.X.; Jiang, X.W.; Sun, X.T.; Liu, T.Y.; Li, R.L.; Wu, Q.; Zhang, Y.S.; Zhao, Q.C. Novel ADAM-17 inhibitor ZLDI-8 inhibits the proliferation and metastasis of chemo-resistant non-small-cell lung cancer by reversing Notch and epithelial mesenchymal transition in vitro and in vivo. Pharmacol. Res. 2019, 148, 104406. [Google Scholar] [CrossRef]
- Li, D.D.; Zhao, C.H.; Ding, H.W.; Wu, Q.; Ren, T.S.; Wang, J.; Chen, C.Q.; Zhao, Q.C. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-Fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 2018, 51, e12480. [Google Scholar] [CrossRef]
- Moss, M.L.; Minond, D. Recent Advances in ADAM17 Research: A Promising Target for Cancer and Inflammation. Mediat. Inflamm. 2017, 2017, 9673537. [Google Scholar] [CrossRef]
- Yang, L.; Bhattacharya, A.; Li, Y.; Sexton, S.; Ling, X.; Li, F.; Zhang, Y. Depleting receptor tyrosine kinases EGFR and HER2 overcomes resistance to EGFR inhibitors in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 184. [Google Scholar] [CrossRef]
- Astudillo, L.; Da Silva, T.G.; Wang, Z.; Han, X.; Jin, K.; VanWye, J.; Zhu, X.; Weaver, K.; Oashi, T.; Lopes, P.E.M.; et al. The Small Molecule IMR-1 Inhibits the Notch Transcriptional Activation Complex to Suppress Tumorigenesis. Cancer Res. 2016, 76, 3593–3603. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Yang, D.; Liu, T.; Liu, S.; Lu, X.; Chen, L. Laminin-integrin a6b4 interaction activates notch signaling to facilitate bladder cancer development. BMC Cancer 2022, 22, 558. [Google Scholar] [CrossRef] [PubMed]
- Kamga, P.T.; Bassi, G.; Cassaro, A.; Midolo, M.; Di Trapani, M.; Gatti, A.; Carusone, R.; Resci, F.; Perbellini, O.; Gottardi, M.; et al. Notch signalling drives bone marrow stromal cell-mediated chemoresistance in acute myeloid leukemia. Oncotarget 2016, 7, 21713–21727. [Google Scholar] [CrossRef]
- Kamga, P.T.; Dal Collo, G.; Midolo, M.; Adamo, A.; Delfino, P.; Mercuri, A.; Cesaro, S.; Mimiola, E.; Bonifacio, M.; Andreini, A.; et al. Inhibition of notch signaling enhances chemosensitivity in B-cell precursor acute lymphoblastic leukemia. Cancer Res. 2019, 79, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Lehal, R.; Zaric, J.; Vigolo, M.; Urech, C.; Frismantas, V.; Zangger, N.; Cao, L.; Berger, A.; Chicote, I.; Loubéry, S.; et al. Pharmacological disruption of the Notch transcription factor complex. Proc. Natl. Acad. Sci. USA 2020, 117, 16292–16301. [Google Scholar] [CrossRef]
- Medinger, M.; Junker, T.; Heim, D.; Tzankov, A.; Jermann, P.M.; Bobadilla, M.; Vigolo, M.; Lehal, R.; Vogl, F.D.; Bauer, M.; et al. CB-103: A novel CSL-NICD inhibitor for the treatment of NOTCH-driven T-cell acute lymphoblastic leukemia: A case report of complete clinical response in a patient with relapsed and refractory T-ALL. eJHaem 2022, 3, 1009–1012. [Google Scholar] [CrossRef]
- Bui, T.O.; Angeli, E.; El Bouchtaoui, M.; Gapihan, G.; Dao, V.T.; Paris, J.; Leboeuf, C.; Soussan, M.; Villarese, P.; Ziol, M.; et al. Metastatic clear-cell renal cell carcinoma: A frequent NOTCH1 mutation predictive of response to anti-NOTCH1 CB-103 treatment. Exp. Hematol. Oncol. 2023, 12, 46. [Google Scholar] [CrossRef]
- Pinazza, M.; Ghisi, M.; Minuzzo, S.; Agnusdei, V.; Fossati, G.; Ciminale, V.; Pezzè, L.; Ciribilli, Y.; Pilotto, G.; Venturoli, C.; et al. Histone deacetylase 6 controls Notch3 trafficking and degradation in T-cell acute lymphoblastic leukemia cells. Oncogene 2018, 37, 3839–3851. [Google Scholar] [CrossRef]
- Hurtado, C.; Safarova, A.; Smith, M.; Chung, R.; Bruyneel, A.A.N.; Gomez-Galeno, J.; Oswald, F.; Larson, C.J.; Cashman, J.R.; Ruiz-Lozano, P.; et al. Disruption of NOTCH signaling by a small molecule inhibitor of the transcription factor RBPJ. Sci. Rep. 2019, 9, 10811. [Google Scholar] [CrossRef]
- Yuan, Z.; VanderWielen, B.D.; Giaimo, B.D.; Pan, L.; Collins, C.E.; Turkiewicz, A.; Hein, K.; Oswald, F.; Borggrefe, T.; Kovall, R.A. Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site. Cell Rep. 2019, 26, 845–854. [Google Scholar] [CrossRef]
- Oswald, F.; Rodriguez, P.; Giaimo, B.D.; Antonello, Z.A.; Mira, L.; Mittler, G.; Thiel, V.N.; Collins, K.J.; Tabaja, N.; Cizelsky, W.; et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res. 2016, 44, 4703–4720. [Google Scholar] [CrossRef]
- Feng, S.; Yang, Y.; Lv, J.; Sun, L.; Liu, M. Valproic acid exhibits different cell growth arrest effect in three HPV-positive/negative cervical cancer cells and possibly via inducing Notch1 cleavage and E6 downregulation. Int. J. Oncol. 2016, 49, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Qian, Q.; Sun, G.; Mackey, L.V.; Fuselier, J.A.; Coy, D.H.; Yu, C.Y. Valproic acid induces NET cell growth arrest and enhances tumor suppression of the receptor-targeted peptide-drug conjugate via activating somatostatin receptor type II. J. Drug Target. 2016, 24, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Aljedai, A.; Buckle, A.-M.; Hiwarkar, P.; Syed, F. Potential Role of Notch Signalling in CD34+ Chronic Myeloid Leukaemia Cells: Cross-Talk between Notch and BCR-ABL. PLoS ONE 2015, 10, e0123016. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, J.; Liang, Q.; Sun, G. Valproic acid reverses sorafenib resistance through inhibiting activated Notch/Akt signaling pathway in hepatocellular carcinoma. Fundam. Clin. Pharmacol. 2021, 35, 690–699. [Google Scholar] [CrossRef]
- Singh, D.; Gupta, S.; Singh, I.; Morsy, M.A.; Nair, A.B.; Ahmed, A.S.F. Hidden pharmacological activities of valproic acid: A new insight. Biomed. Pharmacother. 2021, 142, 112021. [Google Scholar] [CrossRef]
- LaFoya, B.; Munroe, J.A.; Albig, A.R. A comparison of resveratrol and other polyphenolic compounds on Notch activation and endothelial cell activity. PLoS ONE 2019, 14, e0210607. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Q.; Li, Y.; Zhao, D.X.; Gu, R. Dual mechanism of action of resveratrol in notch signaling pathway activation in osteosarcoma. Trop. J. Pharm. Res. 2016, 15, 101–106. [Google Scholar] [CrossRef]
- Pinchot, S.N.; Jaskula-Sztul, R.; Ning, L.; Peters, N.R.; Cook, M.R.; Kunnimalaiyaan, M.; Chen, H. Identification and validation of Notch pathway activating compounds through a novel high-throughput screening method. Cancer 2011, 117, 1386–1398. [Google Scholar] [CrossRef]
- Truong, M.; Cook, M.R.; Pinchot, S.N.; Kunnimalaiyaan, M.; Chen, H. Resveratrol induces Notch2-mediated apoptosis and suppression of neuroendocrine markers in medullary thyroid cancer. Ann. Surg. Oncol. 2011, 18, 1506–1511. [Google Scholar] [CrossRef]
- Yu, X.M.; Jaskula-Sztul, R.; Ahmed, K.; Harrison, A.D.; Kunnimalaiyaan, M.; Chen, H. Resveratrol induces differentiation markers expression in anaplastic thyroid carcinoma via activation of Notch1 signaling and suppresses cell growth. Mol. Cancer Ther. 2013, 12, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Yang, W.; Han, J.; Cheng, R.; Li, L. Effects of Notch signaling components from breast cancer cells treated in culture with resveratrol. Res. Vet. Sci. 2020, 132, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.X.; Li, H.; Wu, M.L.; Liu, X.Y.; Zhong, M.J.; Chen, X.Y.; Liu, J.; Zhang, Y. Inhibition of STAT3 signaling as critical molecular event in resveratrol-suppressed ovarian cancer cells. J. Ovarian Res. 2015, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Park, J.H.; Woo, J.S. Resveratrol induces cell death through ROS-dependent downregulation of Notch1/PTEN/Akt signaling in ovarian cancer cells. Mol. Med. Rep. 2019, 19, 3353–3360. [Google Scholar] [CrossRef]
- Venturelli, S.; Berger, A.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Schleicher, S.; Mayer, M.; Weiss, T.S.; et al. Resveratrol as a Pan-HDAC Inhibitor Alters the Acetylation Status of Jistone Proteins in Human-Derived Hepatoblastoma Cells. PLoS ONE 2013, 8, e73097. [Google Scholar] [CrossRef]
- Ren, B.; Kwah, M.X.Y.; Liu, C.; Ma, Z.; Shanmugam, M.K.; Ding, L.; Xiang, X.; Ho, P.C.L.; Wang, L.; Ong, P.S.; et al. Resveratrol for cancer therapy: Challenges and future perspectives. Cancer Lett. 2021, 515, 63–72. [Google Scholar] [CrossRef]
- Koyama, D.; Kikuchi, J.; Hiraoka, N.; Wada, T.; Kurosawa, H.; Chiba, S.; Furukawa, Y. Proteasome inhibitors exert cytotoxicity and increase chemosensitivity via transcriptional repression of Notch1 in T-cell acute lymphoblastic leukemia. Leukemia 2014, 28, 1216–1226. [Google Scholar] [CrossRef]
- Yu, P.; Petrus, M.N.; Ju, W.; Zhang, M.; Conlon, K.C.; Nakagawa, M.; Maeda, M.; Bamford, R.N.; Waldmann, T.A. Augmented efficacy with the combination of blockade of the Notch-1 pathway, bortezomib and romidepsin in a murine MT-1 adult T-cell leukemia model. Leukemia 2015, 29, 556–566. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Dudimah, D.F.; Pellom, S.T.; Uzhachenko, R.V.; Carbone, D.P.; Dikov, M.M.; Shanker, A. Bortezomib enhances expression of effector molecules in antitumor CD8+T lymphocytes by promoting Notch-nuclear factor-κB crosstalk. Oncotarget 2015, 6, 32439–32455. [Google Scholar] [CrossRef]
- Clementz, A.G.; Osipo, C. Notch versus the proteasome: What is the target of γ-secretase inhibitor-I? Breast Cancer Res. 2009, 11, 110. [Google Scholar] [CrossRef]
- Wu, M.H.; Lu, R.Y.; Yu, S.J.; Tsai, Y.Z.; Lin, Y.C.; Bai, Z.Y.; Liao, R.Y.; Hsu, Y.C.; Chen, C.C.; Cai, B.H. PTC124 Rescues Nonsense Mutation of Two Tumor Suppressor Genes NOTCH1 and FAT1 to Repress HNSCC Cell Proliferation. Biomedicines 2022, 10, 2948. [Google Scholar] [CrossRef] [PubMed]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.; Sonenberg, N. Proposing a mechanism of action for ataluren. Proc. Natl. Acad. Sci. USA 2016, 113, 12353–12355. [Google Scholar] [CrossRef] [PubMed]
- Tutone, M.; Pibiri, I.; Lentini, L.; Pace, A.; Almerico, A.M. Deciphering the Nonsense Readthrough Mechanism of Action of Ataluren: An in Silico Compared Study. ACS Med. Chem. Lett. 2019, 10, 522–527. [Google Scholar] [CrossRef]
- Huang, S.; Bhattacharya, A.; Ghelfi, M.D.; Li, H.; Fritsch, C.; Chenoweth, D.M.; Goldman, Y.E.; Cooperman, B.S. Ataluren binds to multiple protein synthesis apparatus sites and competitively inhibits release factor-dependent termination. Nat. Commun. 2022, 13, 2413. [Google Scholar] [CrossRef]
- Du, M.; Liu, X.; Welch, E.M.; Hirawat, S.; Peltz, S.W.; Bedwell, D.M. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc. Natl. Acad. Sci. USA 2008, 105, 2064–2069. [Google Scholar] [CrossRef]
- Rui, M.; Cai, M.; Zhou, Y.; Zhang, W.; Gao, L.; Mi, K.; Ji, W.; Wang, D.; Feng, C. Identification of Potential RBPJ-Specific Inhibitors for Blocking Notch Signaling in Breast Cancer Using a Drug Repurposing Strategy. Pharmaceuticals 2022, 15, 556. [Google Scholar] [CrossRef]
- Sen, P.; Kandasamy, T.; Ghosh, S.S. Multi-targeting TACE/ADAM17 and gamma-secretase of notch signalling pathway in TNBC via drug repurposing approach using Lomitapide. Cell. Signal. 2023, 102, 110529. [Google Scholar] [CrossRef]
- Dong, Y.; Li, A.; Wang, J.; Weber, J.D.; Michel, L.S. Synthetic lethality through combined notch-epidermal growth factor receptor pathway inhibition in basal-like breast cancer. Cancer Res. 2010, 70, 5465–5474. [Google Scholar] [CrossRef]
- Ferrarotto, R.; Eckhardt, G.; Patnaik, A.; LoRusso, P.; Faoro, L.; Heymach, J.V.; Kapoun, A.M.; Xu, L.; Munster, P. A phase I dose-escalation and dose-expansion study of brontictuzumab in subjects with selected solid tumors. Ann. Oncol. 2018, 29, 1561–1568. [Google Scholar] [CrossRef]
- Xie, M.; Wei, S.; Wu, X.; Li, X.; You, Y.; He, C. Alterations of Notch pathway in patients with adenoid cystic carcinoma of the trachea and its impact on survival. Lung Cancer 2018, 121, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Ferrarotto, R.; Mitani, Y.; Diao, L.; Guijarro, I.; Wang, J.; Zweidler-McKay, P.; Bell, D.; William, W.N.; Glisson, B.S.; Wick, M.J.; et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J. Clin. Oncol. 2017, 35, 352–360. [Google Scholar] [CrossRef] [PubMed]



| Study Title | NCT Number | Status | Conditions | Interventions |
|---|---|---|---|---|
| A Study Of AL101 in Patients with Adenoid Cystic Carcinoma (ACC) Bearing Activating Notch Mutations | NCT03691207 | Unknown status | Adenoid Cystic Carcinoma | AL101 |
| AL101 Before Surgery for the Treatment of Notch-Activated Adenoid Cystic Cancer | NCT04973683 | Recruiting | Adenoid Cystic Carcinoma | AL101 |
| A Study of AL101 Monotherapy in Patients with Notch-Activated Triple-Negative Breast Cancer | NCT04461600 | Active, not recruiting | Triple-Negative Breast Cancer | AL101 |
| A Study of AL102 in Patients with Progressing Desmoid Tumors | NCT04871282 | Recruiting | Desmoid Tumor | AL102 |
| Single-arm Study with Bimiralisib in Patients With HNSCC Harboring NOTCH1 Loss of Function Mutations | NCT03740100 | Terminated | HNSCC | Bimiralisib (PI3Kinase inhibitor) |
| Study to Evaluate the Safety and Tolerability of Weekly Intravenous (IV) Doses of BMS-906024 in Subjects with Acute T-cell Lymphoblastic Leukemia or T-cell Lymphoblastic Lymphoma | NCT01363817 | Completed | Acute T-cel Lymphoblastic Leukemia, Precursor T-Cell Leukemia | BMS-906024 (γ-secretase inhibitor) |
| Study to Evaluate the Safety and Tolerability of IV Doses of BMS-906024 in Subjects with Advanced or Metastatic Solid Tumors | NCT01292655 | Completed | Cancer | BMS-906024 |
| Study to Evaluate Safety and Tolerability of BMS-906024 in Combination with Chemotherapy | NCT01653470 | Completed | Solid cancers | BMS-906024, Paclitaxel, 5-Fluorouracil (5FU), Carboplatin |
| Phase I Ascending Multiple-Dose Study of BMS-986115 in Subjects with Advanced Solid Tumors | NCT01986218 | Terminated | Various Advanced Cancers | BMS-986115 (γ-secretase inhibitor) |
| Compassionate Use of Brontictuzumab for Adenoid Cystic Carcinoma (ACC) | NCT02662608 | Completed | Adenoid Cystic Carcinoma | Brontictuzumab (monoclonal antibody that targets Notch1) |
| CB-103 Plus NSAI in Luminal Advanced Breast Cancer | NCT04714619 | Active, not recruiting | Advanced Breast Cancer | CB-103, NSAIDs |
| Study of CB-103 in Adult Patients with Advanced or Metastatic Solid Tumors and Hematological Malignancies | NCT03422679 | Terminated | Breast Cancer, Colorectal, ACC, Osteosarcoma, HCC | CB-103 |
| A Phase 1/2 Study CB-103 with or without Venetoclax in Patients with NOTCH ACC | NCT05774899 | Recruiting | Adenoid Cystic Carcinoma | CB-103, Venetoclax, Lenvatinib |
| Study in Patients with Advanced Cancers Associated with Expression of DLL3 | NCT04471727 | Recruiting | Small-cell Lung Cancer | HPN328 (a DLL3-targeting T-cell engager), Atezolizumab |
| A Study of LY3039478 in Japanese Participants with Advanced Solid Tumors | NCT02836600 | Active, not recruiting | Advanced Solid Tumor | Crenigacestat/LY3039478 |
| A Study of LY3039478 in Participants with Advanced or Metastatic Solid Tumors | NCT02784795 | Completed | Solid Tumors: Breast Cancer, Colon Cancer | Crenigacestat/LY3039478, Taladegib, Abemaciclib, Cisplatin, Gemcitabine, Carboplatin |
| Notch Inhibitor in Advanced Cancer | NCT01158404 | Completed | Advanced Cancer | LY900009 (γ-secretase inhibitor) |
| A Phase 1 Study to Evaluate the Safety, Tolerability, and Pharmacokinetics of MEDI0639 in Advanced Solid Tumors | NCT01577745 | Completed | Solid Tumors | MEDI0639 (DLL4-targeting antibody) |
| A Notch Signaling Pathway Inhibitor for Patients with Advanced Breast Cancer (0752-014) | NCT00106145 | Completed | Advanced Breast Cancer, other Solid Tumors | MK-0752 |
| A Notch Signaling Pathway Inhibitor for Patients with T-cell Acute Lymphoblastic Leukemia/Lymphoma (ALL) (0752-013) | NCT00100152 | Terminated | Myelogenous Leukemia, Chronic Lymphocytic Leukemia, Lymphoblastic Acute T-cell Leukemia | MK-0752 |
| Phase I/II Study of MK-0752 Followed by Docetaxel in Advanced or Metastatic Breast Cancer | NCT00645333 | Completed | Metastatic Breast Cancer | MK-0752, Docetaxel, Pegfilgrastim |
| MK-0752 and Gemcitabine Hydrochloride in Treating Patients with Stage III and IV Pancreatic Cancer that Cannot be Removed by Surgery | NCT01098344 | Completed | Pancreatic Cancer | MK-0752, gemcitabine |
| Nirogacestat for Adults with Desmoid Tumor/Aggressive Fibromatosis (DT/AF) | NCT03785964 | Active, not recruiting | Desmoid Tumor, Aggressive Fibromatosis | Nirogacestat |
| Individual Patient Compassionate Use of Nirogacestat | NCT05041036 | Available | Desmoid Tumors, NOTCH Gene Mutation Positive Tumors | Nirogacestat |
| γ-secretase inhibitor PF-03084014 in Treating Patients with AIDS-Associated Kaposi Sarcoma | NCT02137564 | Withdrawn | AIDS-related Kaposi Sarcoma | PF-03084014 |
| Biomarker Research Study for PF-03084014 in Chemo-resistant Triple-negative Breast Cancer | NCT02338531 | Withdrawn | Breast Cancer | PF-03084014 |
| A Study Evaluating The PF-03084014 in Combination with Docetaxel in Patients with Advanced Breast Cancer | NCT01876251 | Terminated | Breast Cancer Metastatic | PF-03084014 |
| Phase II Trial of the γ-Secretase Inhibitor PF-03084014 in Adults With Desmoid Tumors/Aggressive Fibromatosis | NCT01981551 | Active, not recruiting | Desmoid Tumors, Aggressive Fibromatosis | PF-03084014 |
| A Trial in Patients with Advanced Cancer and Leukemia | NCT00878189 | Completed | Advanced Cancer And Leukemia | PF-03084014 |
| A Study Evaluating PF-03084014 in Patients with Advanced Breast Cancer with Or without Notch Alterations | NCT02299635 | Terminated | Triple-Negative Breast Neoplasms | PF-03084014 |
| γ-secretase inhibitor RO4929097 in Previously Treated Metastatic Pancreas Cancer | NCT01232829 | Completed | Adenocarcinoma of the Pancreas, Recurrent Pancreatic Cancer | RO4929097 |
| RO4929097 Before Surgery in Treating Patients with Pancreatic Cancer | NCT01192763 | Terminated | Adenocarcinoma of the Pancreas, Stage IA, IB Pancreatic Cancer | RO4929097 |
| Vismodegib and γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Treating Patients with Advanced or Metastatic Sarcoma | NCT01154452 | Completed | Adult Alveolar Soft Part Sarcoma, Angiosarcoma, Desmoplastic Small Round Cell Tumor | RO4929097 |
| RO4929097 in Treating Patients With Recurrent Invasive Gliomas | NCT01269411 | Terminated | Adult Anaplastic Oligodendroglioma, Brain Stem Glioma, Giant Cell Glioblastoma | RO4929097 |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Treating Patients with Recurrent or Progressive Glioblastoma | NCT01122901 | Terminated | Adult Giant Cell Glioblastoma, Gliosarcoma | RO4929097 |
| γ-secretase inhibitor RO4929097 in Treating Young Patients with Relapsed or Refractory Solid Tumors, CNS Tumors, Lymphoma, or T-Cell Leukemia | NCT01088763 | Terminated | Childhood Atypical Teratoid/Rhabdoid Tumor, Choriocarcinoma, Germinoma | RO4929097 |
| A Study of RO4929097 in Patients with Advanced Renal Cell Carcinoma that Have Failed Vascular Endothelial Growth Factor (VEGF)/Vascular Endothelial Growth Factor Receptor (VEGFR) Therapy | NCT01141569 | Completed | Clear Cell Renal Cell Carcinoma, recurrent, and Stage IV Renal Cell Cancer | RO4929097 |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Treating Patients with Advanced, Metastatic, or Recurrent Triple-Negative Invasive Breast Cancer | NCT01151449 | Terminated | Triple-Negative Breast Cancer | RO4929097 |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Treating Patients with Stage IV Melanoma | NCT01120275 | Terminated | Malignant Melanoma | RO4929097 |
| RO4929097 in Treating Patients with Metastatic Colorectal Cancer | NCT01116687 | Completed | Recurrent Colon, Rectal Cancer, Stage IV Colon Cancer | RO4929097 |
| RO4929097 in Treating Patients with Recurrent and/or Metastatic Epithelial Ovarian Cancer, Fallopian Tube Cancer, or Primary Peritoneal Cancer | NCT01175343 | Completed | Fallopian Tube Carcinoma, Ovarian Carcinoma, Primary Peritoneal Carcinoma | RO4929097 |
| RO4929097 in Treating Patients with Advanced Non-Small Cell Lung Cancer who have Recently Completed Treatment with Front-Line Chemotherapy | NCT01193868 | Terminated | Recurrent Non-Small Cell Lung Cancer, Stage IIIB, Stage IV | RO4929097 |
| RO4929097 in Treating Patients with Stage IIIB, Stage IIIC, or Stage IV Melanoma that Can be Removed by Surgery | NCT01216787 | Withdrawn | Melanoma Stage IIIB, IIIC, IV | RO4929097 |
| γ-secretase inhibitor RO4929097 in Treating Patients with Metastatic or Unresectable Solid Malignancies | NCT01096355 | Completed | Unspecified Adult Solid Tumor | RO4929097 |
| RO4929097 After Autologous Stem Cell Transplant in Treating Patients with Multiple Myeloma | NCT01251172 | Withdrawn | Plasma Cell Myeloma Stage I, II, III | RO4929097, Autologous Hematopoietic Stem Cell Transplantation |
| RO4929097 and Bevacizumab in Treating Patients with Progressive or Recurrent Malignant Glioma | NCT01189240 | Terminated | Adult Anaplastic Astrocytoma, Oligodendroglioma, Giant Cell Glioblastoma | RO4929097, bevacizumab |
| Combination Chemotherapy and Bevacizumab with or without RO4929097 in Treating Patients with Metastatic Colorectal Cancer | NCT01270438 | Withdrawn | Adenocarcinoma of the Colon, Rectum | RO4929097, bevacizumab, FOLFOX regimen, |
| Bicalutamide and RO4929097 in Treating Patients with Previously Treated Prostate Cancer | NCT01200810 | Terminated | Adenocarcinoma of the Prostate | RO4929097, bicalutamide |
| RO4929097 and Capecitabine in Treating Patients with Refractory Solid Tumors | NCT01158274 | Completed | Adult Grade III Lymphomatoid Granulomatosis, Adult Nasal Type Extranodal NK/T-cell Lymphoma, AIDS-related Diffuse Large Cell Lymphoma | RO4929097, capecitabine |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097, Paclitaxel, and Carboplatin Before Surgery in Treating Patients with Stage II or Stage III Triple-Negative Breast Cancer | NCT01238133 | Terminated | Triple-Negative Breast Cancer | RO4929097, Carboplatin |
| Phase I Study of Cetuximab with RO4929097 in Metastatic Colorectal Cancer | NCT01198535 | Terminated | Colon Mucinous Adenocarcinoma, Colon Signet Ring Cell Adenocarcinoma, Rectal Mucinous Adenocarcinoma | RO4929097, Cetuximab |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Combination with Cisplatin, Vinblastine, and Temozolomide in Treating Patients with Recurrent or Metastatic Melanoma | NCT01196416 | Completed | Recurrent Melanoma, Stage IV Skin Melanoma | RO4929097, Cisplatin |
| RO4929097 in Children with Relapsed/Refractory Solid or CNS Tumors, Lymphoma, or T-Cell Leukemia | NCT01236586 | Withdrawn | Lymphoma | RO4929097, Dexamethasone |
| RO4929097 and Erlotinib Hydrochloride in Treating Patients with Stage IV or Recurrent Non-Small Cell Lung Cancer | NCT01193881 | Terminated | Recurrent Non-Small Cell Lung Carcinoma, Stage IV Non-Small Cell Lung Cancer | RO4929097, Erlotinib Hydrochloride |
| γ-secretase Inhibitor RO4929097 and Gemcitabine Hydrochloride in Treating Patients with Advanced Solid Tumors | NCT01145456 | Completed | Adenocarcinoma of the Pancreas, Recurrent Pancreatic Cancer, Stage III | RO4929097, gemcitabine |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 in Treating Patients with Advanced Solid Tumors | NCT01218620 | Completed | Adult Solid Neoplasm | RO4929097, Ketoconazole, Rifampicin |
| RO4929097 and Letrozole in Treating Post-Menopausal Women with Hormone-Receptor-Positive Stage II or Stage III Breast Cancer | NCT01208441 | Terminated | Estrogen-Receptor-positive and Progesterone-Receptor-positive Breast cancer, HER2-negative Breast Cancer | RO4929097, letrozole |
| γ-secretase/Notch Signaling Pathway Inhibitor RO4929097 and Temsirolimus in Treating Patients with Advanced Solid Tumors | NCT01198184 | Completed | Endometrial Papillary Serous Carcinoma, Recurrent Endometrial Carcinoma, Recurrent Renal Cell Cancer | RO4929097, temsirolimus |
| RO4929097 and Vismodegib in Treating Patients with Breast Cancer that is Metastatic or Cannot be Removed by Surgery | NCT01071564 | Terminated | Triple-Negative Breast Cancer | RO4929097, Vismodegib |
| RO4929097 and Whole-Brain Radiation Therapy or Stereotactic Radiosurgery in Treating Patients with Brain Metastases From Breast Cancer | NCT01217411 | Terminated | Triple-Negative Breast Cancer | RO4929097, Whole-brain radiation therapy (WBRT), Stereotactic radiosurgery (SRS) |
| RO4929097, Temozolomide, and Radiation Therapy in Treating Patients with Newly Diagnosed Malignant Glioma | NCT01119599 | Completed | Acoustic Schwannoma, Anaplastic Astrocytoma, Anaplastic Meningioma | RO4929097, 3-Dimensional Conformal and Intensity-Modulated Radiation Therapy |
| A Phase 1b/2 Study of OMP-59R5 (Tarextumab) in Combination with Nab-Paclitaxel and Gemcitabine in Subjects with Previously Untreated Stage IV Pancreatic Cancer | NCT01647828 | Completed | Pancreatic Cancer Stage IV | Tarextumab (OMP-59R5, a NOCTH2/3 targeting antibody), Gemcitabine |
| A Study of Tarlatamab in Participants with Neuroendocrine Prostate Cancer | NCT04702737 | Active, not recruiting | Neuroendocrine Prostate Cancer | Tarlatamab (a DLL3-targeting T-cell engager) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czerwonka, A.; Kałafut, J.; Nees, M. Modulation of Notch Signaling by Small-Molecular Compounds and Its Potential in Anticancer Studies. Cancers 2023, 15, 4563. https://doi.org/10.3390/cancers15184563
Czerwonka A, Kałafut J, Nees M. Modulation of Notch Signaling by Small-Molecular Compounds and Its Potential in Anticancer Studies. Cancers. 2023; 15(18):4563. https://doi.org/10.3390/cancers15184563
Chicago/Turabian StyleCzerwonka, Arkadiusz, Joanna Kałafut, and Matthias Nees. 2023. "Modulation of Notch Signaling by Small-Molecular Compounds and Its Potential in Anticancer Studies" Cancers 15, no. 18: 4563. https://doi.org/10.3390/cancers15184563
APA StyleCzerwonka, A., Kałafut, J., & Nees, M. (2023). Modulation of Notch Signaling by Small-Molecular Compounds and Its Potential in Anticancer Studies. Cancers, 15(18), 4563. https://doi.org/10.3390/cancers15184563






