PD-1/PD-L1 Inhibitors Response in Triple-Negative Breast Cancer: Can Long Noncoding RNAs Be Associated?
Abstract
:Simple Summary
Abstract
1. Introduction
2. TNBC Immune Subtypes and Tumor Microenvironment (TME)
3. LncRNAs and Immune Response: The PD-1/PD-L1 Pathway
3.1. lncRNAs and PD-1/PDL-1 Axis in Breast Cancer
3.2. lncRNAs and PD-1/PDL-1 Axis in Other Cancer Types
3.3. lncRNAs Modulated by Treatment with ICI
4. Tying the Knots: Immunotherapy and TNBC
5. Final Considerations
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus Chemotherapy versus Placebo plus Chemotherapy for Previously Untreated Locally Recurrent Inoperable or Metastatic Triple-Negative Breast Cancer (KEYNOTE-355): A Randomised, Placebo-Controlled, Double-Blind, Phase 3 Clinical Trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Adams, S.; Barrios, C.H.; Diéras, V.; Iwata, H.; Loi, S.; Rugo, H.S.; Schneeweiss, A.; Winer, E.P.; Patel, S.; et al. First-Line Atezolizumab plus Nab-Paclitaxel for Unresectable, Locally Advanced, or Metastatic Triple-Negative Breast Cancer: IMpassion130 Final Overall Survival Analysis. Ann. Oncol. 2021, 32, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus Nab-Paclitaxel as First-Line Treatment for Unresectable, Locally Advanced or Metastatic Triple-Negative Breast Cancer (IMpassion130): Updated Efficacy Results from a Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A. Identification and Use of Biomarkers in Treatment Strategies for Triple-Negative Breast Cancer Subtypes. J. Pathol. 2014, 232, 142–150. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023. [Google Scholar] [CrossRef]
- Zhou, Y.; Yue, Y.; Fan, S.; Jia, Q.; Ding, X. Advances in Pathophysiology of Triple-Negative Breast Cancer: The Potential of LncRNAs for Clinical Diagnosis, Treatment, and Prognostic Monitoring. Mol. Biotechnol. 2021, 63, 1093–1102. [Google Scholar] [CrossRef]
- He, Y.; Xiao, B.; Lei, T.; Xuan, J.; Zhu, Y.; Kuang, Z.; Liu, J.; He, J.; Li, L.; Sun, Z. LncRNA T376626 Is a Promising Serum Biomarker and Promotes Proliferation, Migration, and Invasion via Binding to LAMC2 in Triple-Negative Breast Cancer. Gene 2023, 860, 147227. [Google Scholar] [CrossRef]
- Xiu, Y.; Cao, S.; Jiang, R.; Zhou, Y. LncRNA LINC01315 Promotes Malignancy of Triple-Negative Breast Cancer and Predicts Poor Outcomes by Modulating MicroRNA-876-5p/GRK5. Bioengineered 2022, 13, 10001–10009. [Google Scholar] [CrossRef]
- Liu, J.; Yu, H.; Cui, H.; Wei, F.; Yan, T.; Li, T.; Liu, Y.; Chu, J. LncRNA LINC000466 Predicts the Prognosis and Promotes the Progression of Triple-negative Breast Cancer via Modulating MiR-539-5p. Clin. Breast Cancer 2022, 22, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, G.; Ma, X.; Liu, L.; Liu, J.; Yin, Y.; Li, H.; Chen, Y.; Zhang, X.; Zhang, L.; et al. LncRNA TINCR Impairs the Efficacy of Immunotherapy against Breast Cancer by Recruiting DNMT1 and Downregulating MiR-199a-5p via the STAT1–TINCR-USP20-PD-L1 Axis. Cell Death Dis. 2023, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-Negative Breast Cancer: Challenges and Opportunities of a Heterogeneous Disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-Infiltrating Lymphocytes and Prognosis in Different Subtypes of Breast Cancer: A Pooled Analysis of 3771 Patients Treated with Neoadjuvant Therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Saleh, S.M.I.; Bertos, N.; Gruosso, T.; Gigoux, M.; Souleimanova, M.; Zhao, H.; Omeroglu, A.; Hallett, M.T.; Park, M. Identification of Interacting Stromal Axes in Triple-Negative Breast Cancer. Cancer Res. 2017, 77, 4673–4683. [Google Scholar] [CrossRef] [PubMed]
- Jézéquel, P.; Kerdraon, O.; Hondermarck, H.; Guérin-Charbonnel, C.; Lasla, H.; Gouraud, W.; Canon, J.L.; Gombos, A.; Dalenc, F.; Delaloge, S.; et al. Identification of Three Subtypes of Triple-Negative Breast Cancer with Potential Therapeutic Implications. Breast Cancer Res. 2019, 21, 65. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.D.; Liu, Y.R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440. [Google Scholar] [CrossRef]
- Quist, J.; Mirza, H.; Cheang, M.C.U.; Telli, M.L.; O’Shaughnessy, J.A.; Lord, C.J.; Tutt, A.N.J.; Grigoriadis, A. A Four-Gene Decision Tree Signature Classification of Triple-Negative Breast Cancer: Implications for Targeted Therapeutics. Mol. Cancer Ther. 2019, 18, 204–212. [Google Scholar] [CrossRef]
- He, Y.; Jiang, Z.; Chen, C.; Wang, X. Classification of Triple-Negative Breast Cancers Based on Immunogenomic Profiling. J. Exp. Clin. Cancer Res. 2018, 37, 327. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, D.; Zhao, S.; Suo, C.; Shi, J.; Xue, M.Z.; Ruan, M.; Wang, H.; Zhao, J.; Li, Q.; et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin. Cancer Res. 2019, 25, 5002–5014. [Google Scholar] [CrossRef]
- Zheng, H.; Siddharth, S.; Parida, S.; Wu, X.; Sharma, D. Tumor Microenvironment: Key Players in Triple Negative Breast Cancer Immunomodulation. Cancers 2021, 13, 3357. [Google Scholar] [CrossRef] [PubMed]
- Prado-Vázquez, G.; Gámez-Pozo, A.; Trilla-Fuertes, L.; Arevalillo, J.M.; Zapater-Moros, A.; Ferrer-Gómez, M.; Díaz-Almirón, M.; López-Vacas, R.; Navarro, H.; Maín, P.; et al. A Novel Approach to Triple-Negative Breast Cancer Molecular Classification Reveals a Luminal Immune-Positive Subgroup with Good Prognoses. Sci. Rep. 2019, 9, 1538. [Google Scholar] [CrossRef] [PubMed]
- DiNome, M.L.; Orozco, J.I.J.; Matsuba, C.; Manughian-Peter, A.O.; Ensenyat-Mendez, M.; Chang, S.C.; Jalas, J.R.; Salomon, M.P.; Marzese, D.M. Clinicopathological Features of Triple-Negative Breast Cancer Epigenetic Subtypes. Ann. Surg. Oncol. 2019, 26, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zou, Y.; Xie, X.; Liang, J.Y.; Yang, A.; Yu, K.; Wang, J.; Tang, H.; Xie, X. Development and Validation of a Stromal Immune Phenotype Classifier for Predicting Immune Activity and Prognosis in Triple-Negative Breast Cancer. Int. J. Cancer 2020, 147, 542–553. [Google Scholar] [CrossRef]
- Romero-Cordoba, S.; Meneghini, E.; Sant, M.; Iorio, M.V.; Sfondrini, L.; Paolini, B.; Agresti, R.; Tagliabue, E.; Bianchi, F. Decoding Immune Heterogeneity of Triple Negative Breast Cancer and Its Association with Systemic Inflammation. Cancers 2019, 11, 911. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef]
- Eptaminitaki, G.C.; Wolff, N.; Stellas, D.; Sifakis, K.; Baritaki, S. Long Non-Coding RNAs (LncRNAs) in Response and Resistance to Cancer Immunosurveillance and Immunotherapy. Cells 2021, 10, 3313. [Google Scholar] [CrossRef]
- Jiang, W.; Pan, S.; Chen, X.; Wang, Z.W.; Zhu, X. The Role of LncRNAs and CircRNAs in the PD-1/PD-L1 Pathway in Cancer Immunotherapy. Mol. Cancer 2021, 20, 116. [Google Scholar] [CrossRef]
- Guo, Y.; Xie, Y.; Luo, Y. The Role of Long Non-Coding RNAs in the Tumor Immune Microenvironment. Front. Immunol. 2022, 13, 851004. [Google Scholar] [CrossRef]
- Li, G.; Kryczek, I.; Nam, J.; Li, X.; Li, S.; Li, J.; Wei, S.; Grove, S.; Vatan, L.; Zhou, J.; et al. LIMIT Is an Immunogenic LncRNA in Cancer Immunity and Immunotherapy. Nat. Cell Biol. 2021, 23, 526–537. [Google Scholar] [CrossRef]
- Pan, X.; Li, C.; Feng, J. The Role of LncRNAs in Tumor Immunotherapy. Cancer Cell Int. 2023, 23, 30. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced Expression of PD-1, a Novel Member of the Immunoglobulin Gene Superfamily, upon Programmed Cell Death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, R.J.; Freeman, G.J.; Sharpe, A.H. The B7 Family Revisited. Annu. Rev. Immunol. 2005, 23, 515–548. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D.; et al. PD-1 Expression by Tumour-Associated Macrophages Inhibits Phagocytosis and Tumour Immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef]
- Collins, M.; Ling, V.; Carreno, B.M. The B7 Family of Immune-Regulatory Ligands. Genome Biol. 2005, 6, 223. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Robert, C. A Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.Y.; Andre, F.; et al. ESMO Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and Its Relationship with PD-1/PD-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef]
- Davis, A.A.; Patel, V.G. The Role of PD-L1 Expression as a Predictive Biomarker: An Analysis of All US Food and Drug Administration (FDA) Approvals of Immune Checkpoint Inhibitors. J. Immunother. Cancer 2019, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Samir, A.; Tawab, R.A.; Eltayebi, H.M. Long Non-Coding RNAs XIST and MALAT1 Hijack the PD-L1 Regulatory Signaling Pathway in Breast Cancer Subtypes. Oncol. Lett. 2021, 22, 593. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, Prognostic, and Therapeutic Significance of Long Non-Coding RNA MALAT1 in Cancer. Biochim. Biophys. Acta-Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-Coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef] [PubMed]
- Hamed, M.M.; Handoussa, H.; Hussein, N.H.; Eissa, R.A.; Abdel-Aal, L.K.; El Tayebi, H.M. Oleuropin Controls MiR-194/XIST/PD-L1 Loop in Triple Negative Breast Cancer: New Role of Nutri-Epigenetics in Immune-Oncology. Life Sci. 2021, 277, 119353. [Google Scholar] [CrossRef]
- Salama, E.A.; Adbeltawab, R.E.; El Tayebi, H.M. XIST and TSIX: Novel Cancer Immune Biomarkers in PD-L1-Overexpressing Breast Cancer Patients. Front. Oncol. 2020, 9, 1459. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Chen, M.; Chen, H.; Zhong, Q.; Liang, L.; Li, B. LncRNA TCL6 Correlates with Immune Cell Infiltration and Indicates Worse Survival in Breast Cancer. Breast Cancer 2020, 27, 573–585. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, N.; Song, P.; Fu, Y.; Ren, Y.; Li, Z.; Wang, J. LncRNA GATA3-AS1 Facilitates Tumour Progression and Immune Escape in Triple-Negative Breast Cancer through Destabilization of GATA3 but Stabilization of PD-L1. Cell Prolif. 2020, 53, e12855. [Google Scholar] [CrossRef]
- Hu, Q.; Ye, Y.; Chan, L.C.; Li, Y.; Liang, K.; Lin, A.; Egranov, S.D.; Zhang, Y.; Xia, W.; Gong, J.; et al. Oncogenic LncRNA Downregulates Cancer Cell Antigen Presentation and Intrinsic Tumor Suppression. Nat. Immunol. 2019, 20, 835–851. [Google Scholar] [CrossRef]
- Tan, M.; Huang, G.; Chen, J.; Yi, J.; Liu, X.; Liao, N.; Hu, Y.; Zhou, W.; Guo, Q. Construction and Validation of an Eight Pyroptosis-Related LncRNA Risk Model for Breast Cancer. Am. J. Transl. Res. 2022, 14, 2779–2800. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, F.; Yu, X.; Guan, S.; Suo, H.; Tao, Z.; Qiu, Y.; Wu, Y.; Cao, Y.; Jin, F. Immune-Related LncRNAs as Predictors of Survival in Breast Cancer: A Prognostic Signature. J. Transl. Med. 2020, 18, 442. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Wang, X.; Yang, Q. Identification of a Six-Immune-Related Long Non-Coding RNA Signature for Predicting Survival and Immune Infiltrating Status in Breast Cancer. Front. Genet. 2020, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mi, M.; Li, X.; Zheng, X.; Wu, G.; Zhang, L. A LncRNA Prognostic Signature Associated with Immune Infiltration and Tumour Mutation Burden in Breast Cancer. J. Cell. Mol. Med. 2020, 24, 12444–12456. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Peng, X.; Shen, C. Identification and Validation of Immune-Related LncRNA Prognostic Signature for Breast Cancer. Genomics 2020, 112, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Mathias, C.; Muzzi, J.C.D.; Antunes, B.B.; Gradia, D.F.; Castro, M.A.A.; Carvalho de Oliveira, J. Unraveling Immune-Related LncRNAs in Breast Cancer Molecular Subtypes. Front. Oncol. 2021, 11, 692170. [Google Scholar] [CrossRef]
- Sivanandam, V.; LaRocca, C.J.; Chen, N.G.; Fong, Y.; Warner, S.G. Oncolytic Viruses and Immune Checkpoint Inhibition: The Best of Both Worlds. Mol. Ther.-Oncolytics 2019, 13, 93–106. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef]
- NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Melanoma: Cutaneous Version 2.2023. Available online: https://jnccn.org/view/journals/jnccn/19/4/article-p364.xml (accessed on 8 August 2023).
- Seyhan, A.A.; Carini, C. Insights and Strategies of Melanoma Immunotherapy: Predictive Biomarkers of Response and Resistance and Strategies to Improve Response Rates. Int. J. Mol. Sci. 2023, 24, 41. [Google Scholar] [CrossRef]
- Zhou, J.G.; Liang, B.; Liu, J.G.; Jin, S.H.; He, S.S.; Frey, B.; Gu, N.; Fietkau, R.; Hecht, M.; Ma, H.; et al. Identification of 15 LncRNAs Signature for Predicting Survival Benefit of Advanced Melanoma Patients Treated with Anti-PD-1 Monotherapy. Cells 2021, 10, 977. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, Z.; Bao, S.; Hou, P.; Yan, C.; Su, J.; Sun, J. Computational Recognition of LncRNA Signature of Tumor-Infiltrating B Lymphocytes with Potential Implications in Prognosis and Immunotherapy of Bladder Cancer. Brief. Bioinform. 2021, 22, bbaa047. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.; Li, A.; Chen, Y.; Ou, Q.; He, Z.; Zhang, Y.; Liu, R.; Yao, H.; Song, E. Association of Long Noncoding RNA Biomarkers With Clinical Immune Subtype and Prediction of Immunotherapy Response in Patients With Cancer. JAMA Netw. Open 2020, 3, e202149. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors With Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Lemery, S.; Keegan, P.; Pazdur, R. First FDA Approval Agnostic of Cancer Site—When a Biomarker Defines the Indication. N. Engl. J. Med. 2017, 377, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Colon Cancer Version 1.2023. NCCN Guidel. 2023. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1428 (accessed on 8 August 2023).
- Li, J.; Han, T.; Wang, X.; Wang, Y.; Chen, X.; Chen, W.; Yang, Q. Identification of Prognostic Immune-Related LncRNA Signature Predicting the Overall Survival for Colorectal Cancer. Sci. Rep. 2023, 13, 1333. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Pei, L.; Yang, F.; Zhang, C. A Robust Immune-Related LncRNA Signature for the Prognosis of Human Colorectal Cancer. Biosci. Rep. 2022, 42, BSR20220078. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, L.; Weng, S.; Guo, C.; Dang, Q.; Xu, H.; Wang, L.; Lu, T.; Zhang, Y.; Sun, Z.; et al. Machine Learning-Based Integration Develops an Immune-Derived LncRNA Signature for Improving Outcomes in Colorectal Cancer. Nat. Commun. 2022, 13, 816. [Google Scholar] [CrossRef]
- Ding, C.; Shan, Z.; Li, M.; Xia, Y.; Jin, Z. Exploration of the Associations of Lncrna Expression Patterns with Tumor Mutation Burden and Prognosis in Colon Cancer. OncoTargets Ther. 2021, 14, 2893–2909. [Google Scholar] [CrossRef]
- Kathuria, H.; Millien, G.; McNally, L.; Gower, A.C.; Tagne, J.B.; Cao, Y.; Ramirez, M.I. NKX2-1-AS1 Negatively Regulates CD274/PD-L1, Cell-Cell Interaction Genes, and Limits Human Lung Carcinoma Cell Migration. Sci. Rep. 2018, 8, 14418. [Google Scholar] [CrossRef]
- Cheng, G.; Li, Y.; Liu, Z.; Song, X. LncRNA PSMA3-AS1 Promotes the Progression of Non-Small Cell Lung Cancer through Targeting MiR-17-5p/PD-L1. Adv. Clin. Exp. Med. 2021, 30, 1043–1050. [Google Scholar] [CrossRef]
- Du, Z.; Niu, S.; Wang, J.; Wu, J.; Li, S.; Yi, X. SChLAP1 Contributes to Non-Small Cell Lung Cancer Cell Progression and Immune Evasion through Regulating the AUF1/PD-L1 Axis. Autoimmunity 2021, 54, 225–233. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Z.; Xu, S.; Zhang, Q. LncRNA SOX2-OT/MiR-30d-5p/PDK1 Regulates PD-L1 Checkpoint Through the MTOR Signaling Pathway to Promote Non-Small Cell Lung Cancer Progression and Immune Escape. Front. Genet. 2021, 12, 674856. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Z.; Bao, S.; Yan, C.; Hou, P.; Wu, N.; Su, J.; Xu, L.; Zhou, M. Identification of Tumor Immune Infiltration-Associated LncRNAs for Improving Prognosis and Immunotherapy Response of Patients with Non-Small Cell Lung Cancer. J. Immunother. Cancer 2020, 8, e000110. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Jiao, Z.; Lu, G.; Yao, B.; Wang, T.; Rong, W.; Xu, J.; Fan, T.; Sun, X.; Yang, R.; et al. PD-L1 LncRNA Splice Isoform Promotes Lung Adenocarcinoma Progression via Enhancing c-Myc Activity. Genome Biol. 2021, 22, 104. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Wang, W.; Gu, C.; Chen, C.; Zeng, B.; Yang, Y.; Ji, P.; Sun, J.; Wu, J.; Lu, W.; et al. Long Non-Coding RNA HOTTIP Enhances IL-6 Expression to Potentiate Immune Escape of Ovarian Cancer Cells by Upregulating the Expression of PD-L1 in Neutrophils. J. Exp. Clin. Cancer Res. 2019, 38, 411. [Google Scholar] [CrossRef]
- Wang, C.J.; Zhu, C.C.; Xu, J.; Wang, M.; Zhao, W.Y.; Liu, Q.; Zhao, G.; Zhang, Z.Z. The LncRNA UCA1 Promotes Proliferation, Migration, Immune Escape and Inhibits Apoptosis in Gastric Cancer by Sponging Anti-Tumor MiRNAs. Mol. Cancer 2019, 18, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Zhang, M.M.; Liu, C.; Kang, Y.; Wang, J.O.; Yang, X.H. Long Noncoding RNA LINC00473 Drives the Progression of Pancreatic Cancer via Upregulating Programmed Death-Ligand 1 by Sponging MicroRNA-195-5p. J. Cell. Physiol. 2019, 234, 23176–23189. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Z.X.; Chen, Y.X.; Wu, H.X.; Yin, L.; Zhao, Q.; Luo, H.Y.; Zeng, Z.L.; Qiu, M.Z.; Xu, R.H. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, Z.; Zhang, D.; Li, H.; Liu, X.; Zhu, K.; Zhang, H.; Wang, Z.; Zhou, P.; Ren, J.; et al. TIGER: A Web Portal of Tumor Immunotherapy Gene Expression Resource. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, B. Efficacy of PD-1 or PD-L1 Inhibitors and PD-L1 Expression Status in Cancer: Meta-Analysis. BMJ 2018, 362, k3529. [Google Scholar] [CrossRef]
- Grossman, J.E.; Vasudevan, D.; Joyce, C.E.; Hildago, M. Is PD-L1 a Consistent Biomarker for Anti-PD-1 Therapy? The Model of Balstilimab in a Virally-Driven Tumor. Oncogene 2021, 40, 1393–1395. [Google Scholar] [CrossRef]
- Su, X.; Yu, Z.; Zhang, Y.; Chen, J.; Wei, L.; Sun, L. Construction and Analysis of the Dysregulated CeRNA Network and Identification of Risk Long Noncoding RNAs in Breast Cancer. Front. Genet. 2021, 12, 664393. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wu, D.; Zhang, Z.; Li, S. Regulatory Effect of the MAFG-AS1/MiR-150-5p/MYB Axis on the Proliferation and Migration of Breast Cancer Cells. Int. J. Oncol. 2021, 58, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Zhang, S.; Sun, H.; Wu, Y.; Yan, M. LncRNA MAFG-AS1 Affects the Tumorigenesis of Breast Cancer Cells via the MiR-574-5p/SOD2 Axis. Biochem. Biophys. Res. Commun. 2021, 560, 119–125. [Google Scholar] [CrossRef]
- Li, H.; Zhang, G.Y.; Pan, C.H.; Zhang, X.Y.; Su, X.Y. LncRNA MAFG-AS1 Promotes the Aggressiveness of Breast Carcinoma through Regulating MiR-339-5p/MMP15. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2838–2846. [Google Scholar] [CrossRef]
- Ding, M.; Fu, Y.; Guo, F.; Chen, H.; Fu, X.; Tan, W.; Zhang, H. Long Non-Coding RNA MAFG-AS1 Knockdown Blocks Malignant Progression in Breast Cancer Cells by Inactivating JAK2/STAT3 Signaling Pathway via MAFG-AS1/MiR-3196/TFAP2A Axis. Int. J. Clin. Exp. Pathol. 2020, 13, 2455–2473. [Google Scholar] [PubMed]
- Feng, J.; Wen, T.; Li, Z.; Feng, L.; Zhou, L.; Yang, Z.; Xu, L.; Shi, S.; Hou, K.; Shen, J.; et al. Cross-Talk between the ER Pathway and the LncRNA MAFG-AS1/MiR-339-5p/ CDK2 Axis Promotes Progression of ER+ Breast Cancer and Confers Tamoxifen Resistance. Aging (Albany NY) 2020, 12, 20658–20683. [Google Scholar] [CrossRef]
- Wang, L.; Luan, T.; Zhou, S.; Lin, J.; Yang, Y.; Liu, W.; Tong, X.; Jiang, W. LncRNA HCP5 Promotes Triple Negative Breast Cancer Progression as a CeRNA to Regulate BIRC3 by Sponging MiR-219a-5p. Cancer Med. 2019, 8, 4389–4403. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, H.; Ye, M.; Wang, B.; Zhang, Y.; Sheng, J.; Meng, T.; Chen, H. Downregulation of Long Noncoding RNA HCP5 Contributes to Cisplatin Resistance in Human Triple-Negative Breast Cancer via Regulation of PTEN Expression. Biomed. Pharmacother. 2020, 115, 108869. [Google Scholar] [CrossRef]
- Li, J.; Gao, C.; Liu, C.; Zhou, C.; Ma, X.; Li, H.; Li, J.; Wang, X.; Qi, L.; Yao, Y.; et al. Four LncRNAs Associated with Breast Cancer Prognosis Identified by Coexpression Network Analysis. J. Cell. Physiol. 2019, 234, 14019–14030. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Q.; Kang, Y.; Liu, J.; Yin, Y.; Liu, L.; Wu, H.; Li, S.; Sui, S.; Shen, M.; et al. Long Noncoding RNAs Control the Modulation of Immune Checkpoint Molecules in Cancer. Cancer Immunol. Res. 2020, 8, 937–951. [Google Scholar] [CrossRef]
- Puig-Saus, C.; Sennino, B.; Peng, S.; Wang, C.L.; Pan, Z.; Yuen, B.; Purandare, B.; An, D.; Quach, B.B.; Nguyen, D.; et al. Neoantigen-Targeted CD8+ T Cell Responses with PD-1 Blockade Therapy. Nature 2023, 615, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Kulski, J.K. Long Noncoding RNA HCP5, a Hybrid HLA Class I Endogenous Retroviral Gene: Structure, Expression, and Disease Associations. Cells 2019, 8, 480. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, M.; Zhang, Y.; Cai, Y.; Zhao, G. Molecular Mechanism of Activated T Cells in Breast Cancer. OncoTargets Ther. 2018, 11, 5015–5024. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Feng, Y.; He, X.L.; Wang, Y. Effects of LncRNA-UCA1 Targeting MiR-204-5p on the Proliferation, Migration, Apoptosis and Immune Escape of Endometrial Carcinoma Cells. Zhonghua Zhong Liu Za Zhi 2023, 45, 56–63. [Google Scholar] [CrossRef] [PubMed]
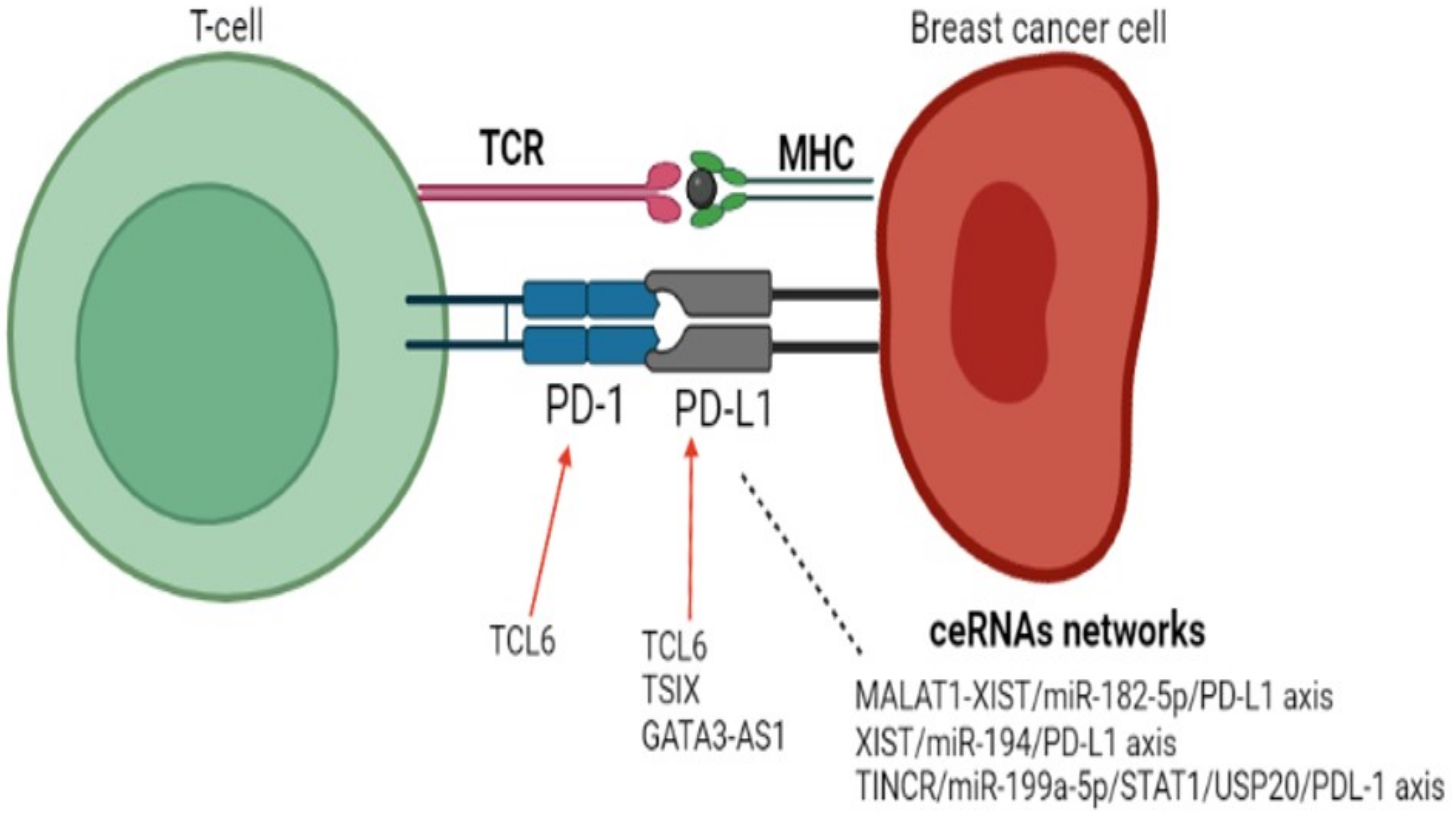
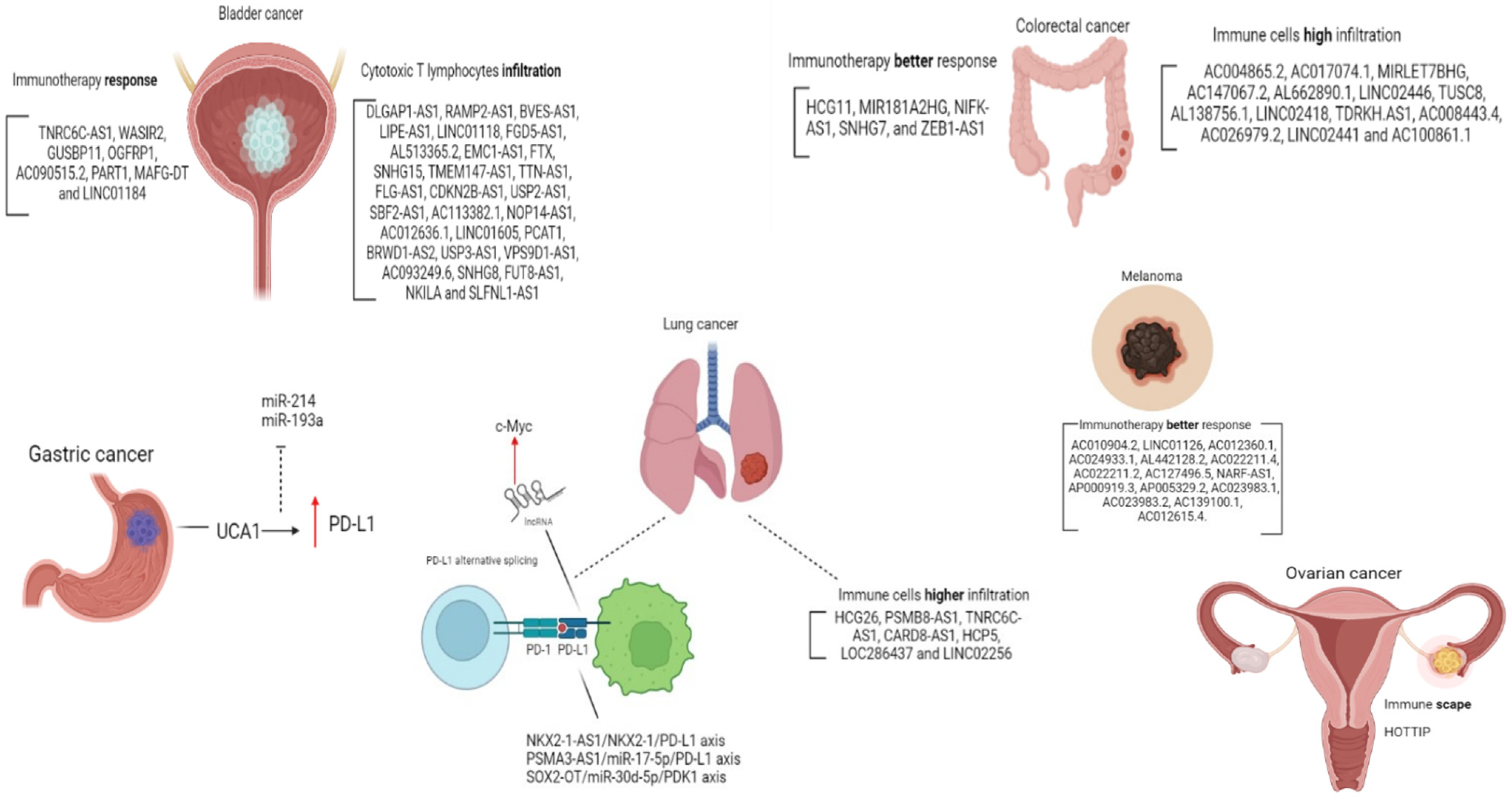

| Subtype | Clinical Significance | Reference |
|---|---|---|
| The described IM subtype was related with better prognosis | [5] |
| Signature classified patients according to outcomes | [15] |
| C2 and C3 were sensitive to therapy combating immunosuppression | [16] |
| High level of CD8+ and CD4+ immune signatures in MC6 subtype. | [18] |
| Immunity H subtype was related with better prognosis as it shows more immune cells expression | [19] |
| Claudin-h and immune-positive subtypes have better prognosis | [22] |
| Epi-CL-D showed a positive regulation of T lymphocyte cytotoxicity, enriched response to interferon-beta and antigen processing | [23] |
| Phenotype A exhibited an enrichment of immune-related pathways | [24] |
| Systemic inflammation parameters are informative markers | [25] |
| C2 immune subtype is related with immune activation | [26] |
| lncRNAs | Measured Outcome | Reference |
|---|---|---|
| OTUD6B-AS1, AL122010.1, AC136475.2, AL161646.1, AC245297.3, LINC00578, LINC01871, AP000442.2 | Overall survival | [51] |
| MAPT-IT1, SLC26A4-AS1, VPS9D1-AS1, PCAT18, LINC01234, SPATA41, LINC01215 | Overall survival | [53] |
| AC116366.1, AC244502.1, AC100810.1, MIAT, AC093297.2, AL356417.2 | Overall survival | [52] |
| LINC01010, AP005131.6, AC004847.1, AL591686.1, LINC00668, LINC02418, AL356515.1, AC027514.1, AL772337.1, AL161646.2, AC243773.2 | Overall survival and immune cell infiltration | [54] |
| lncRNA | logFC | p-Value |
|---|---|---|
| NARF-AS1 | 0.38 | 1.64 × 10−33 |
| WASIR2 | 0.57 | 6.54 × 10−10 |
| GUSBP11 | −0.17 | 0.003 |
| OGFRP1 | 0.55 | 6.28 × 10−31 |
| PART1 | 0.87 | 1.40 × 10−10 |
| MILIP (MAFG-DT) | 1.28 | 9.20 × 10−40 |
| LINC01184 | −0.44 | 5.09 × 10−11 |
| UCA1 | 0.87 | 8.50 × 10−13 |
| FGD5-AS1 | −0.21 | 0.004 |
| NKX2-1-AS1 | 0.36 | 5.70 × 10−26 |
| HCP5 | 0.68 | 1.48 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathias, C.; Kozak, V.N.; Magno, J.M.; Baal, S.C.S.; dos Santos, V.H.A.; Ribeiro, E.M.d.S.F.; Gradia, D.F.; Castro, M.A.A.; Carvalho de Oliveira, J. PD-1/PD-L1 Inhibitors Response in Triple-Negative Breast Cancer: Can Long Noncoding RNAs Be Associated? Cancers 2023, 15, 4682. https://doi.org/10.3390/cancers15194682
Mathias C, Kozak VN, Magno JM, Baal SCS, dos Santos VHA, Ribeiro EMdSF, Gradia DF, Castro MAA, Carvalho de Oliveira J. PD-1/PD-L1 Inhibitors Response in Triple-Negative Breast Cancer: Can Long Noncoding RNAs Be Associated? Cancers. 2023; 15(19):4682. https://doi.org/10.3390/cancers15194682
Chicago/Turabian StyleMathias, Carolina, Vanessa Nascimento Kozak, Jessica Maria Magno, Suelen Cristina Soares Baal, Victor Henrique Apolonio dos Santos, Enilze Maria de Souza Fonseca Ribeiro, Daniela Fiori Gradia, Mauro Antonio Alves Castro, and Jaqueline Carvalho de Oliveira. 2023. "PD-1/PD-L1 Inhibitors Response in Triple-Negative Breast Cancer: Can Long Noncoding RNAs Be Associated?" Cancers 15, no. 19: 4682. https://doi.org/10.3390/cancers15194682
APA StyleMathias, C., Kozak, V. N., Magno, J. M., Baal, S. C. S., dos Santos, V. H. A., Ribeiro, E. M. d. S. F., Gradia, D. F., Castro, M. A. A., & Carvalho de Oliveira, J. (2023). PD-1/PD-L1 Inhibitors Response in Triple-Negative Breast Cancer: Can Long Noncoding RNAs Be Associated? Cancers, 15(19), 4682. https://doi.org/10.3390/cancers15194682









