Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA Isolation
2.3. TP53 Genotyping
2.4. Cytoplasmatic Immunoglobulin and FISH (clg-FISH) Method
2.5. RNA Isolation and TP53 Expression
2.6. Statistical Analysis
3. Results
3.1. Frequencies of Alleles and Genotypes and Their Associations with MM Risk
3.2. TP53 Variants and Risk of Death and MM
3.3. Association of Studied Variant and TP53 Expression with Clinical/Laboratory Values
3.4. Survival of MM Patients Taking into Account Type of Treatment and Studied Variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sonneveld, P.; Avet-Loiseau, H.; Lonial, S.; Usmani, S.; Siegel, D.; Anderson, K.C.; Chng, W.-J.; Moreau, P.; Attal, M.; Kyle, R.A.; et al. Treatment of Multiple Myeloma with High-Risk Cytogenetics: A Consensus of the International Myeloma Working Group. Blood 2016, 127, 2955–2962. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V. Multiple Myeloma: 2020 Update on Diagnosis, Risk-stratification and Management. Am. J. Hematol. 2020, 95, 548–567. [Google Scholar] [CrossRef] [PubMed]
- Bergsagel, P.L.; Mateos, M.-V.; Gutierrez, N.C.; Rajkumar, S.V.; San Miguel, J.F. Improving Overall Survival and Overcoming Adverse Prognosis in the Treatment of Cytogenetically High-Risk Multiple Myeloma. Blood 2013, 121, 884–892. [Google Scholar] [CrossRef]
- Hanamura, I. Multiple Myeloma with High-Risk Cytogenetics and Its Treatment Approach. Int. J. Hematol. 2022, 115, 762–777. [Google Scholar] [CrossRef]
- Antoun, S.; Atallah, D.; Tahtouh, R.; Alaaeddine, N.; Moubarak, M.; Khaddage, A.; Ayoub, E.N.; Chahine, G.; Hilal, G. Different TP53 Mutants in P53 Overexpressed Epithelial Ovarian Carcinoma Can Be Associated Both with Altered and Unaltered Glycolytic and Apoptotic Profiles. Cancer Cell Int. 2018, 18, 14. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting P53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [PubMed]
- Cardona-Benavides, I.J.; de Ramón, C.; Gutiérrez, N.C. Genetic Abnormalities in Multiple Myeloma: Prognostic and Therapeutic Implications. Cells 2021, 10, 336. [Google Scholar] [CrossRef]
- Shah, V.; Sherborne, A.L.; Walker, B.A.; Johnson, D.C.; Boyle, E.M.; Ellis, S.; Begum, D.B.; Proszek, P.Z.; Jones, J.R.; Pawlyn, C.; et al. Prediction of Outcome in Newly Diagnosed Myeloma: A Meta-Analysis of the Molecular Profiles of 1905 Trial Patients. Leukemia 2018, 32, 102–110. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. A High-Risk, Double-Hit, Group of Newly Diagnosed Myeloma Identified by Genomic Analysis. Leukemia 2019, 33, 159–170. [Google Scholar] [CrossRef]
- An, G.; Li, Z.; Tai, Y.-T.; Acharya, C.; Li, Q.; Qin, X.; Yi, S.; Xu, Y.; Feng, X.; Li, C.; et al. The Impact of Clone Size on the Prognostic Value of Chromosome Aberrations by Fluorescence In Situ Hybridization in Multiple Myeloma. Clin. Cancer Res. 2015, 21, 2148–2156. [Google Scholar] [CrossRef]
- Thanendrarajan, S.; Tian, E.; Qu, P.; Mathur, P.; Schinke, C.; van Rhee, F.; Zangari, M.; Rasche, L.; Weinhold, N.; Alapat, D.; et al. The Level of Deletion 17p and Bi-Allelic Inactivation of TP53 Has a Significant Impact on Clinical Outcome in Multiple Myeloma. Haematologica 2017, 102, e364–e367. [Google Scholar] [CrossRef] [PubMed]
- Thakurta, A.; Ortiz, M.; Blecua, P.; Towfic, F.; Corre, J.; Serbina, N.V.; Flynt, E.; Yu, Z.; Yang, Z.; Palumbo, A.; et al. High Subclonal Fraction of 17p Deletion Is Associated with Poor Prognosis in Multiple Myeloma. Blood 2019, 133, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Teoh, P.J.; Chung, T.H.; Sebastian, S.; Choo, S.N.; Yan, J.; Ng, S.B.; Fonseca, R.; Chng, W.J. P53 Haploinsufficiency and Functional Abnormalities in Multiple Myeloma. Leukemia 2014, 28, 2066–2074. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.-C.; Khoury, M.P.; Diot, A.; Baker, L.; Fernandes, K.; Aoubala, M.; Quinlan, P.; Purdie, C.A.; Jordan, L.B.; Prats, A.-C.; et al. P53 Mutant Breast Cancer Patients Expressing P53γ Have as Good a Prognosis as Wild-Type P53 Breast Cancer Patients. Breast Cancer Res. 2011, 13, R7. [Google Scholar] [CrossRef] [PubMed]
- Anensen, N.; Oyan, A.M.; Bourdon, J.-C.; Kalland, K.H.; Bruserud, O.; Gjertsen, B.T. A Distinct P53 Protein Isoform Signature Reflects the Onset of Induction Chemotherapy for Acute Myeloid Leukemia. Clin. Cancer Res. 2006, 12, 3985–3992. [Google Scholar] [CrossRef]
- Boldrup, L.; Bourdon, J.-C.; Coates, P.J.; Sjöström, B.; Nylander, K. Expression of P53 Isoforms in Squamous Cell Carcinoma of the Head and Neck. Eur. J. Cancer 2007, 43, 617–623. [Google Scholar] [CrossRef]
- Martello, M.; Poletti, A.; Borsi, E.; Solli, V.; Dozza, L.; Barbato, S.; Zamagni, E.; Tacchetti, P.; Pantani, L.; Mancuso, K.; et al. Clonal and Subclonal TP53 Molecular Impairment Is Associated with Prognosis and Progression in Multiple Myeloma. Blood Cancer J. 2022, 12, 15. [Google Scholar] [CrossRef]
- Stracquadanio, G.; Wang, X.; Wallace, M.D.; Grawenda, A.M.; Zhang, P.; Hewitt, J.; Zeron-Medina, J.; Castro-Giner, F.; Tomlinson, I.P.; Goding, C.R.; et al. The Importance of P53 Pathway Genetics in Inherited and Somatic Cancer Genomes. Nat. Rev. Cancer 2016, 16, 251–265. [Google Scholar] [CrossRef]
- Kang, H.-J.; Feng, Z.; Sun, Y.; Atwal, G.; Murphy, M.E.; Rebbeck, T.R.; Rosenwaks, Z.; Levine, A.J.; Hu, W. Single-Nucleotide Polymorphisms in the P53 Pathway Regulate Fertility in Humans. Proc. Natl. Acad. Sci. USA 2009, 106, 9761–9766. [Google Scholar] [CrossRef]
- De Souza, C.; Madden, J.; Koestler, D.C.; Minn, D.; Montoya, D.J.; Minn, K.; Raetz, A.G.; Zhu, Z.; Xiao, W.-W.; Tahmassebi, N.; et al. Effect of the P53 P72R Polymorphism on Mutant TP53 Allele Selection in Human Cancer. J. Natl. Cancer Inst. 2021, 113, 1246–1257. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Alharbi, R.A. Genetic Association of TP53 Pro72Arg Polymorphism (Rs1042522) in Leukemia: An Updated Meta-Analysis of 10 Case-Control Studies. Hum. Gene 2022, 34, 201130. [Google Scholar] [CrossRef]
- Baptiste, N.; Friedlander, P.; Chen, X.; Prives, C. The Proline-Rich Domain of P53 Is Required for Cooperation with Anti-Neoplastic Agents to Promote Apoptosis of Tumor Cells. Oncogene 2002, 21, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Phan, L.; Jin, Y.; Zhang, H.; Qiang, W.; Shekhtman, E.; Shao, D.; Revoe, D.; Villamarin, R.; Ivanchenko, E.; Kimura, M.; et al. ALFA: Allele Frequency Aggregator; National Center for Biotechnology Information, US National Library of Medicine: Rockville Pike Bethesda, MD, USA, 2020; Volume 10. Available online: www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/ (accessed on 1 July 2023).
- Yemelyanova, A.; Vang, R.; Kshirsagar, M.; Lu, D.; Marks, M.A.; Shih, I.M.; Kurman, R.J. Immunohistochemical Staining Patterns of P53 Can Serve as a Surrogate Marker for TP53 Mutations in Ovarian Carcinoma: An Immunohistochemical and Nucleotide Sequencing Analysis. Mod. Pathol. 2011, 24, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, E.D.; Soulitzis, N.; Spandidos, D.A. Association of P53 Codon 72 Polymorphism with Advanced Lung Cancer: The Arg Allele Is Preferentially Retained in Tumours Arising in Arg/Pro Germline Heterozygotes. Br. J. Cancer 2002, 87, 1013–1018. [Google Scholar] [CrossRef]
- Dumont, P.; Leu, J.I.-J.; Della Pietra, A.C.; George, D.L.; Murphy, M. The Codon 72 Polymorphic Variants of P53 Have Markedly Different Apoptotic Potential. Nat. Genet. 2003, 33, 357–365. [Google Scholar] [CrossRef]
- Siddique, M.; Sabapathy, K. Trp53-Dependent DNA-Repair Is Affected by the Codon 72 Polymorphism. Oncogene 2006, 25, 3489–3500. [Google Scholar] [CrossRef]
- Miller, D.P.; Liu, G.; De Vivo, I.; Lynch, T.J.; Wain, J.C.; Su, L.; Christiani, D.C. Combinations of the Variant Genotypes of GSTP1, GSTM1, and P53 Are Associated with an Increased Lung Cancer Risk. Cancer Res. 2002, 62, 2819–2823. [Google Scholar]
- Pim, D.; Banks, L. P53 Polymorphic Variants at Codon 72 Exert Different Effects on Cell Cycle Progression. Int. J. Cancer 2004, 108, 196–199. [Google Scholar] [CrossRef]
- Zmorzyński, S.; Popek-Marciniec, S.; Styk, W.; Wojcierowska-Litwin, M.; Korszeń-Pilecka, I.; Szudy-Szczyrek, A.; Chocholska, S.; Hus, M.; Filip, A.A. The Impact of the NOD2/CARD15 Variant (3020insC) and PSMA6 Polymorphism (-8C > G) on the Development and Outcome of Multiple Myeloma. Biomed Res. Int. 2020, 2020, 7629456. [Google Scholar] [CrossRef]
- Ross, F.M.; Avet-Loiseau, H.; Ameye, G.; Gutierrez, N.C.; Liebisch, P.; O’Connor, S.; Dalva, K.; Fabris, S.; Testi, A.M.; Jarosova, M.; et al. Report from the European Myeloma Network on Interphase FISH in Multiple Myeloma and Related Disorders. Haematologica 2012, 97, 1272–1277. [Google Scholar] [CrossRef]
- Ahmann, G.J.; Jalal, S.M.; Juneau, A.L.; Christensen, E.R.; Hanson, C.A.; Dewald, G.W.; Greipp, P.R. A Novel Three-Color, Clone-Specific Fluorescence in Situ Hybridization Procedure for Monoclonal Gammopathies. Cancer Genet. Cytogenet. 1998, 101, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Dmoszyńska, A.; Chocholska, S. Molecular Biology Methods in the Diagnosis of Multiple Myeloma. In Molecular Aspects of Hematologic Malignancies: Diagnostic Tools and Clinical Applications; Springer: Berlin/Heidelberg, Germany, 2012; pp. 443–449. [Google Scholar]
- Zmorzyński, S.; Popek-Marciniec, S.; Szudy-Szczyrek, A.; Wojcierowska-Litwin, M.; Korszeń-Pilecka, I.; Chocholska, S.; Styk, W.; Hus, M.; Filip, A.A. The Association of GSTT1, GSTM1, and TNF-α Polymorphisms With the Risk and Outcome in Multiple Myeloma. Front. Oncol. 2019, 9, 1056. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Pang, Z. Stability of Methods for Differential Expression Analysis of RNA-Seq Data. BMC Genom. 2019, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential Expression in RNA-Seq: A Matter of Depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Hattori, Y.; Ikeda, Y.; Suzuki, Y.; Ichikawa, D.; Matsushita, M. Codon 72 Polymorphism of TP53 Gene Is a Novel Prognostic Marker for Therapy in Multiple Myeloma. Br. J. Haematol. 2014, 165, 728–731. [Google Scholar] [CrossRef]
- Govindasamy, P.; Sharchil, C.; Mohan, N.; Pandurangan, P.; Tarigopula, A.; Mani, R.; Samuel, C.R. TP53 Gene Alterations Including Codon 72 Polymorphism in Patients with Multiple Myeloma. J. Clin. Diagn. Res. 2018, 12. [Google Scholar] [CrossRef]
- Danthinne, E.S.; Giorgianni, F.E.; Rodgers, R.F. Labels to Prevent the Detrimental Effects of Media on Body Image: A Systematic Review and Metaanalysis. Int. J. Eat. Disord. 2020, 53, 647–661. [Google Scholar] [CrossRef]
- Ahmed, S.; Safwat, G.; Moneer, M.M.; El Ghareeb, A.; El Sherif, A.A.; Loutfy, S.A. Prevalence of TP53 Gene Pro72Arg (Rs1042522) Single Nucleotide Polymorphism among Egyptian Breast Cancer Patients. Egypt. J. Med. Hum. Genet. 2023, 24, 24. [Google Scholar] [CrossRef]
- Mohammed Basabaeen, A.A.; Abdelgader, E.A.; Babekir, E.A.; Abdelrahim, S.O.; Eltayeb, N.H.; Altayeb, O.A.; Fadul, E.A.; Sabo, A.; Ibrahim, I.K. TP53 Gene 72 Arg/Pro (Rs1042522) Single Nucleotide Polymorphism Contribute to Increase the Risk of B-Chronic Lymphocytic Leukemia in the Sudanese Population. Asian Pac. J. Cancer Prev. 2019, 20, 1579–1585. [Google Scholar] [CrossRef]
- Scarfò, L.; Ferreri, A.J.M.; Ghia, P. Chronic Lymphocytic Leukaemia. Crit. Rev. Oncol. Hematol. 2016, 104, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Zhao, Y.; Feng, Z. MicroRNA Control of P53. J. Cell. Biochem. 2017, 118, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Sargolzaei, J.; Etemadi, T.; Alyasin, A. The P53/MicroRNA Network: A Potential Tumor Suppressor with a Role in Anticancer Therapy. Pharmacol. Res. 2020, 160, 105179. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Q. TP53 Mutations, Expression and Interaction Networks in Human Cancers. Oncotarget 2017, 8, 624–643. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Ahn, A.-R.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Ha, G.W.; Lee, M.R.; Chung, M.J. Clinical Significance of P53 Protein Expression and TP53 Variation Status in Colorectal Cancer. BMC Cancer 2022, 22, 940. [Google Scholar] [CrossRef]
- Taghavi, N. Association of P53/P21 Expression with Cigarette Smoking and Prognosis in Esophageal Squamous Cell Carcinoma Patients. World J. Gastroenterol. 2010, 16, 4958. [Google Scholar] [CrossRef]
- Baosen, Z.; Anguang, H.; Jijiang, Z.; Enhua, W. The Study on Relationship between Cigarette Smoking and the P53 Protein and P21 Protein Expression in Non-Small Lung Cancer. Chin. J. Cancer Res. 1996, 8, 187–191. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Silwal-Pandit, L.; Meza-Zepeda, L.A.; Vodak, D.; Vu, P.; Sagerup, C.; Hovig, E.; Myklebost, O.; Børresen-Dale, A.-L.; Brustugun, O.T.; et al. TP53 Mutation Spectrum in Smokers and Never Smoking Lung Cancer Patients. Front. Genet. 2016, 7, 85. [Google Scholar] [CrossRef]
- Rojas, E.A.; Corchete, L.A.; De Ramón, C.; Krzeminski, P.; Quwaider, D.; García-Sanz, R.; Martínez-López, J.; Oriol, A.; Rosiñol, L.; Bladé, J.; et al. Expression of P53 Protein Isoforms Predicts Survival in Patients with Multiple Myeloma. Am. J. Hematol. 2022, 97, 700–710. [Google Scholar] [CrossRef]
- Bourdon, J.-C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. P53 Isoforms Can Regulate P53 Transcriptional Activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef]
- Surget, S.; Khoury, M.P.; Bourdon, J.-C. Uncovering the Role of P53 Splice Variants in Human Malignancy: A Clinical Perspective. OncoTargets Ther. 2013, 7, 57–68. [Google Scholar] [CrossRef]
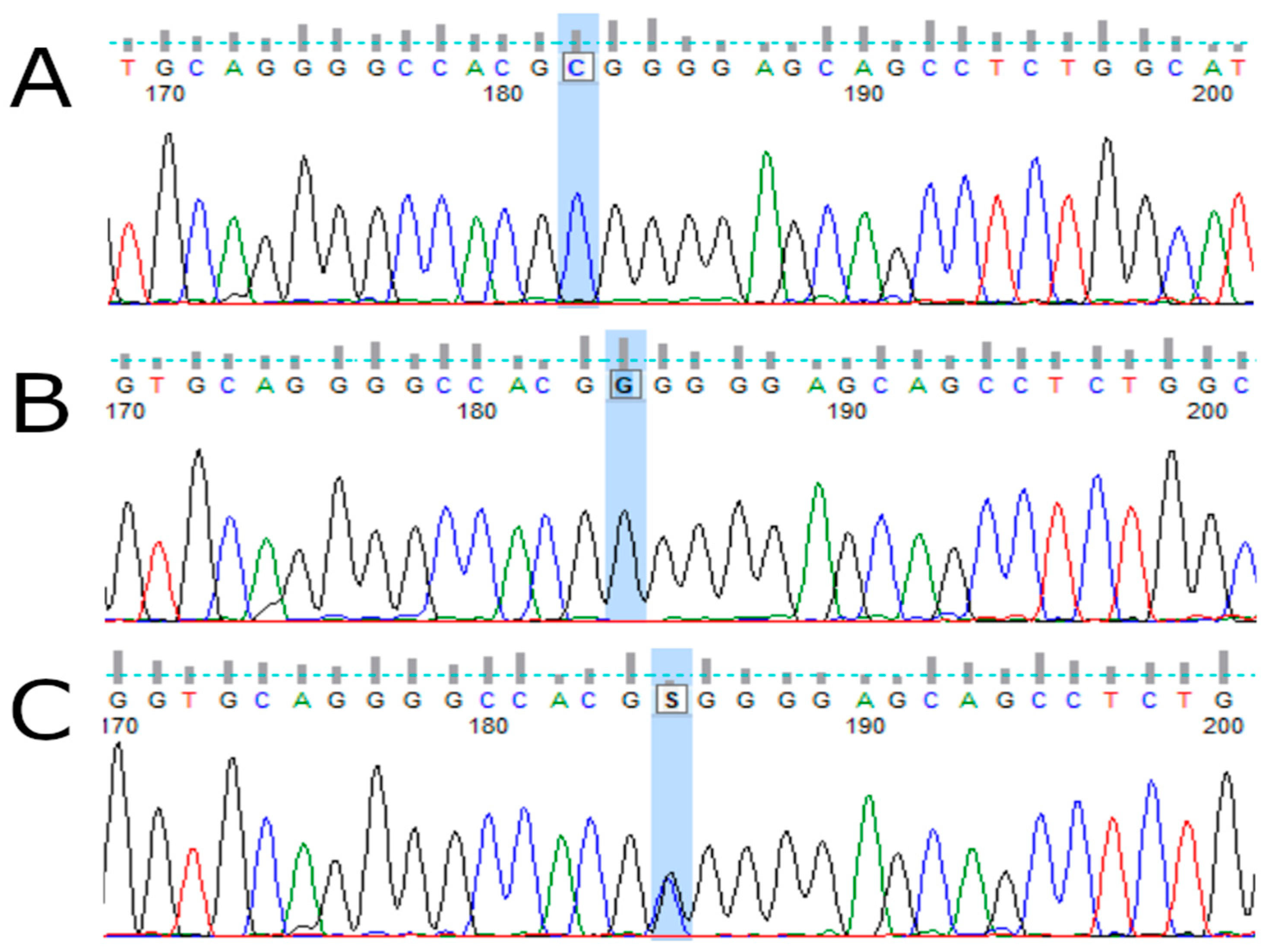
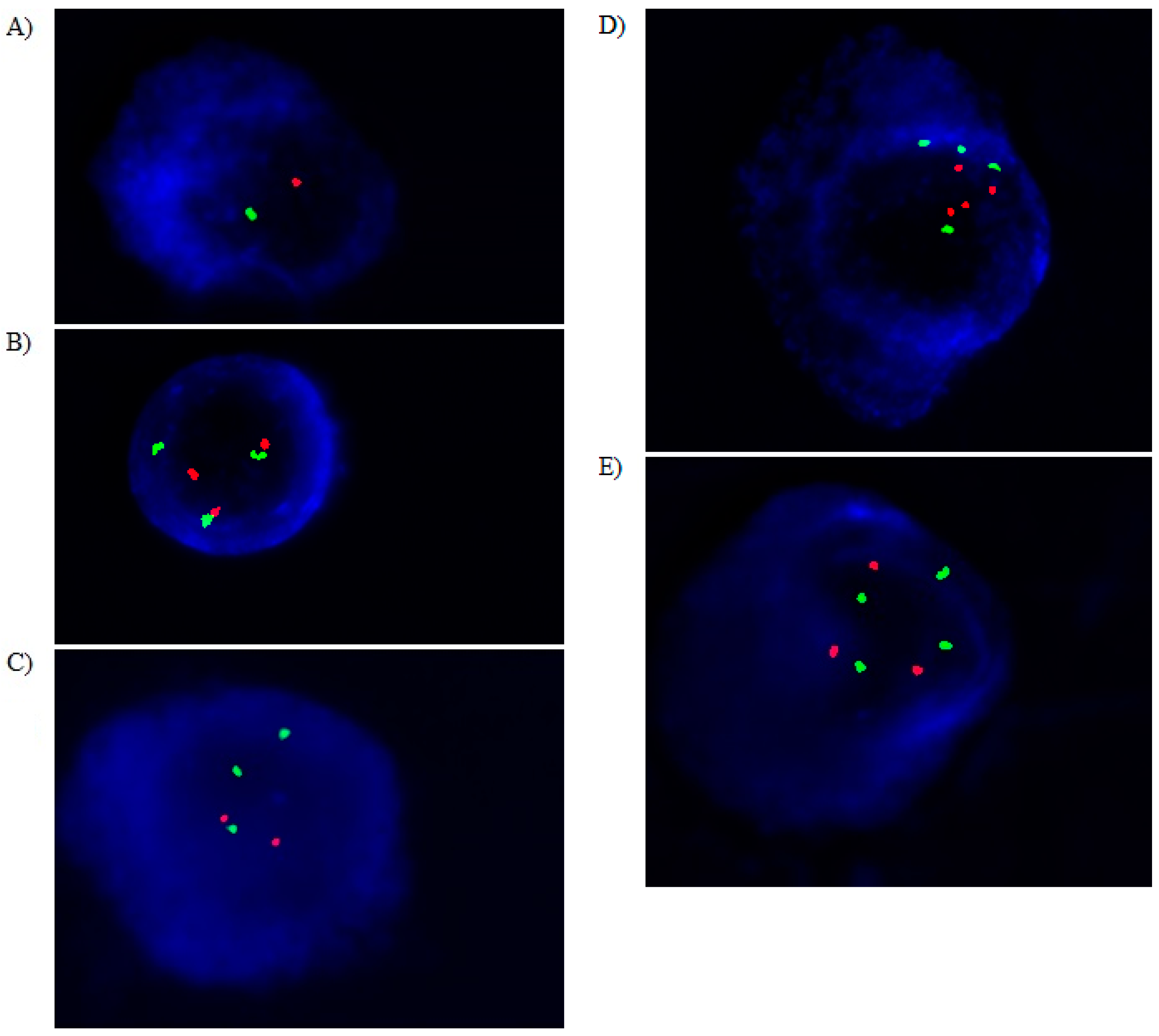
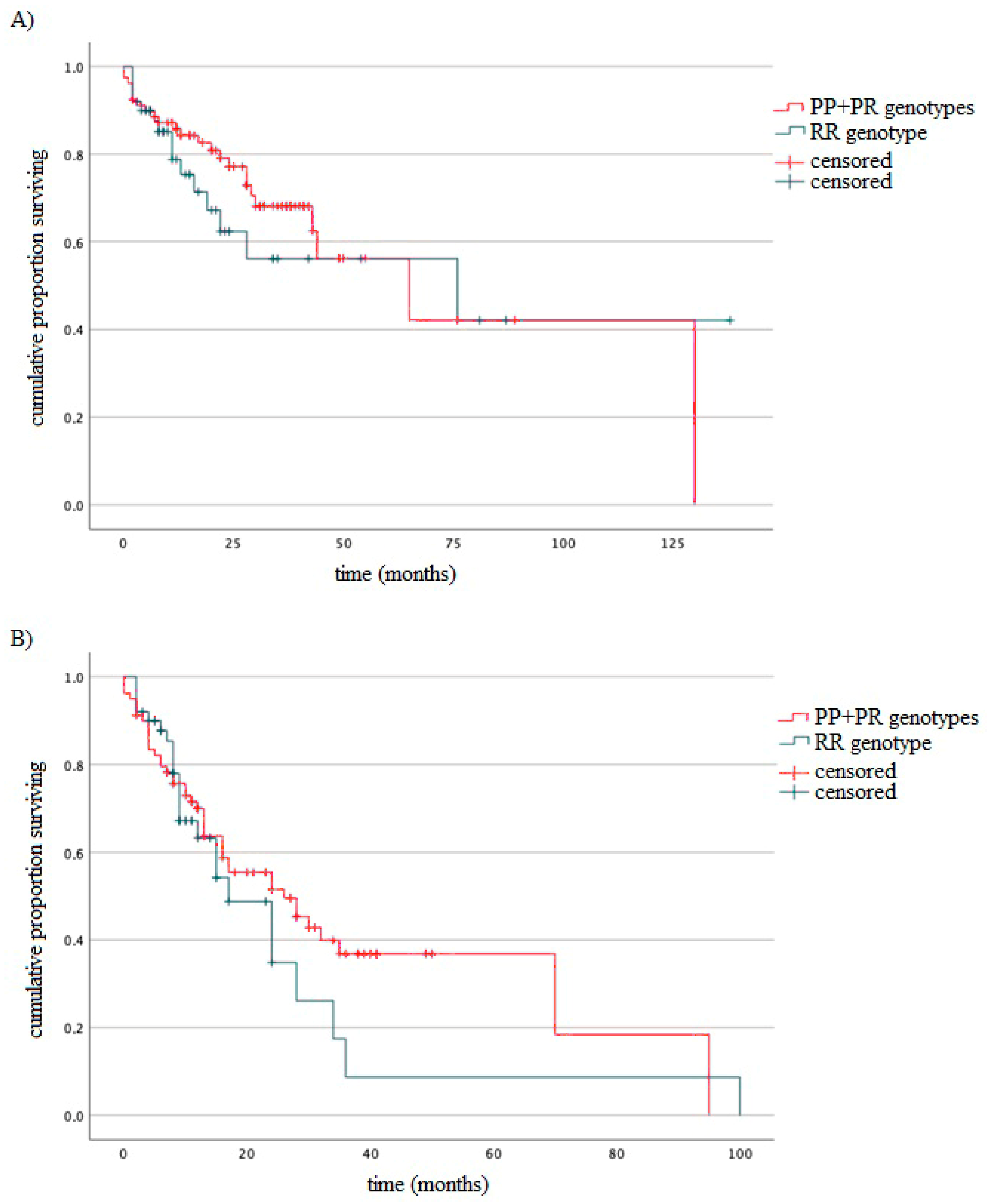
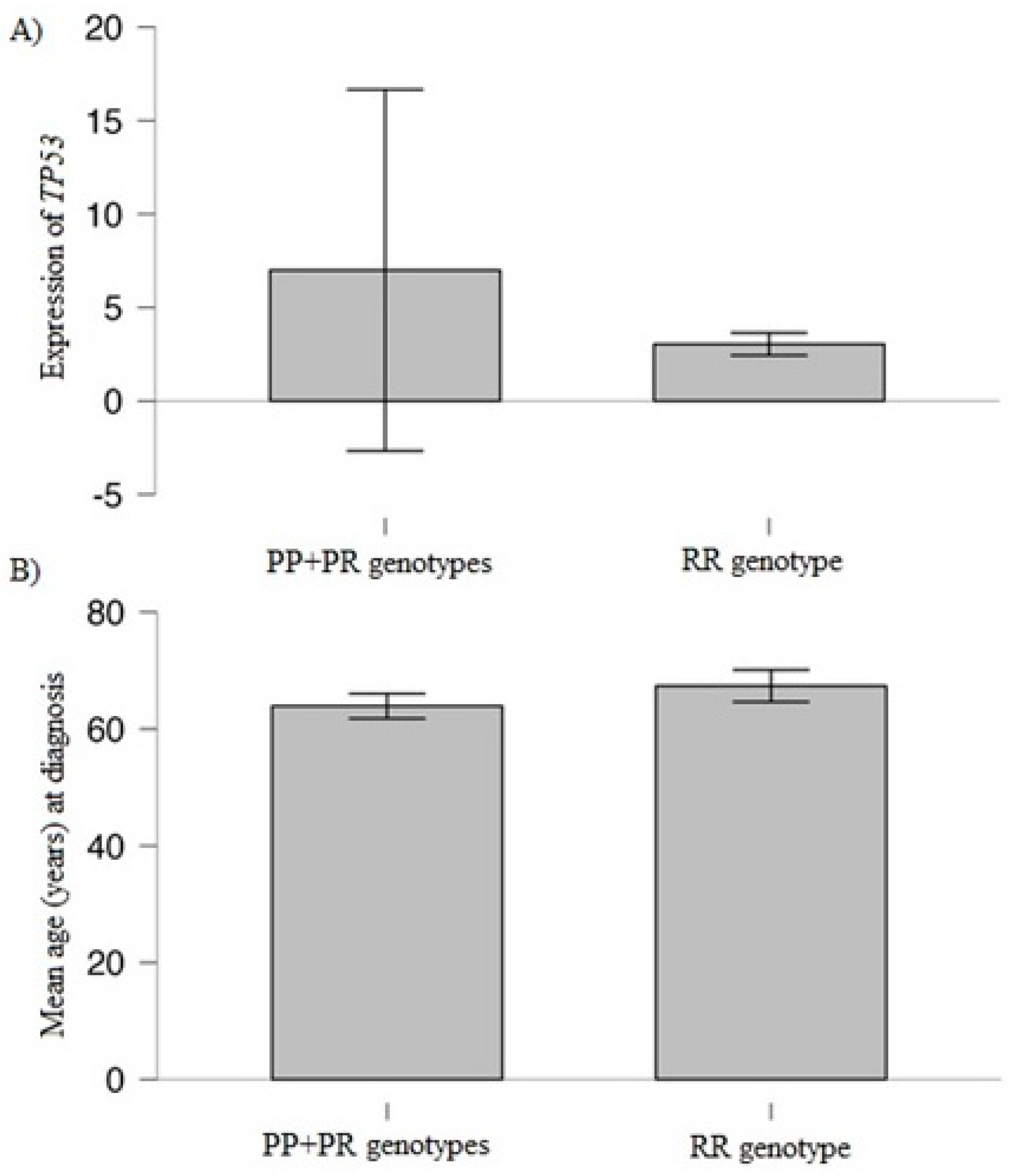
| Variables | MM Patients, n = 129 |
|---|---|
| Age | 65.24 (mean) |
| Sex | |
| Male | 68 |
| Female | 61 |
| Type of MM | |
| IgG | 72 |
| IgA | 28 |
| Light chain | 29 |
| Stage according to the International Staging System | |
| I | 32 |
| II | 37 |
| III | 60 |
| Smoking | |
| Yes | 20 |
| No: Non-smokers | 97 |
| No: Ex-smokers | 12 |
| Exposure to carcinogenic factors | |
| Yes | 26 |
| No | 103 |
| Additionally other type of cancer | |
| Yes | 8 |
| No | 121 |
| Renal failure | |
| Yes | 102 |
| No | 27 |
| The stage of chronic kidney disease (grade) | |
| G1 | 39 |
| G2 | 36 |
| G3A | 18 |
| G3B | 15 |
| G4 | 10 |
| G5 | 11 |
| Anemia grade before treatment (WHO) | |
| Absent | 30 |
| I—mild | 43 |
| II—moderate | 44 |
| III—severe | 12 |
| Structural cytogenetic changes | |
| del(17p13.1) | 15 |
| del(17p13.1) and t(4;16) | 7 |
| del(17p13.1) and t(14;16) | 1 |
| t(4;14) | 12 |
| t(14;16) | 3 |
| Chromosome 17 aneuploidies | |
| Present | 20 |
| Absent | 109 |
| Chemotherapy | |
| CTD 1 | 49 |
| VCD 2 and VD 3 | 43 |
| VTD 4 | 35 |
| Died before chemotherapy | 2 |
| Inclusion Criteria for MM Patients and Healthy Blood Donors | Exclusion Criteria for MM Patients |
|---|---|
|
|
| Additional inclusion criteria for MM patients | Exclusion criteria for healthy donors |
|
|
| Groups | Genotypes of TP53—p.P72R Variants | Total | HWE p Value and χ2 * | ||
|---|---|---|---|---|---|
| - | PP | PR | RR | - | - |
| Control | |||||
| E | 9.92 | 43.15 | 46.92 | 100 | p = 0.07 χ2 = 3.17 |
| O | 15 | 33 | 52 | 100 | |
| Case | |||||
| E | 14.33 | 57.33 | 57.33 | 129 | p = 0.03 χ2 = 4.68 |
| O | 7 | 72 | 50 | 129 | |
| Gene Variants and Alleles | MM, n (%) | Controls, n (%) | Odds Ratio | 95% Cl | p Values |
|---|---|---|---|---|---|
| RR | 50 (38.76%) | 52 (52%) | 1 | - | - |
| PR | 72 (55.81%) | 33 (33%) | 0.44 | 0.25–0.77 | 0.004 |
| PP | 7 (5.42%) | 15 (15%) | 2.06 | 0.77–5.47 | 0.14 |
| PP + PR | 19 (18.81%) | 14 (14%) | 0.70 | 0.32–1.56 | 0.39 |
| Total | 129 (100%) | 100 (100%) | |||
| R | 173 (67%) | 134 (67%) | 1 | - | - |
| P | 85 (33%) | 66 (33%) | 1 | 0.67–1.47 | 1.0 |
| Total | 258 (100%) | 200 (100%) | |||
| Variable | Univariate Cox Analysis for OS | Univariate Cox Analysis for PFS | ||||
|---|---|---|---|---|---|---|
| p Value | HR | 95% Cl | p Value | HR | 95% Cl | |
| ISS | ||||||
| I + II | - | R | - | R | - | |
| III | 0.002 | 2.08 | 1.30–3.32 | <0.001 | 2.22 | 1.26–2.43 |
| Auto-HSCT | ||||||
| Yes | <0.001 | 0.24 | 0.10–0.57 | <0.001 | 0.34 | 0.19–0.61 |
| No | - | R | - | - | R | - |
| TP53 P72R variant | ||||||
| RR | - | R | - | R | ||
| PR + PP | 0.49 | 1.26 | 0.65–2.43 | 0.37 | 1.27 | 0.75–2.14 |
| Variable | Multivariate Cox Analysis for OS | Multivariate Cox Analysis for PFS | ||||
|---|---|---|---|---|---|---|
| p Value | HR | 95% Cl | p Value | HR | 95% Cl | |
| ISS | ||||||
| I + II | - | R | - | R | - | |
| III | 0.04 | 1.66 | 1.02–2.70 | 0.03 | 1.46 | 1.03–2.05 |
| Auto-HSCT | ||||||
| Yes | 0.01 | 0.31 | 0.13–0.77 | 0.005 | 0.40 | 0.22–0.76 |
| No | - | R | - | - | R | - |
| TP53 P72R variant | ||||||
| RR | - | R | - | - | R | - |
| PR + PP | 0.51 | 1.25 | 0.65 | 0.24 | 1.38 | 0.81–2.35 |
| Variables | MM Patients | TP53 rs1042522 | ||
|---|---|---|---|---|
| PP + PR | RR | p-Value | ||
| Mean age (years) * | 65.24 | 63.91 | 67.36 | 0.04 |
| TP53 expression | 8.35 | 7.0 | 3.04 | p < 0.001 |
| Free light chain ratio * | 252.43 | 290.45 | 192.38 | 0.41 |
| % of plasma cells in bone marrow * | 32 | 28.30 | 36.74 | 0.02 |
| Albumins (g/dL) * | 3.62 | 3.64 | 3.56 | 0.78 |
| β2-microglobulin * (mg/L) | 6.81 | 6.82 | 6.77 | 0.74 |
| Calcium * (mM/L) | 2.46 | 2.51 | 2.43 | 0.43 |
| Hemoglobin * (g/dL) | 10.29 | 10.29 | 10.28 | 0.88 |
| Creatinine * (mg/dL) | 1.63 | 1.41 | 1.94 | 0.27 |
| Platelets (K/μL) | 214.71 | 223.92 | 200.16 | 0.27 |
| C-reactive protein * (mg/L) | 14.36 | 14.88 | 13.30 | 0.24 |
| Estimated glomerular filtration rate * mL/min/1.73 m2 | 67.67 | 70.72 | 62.97 | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popek-Marciniec, S.; Styk, W.; Wojcierowska-Litwin, M.; Chocholska, S.; Szudy-Szczyrek, A.; Samardakiewicz, M.; Swiderska-Kolacz, G.; Czerwik-Marcinkowska, J.; Zmorzynski, S. Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment. Cancers 2023, 15, 4747. https://doi.org/10.3390/cancers15194747
Popek-Marciniec S, Styk W, Wojcierowska-Litwin M, Chocholska S, Szudy-Szczyrek A, Samardakiewicz M, Swiderska-Kolacz G, Czerwik-Marcinkowska J, Zmorzynski S. Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment. Cancers. 2023; 15(19):4747. https://doi.org/10.3390/cancers15194747
Chicago/Turabian StylePopek-Marciniec, Sylwia, Wojciech Styk, Magdalena Wojcierowska-Litwin, Sylwia Chocholska, Aneta Szudy-Szczyrek, Marzena Samardakiewicz, Grazyna Swiderska-Kolacz, Joanna Czerwik-Marcinkowska, and Szymon Zmorzynski. 2023. "Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment" Cancers 15, no. 19: 4747. https://doi.org/10.3390/cancers15194747
APA StylePopek-Marciniec, S., Styk, W., Wojcierowska-Litwin, M., Chocholska, S., Szudy-Szczyrek, A., Samardakiewicz, M., Swiderska-Kolacz, G., Czerwik-Marcinkowska, J., & Zmorzynski, S. (2023). Association of Chromosome 17 Aneuploidy, TP53 Deletion, Expression and Its rs1042522 Variant with Multiple Myeloma Risk and Response to Thalidomide/Bortezomib Treatment. Cancers, 15(19), 4747. https://doi.org/10.3390/cancers15194747






