Ex Vivo Functional Assay for Evaluating Treatment Response in Tumor Tissue of Head and Neck Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Tumor Tissue
2.2. Tissue Slice Preparation and Culture
2.3. Staining Protocols
2.4. Scoring of RAD51 Foci (RECAP Assay)
2.5. TUNEL Assay
2.6. Image Acquisition and Analysis
3. Results
3.1. Optimization of Culture Medium for HNSCC
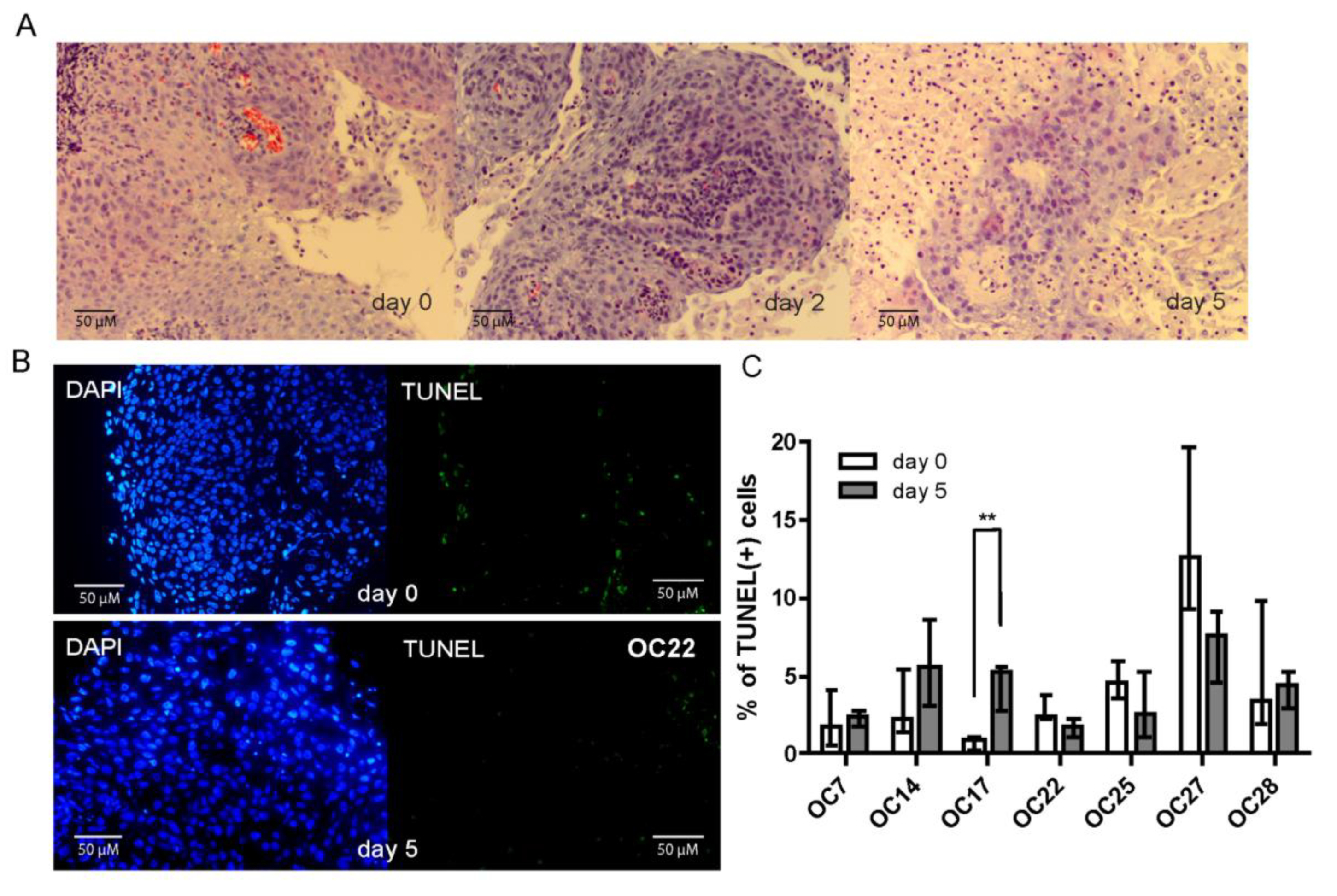
3.2. HNSCC Tumor Tissue Slices Remain Viable during Several Days of Culture
3.3. The Proliferative Capacity of HNSCC Tumor Cells Is Retained during Ex Vivo Culture
3.4. HNSCC Tumor Tissue Slices Display Heterogeneity in Response to IR
3.5. IR-Sensitive HNSCC Tumors Show More Unresolved DNA Damage
3.6. Irradiated HNSCC Tumor Cells form RAD51 Foci
3.7. HNSCC Tumor Tissue Slices Display Heterogeneity in Response to Cisplatin Treatment
4. Discussion
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved Survival of Patients With Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. Gynecol. Oncol. 2008, 100, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Lassen, P.; Lacas, B.; Pignon, J.-P.; Trotti, A.; Zackrisson, B.; Zhang, Q.; Overgaard, J.; Blanchard, P. Prognostic impact of HPV-associated p16-expression and smoking status on outcomes following radiotherapy for oropharyngeal cancer: The MARCH-HPV project. Radiother. Oncol. 2018, 126, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Alterio, D.; Marvaso, G.; Ferrari, A.; Volpe, S.; Orecchia, R.; Jereczek-Fossa, B.A. Modern radiotherapy for head and neck cancer. Semin. Oncol. 2019, 46, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Brook, I. Late side effects of radiation treatment for head and neck cancer. Radiat. Oncol. J. 2020, 38, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Ortigara, G.B.; Bonzanini, L.I.L.; Schulz, R.E.; Ferrazzo, K.L. Late radiation effects in survivors of head and neck cancer: State of the science. Crit. Rev. Oncol. 2021, 162, 103335. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Trotti, A.M.; Harris, J.; Eisbruch, A.; Harari, P.M.; Adelstein, D.J.; Jordan, R.C.K.; Zhao, W.; Sturgis, E.M.; Burtness, B.; et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): A randomised, multicentre, non-inferiority trial. Lancet 2018, 393, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Mehanna, H.; Robinson, M.; Hartley, A.; Kong, A.; Foran, B.; Fulton-Lieuw, T.; Dalby, M.; Mistry, P.; Sen, M.; O’Toole, L. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): An open-label randomised controlled phase 3 trial. Lancet 2019, 393, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henk, J. Controlled trials of synchronous chemotherapy with radiotherapy in head and neck cancer: Overview of radiation morbidity. Clin. Oncol. 1997, 9, 308–312. [Google Scholar] [CrossRef]
- Pignon, J.-P.; le Maître, A.; Maillard, E.; Bourhis, J.; MACH-NC Collaborative Group. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): An update on 93 randomised trials and 17,346 patients. Radiother. Oncol. 2009, 92, 4–14. [Google Scholar] [CrossRef]
- Adelstein, D.J.; Li, Y.; Adams, G.L.; Wagner, H., Jr.; Kish, J.A.; Ensley, J.F.; Schuller, D.E.; Forastiere, A.A. An Intergroup Phase III Comparison of Standard Radiation Therapy and Two Schedules of Concurrent Chemoradiotherapy in Patients With Unresectable Squamous Cell Head and Neck Cancer. J. Clin. Oncol. 2003, 21, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotti, A.; Bellm, L.A.; Epstein, J.B.; Frame, D.; Fuchs, H.J.; Gwede, C.K.; Komaroff, E.; Nalysnyk, L.; Zilberberg, M.D. Mucositis incidence, severity and associated outcomes in patients with head and neck cancer receiving radiotherapy with or without chemotherapy: A systematic literature review. Radiother. Oncol. 2003, 66, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, C.; Eriksen, J.; Jensen, K.; Hansen, C.; Sørensen, B.; Lassen, P.; Alsner, J.; Schack, L.; Overgaard, J.; Grau, C. IMRT—Biomarkers for dose escalation, dose de-escalation and personalized medicine in radiotherapy for head and neck cancer. Oral Oncol. 2018, 86, 91–99. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Molinolo, A.A.; Amornphimoltham, P.; Squarize, C.; Castilho, R.M.; Patel, V.; Gutkind, J.S. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009, 45, 324–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and Comparative Genomic Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas. Clin. Cancer Res. 2015, 21, 632–641. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.; Guthrie, V.; Masica, D.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar]
- Budach, V.; Tinhofer, I. Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: A systematic review. Lancet Oncol. 2019, 20, e313–e326. [Google Scholar] [CrossRef]
- Chia, S.; Low, J.-L.; Zhang, X.; Kwang, X.-L.; Chong, F.-T.; Sharma, A.; Bertrand, D.; Toh, S.Y.; Leong, H.-S.; Thangavelu, M.T.; et al. Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Affolter, A.; Lammert, A.; Kern, J.; Scherl, C.; Rotter, N. Precision Medicine Gains Momentum: Novel 3D Models and Stem Cell-Based Approaches in Head and Neck Cancer. Front. Cell Dev. Biol. 2021, 9, 666515. [Google Scholar] [CrossRef]
- Demers, I.; Donkers, J.; Kremer, B.; Speel, E.J. Ex Vivo Culture Models to Indicate Therapy Response in Head and Neck Squamous Cell Carcinoma. Cells 2020, 9, 2527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; van Weerden, W.M.; de Ridder, C.M.A.; Erkens-Schulze, S.; Schönfeld, E.; Meijer, T.G.; Kanaar, R.; van Gent, D.C.; Nonnekens, J. Ex vivo treatment of prostate tumor tissue recapitulates in vivo therapy response. Prostate 2018, 79, 390–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijer, T.G.; Verkaik, N.S.; Sieuwerts, A.M.; van Riet, J.; Naipal, K.A.; van Deurzen, C.H.; Bakker, M.A.D.; Sleddens, H.F.; Dubbink, H.-J.; Toom, T.D.D.; et al. Functional Ex Vivo Assay Reveals Homologous Recombination Deficiency in Breast Cancer Beyond BRCA Gene Defects. Clin. Cancer Res. 2018, 24, 6277–6287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, S.; Moro, C.F.; Del Chiaro, M.; Pouso, S.; Sebestyén, A.; Löhr, M.; Björnstedt, M.; Verbeke, C.S. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naipal, K.A.; Verkaik, N.S.; Ameziane, N.; van Deurzen, C.H.; ter Brugge, P.; Meijers, M.; Sieuwerts, A.M.; Martens, J.W.; O’Connor, M.J.; Vrieling, H.; et al. Functional Ex Vivo Assay to Select Homologous Recombination–Deficient Breast Tumors for PARP Inhibitor Treatment. Clin. Cancer Res. 2014, 20, 4816–4826. [Google Scholar] [CrossRef] [Green Version]
- Kishan, A.T.N.; Nicole, S.V.; Humberto, S.; van Deurzen Carolien, H.M.; den Bakker Michael, A.; Jan, H.J.H.; Roland, K.; Vreeswijk Maaike, P.G.; Agnes, J.; van Gent, D.C. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 2016, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Van Den Tempel, N.; Naipal, K.A.T.; Raams, A.; Van Gent, D.C.; Franckena, M.; Boormans, J.L.; Kanaar, R. Ex vivo assays to predict enhanced chemosensitization by hyperthermia in urothelial cancer of the bladder. PLoS ONE 2018, 13, e0209101. [Google Scholar] [CrossRef] [Green Version]
- Merz, F.; Gaunitz, F.; Dehghani, F.; Renner, C.; Meixensberger, J.; Gutenberg, A.; Giese, A.; Schopow, K.; Hellwig, C.; Schäfer, M.; et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro-Oncology 2013, 15, 670–681. [Google Scholar] [CrossRef]
- Lauwers, I.; Pachler, K.; Capala, M.; Sijtsema, N.; Hoogeman, M.; Verduijn, G.; Petit, S. MO-0144 Deep learning to detect nuclei and DNA damage foci for ex vivo tissue radiation sensitivity analysis. Radiother. Oncol. 2022, 170, S119–S120. [Google Scholar] [CrossRef]
- Edge, S.; Byrd, D.; Compton, C.; Fritz, A.; Greene, F.; Trotti, A. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Mena, M.; Wang, X.; Tous, S.; Quiros, B.; Clavero, O.; Alejo, M.; Morey, F.; Taberna, M.; Leon Vintro, X.; Lloveras Rubio, B.; et al. Concordance of p16INK4a and E6*I mRNA among HPV-DNA-Positive Oropharyngeal, Laryngeal, and Oral Cavity Carcinomas from the ICO International Study. Cancers 2022, 14, 3787. [Google Scholar]
- Sharkey Ochoa, I.; O’Regan, E.; Toner, M.; Kay, E.; Faul, P.; O’Keane, C.; O’Connor, R.; Mullen, D.; Nur, M.; O’Murchu, E.; et al. The Role of HPV in Determining Treatment, Survival, and Prognosis of Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 4321. [Google Scholar] [CrossRef] [PubMed]
- Van Oijen, M.G.; Medema, R.H.; Slootweg, P.J.; Rijksen, G. Positivity of the Proliferation Marker Ki-67 in Noncycling Cells. Am. J. Clin. Pathol. 1998, 110, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Agami, R.; Bernards, R. Distinct Initiation and Maintenance Mechanisms Cooperate to Induce G1 Cell Cycle Arrest in Response to DNA Damage. Cell 2000, 102, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.H.; Hales, C.N.; Ozanne, S.E. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007, 35, 7417–7428. [Google Scholar] [CrossRef]
- Fitsiou, E.; Soto-Gamez, A.; Demaria, M. Biological functions of therapy-induced senescence in cancer. Semin. Cancer Biol. 2022, 81, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Piskorz, W.M.; Cechowska-Pasko, M. Senescence of Tumor Cells in Anticancer Therapy—Beneficial and Detrimental Effects. Int. J. Mol. Sci. 2022, 23, 11082. [Google Scholar] [CrossRef]
- De Zio, D.; Cianfanelli, V.; Cecconi, F. New Insights into the Link Between DNA Damage and Apoptosis. Antioxidants Redox Signal. 2013, 19, 559–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013, 332, 237–248. [Google Scholar] [CrossRef]
- Wang, J.Y.J. DNA damage and apoptosis. Cell Death Differ. 2001, 8, 1047–1048. [Google Scholar] [CrossRef] [Green Version]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Black, S.J.; Kashkina, E.; Kent, T.; Pomerantz, R.T. DNA Polymerase θ: A Unique Multifunctional End-Joining Machine. Genes 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Tribius, S.; Reemts, E.; Prosch, C.; Raguse, M.; Petersen, C.; Kruell, A.; Singer, S.; Bergelt, C. Global quality of life during the acute toxicity phase of multimodality treatment for patients with head and neck cancer: Can we identify patients most at risk of profound quality of life decline? Oral Oncol. 2012, 48, 898–904. [Google Scholar]
- Ladan, M.M.; Meijer, T.G.; Verkaik, N.S.; Komar, Z.M.; van Deurzen, C.H.M.; Bakker, M.A.D.; Kanaar, R.; van Gent, D.C.; Jager, A. Functional Ex Vivo Tissue-Based Chemotherapy Sensitivity Testing for Breast Cancer. Cancers 2022, 14, 1252. [Google Scholar] [CrossRef]
- Suckert, T.; Rassamegevanon, T.; Müller, J.; Dietrich, A.; Graja, A.; Reiche, M.; Löck, S.; Krause, M.; Beyreuther, E.; Von Neubeck, C. Applying Tissue Slice Culture in Cancer Research—Insights from Preclinical Proton Radiotherapy. Cancers 2020, 12, 1589. [Google Scholar] [CrossRef]
- Meneceur, S.; Löck, S.; Gudziol, V.; Hering, S.; Bütof, R.; Rehm, M.; Baumann, M.; Krause, M.; von Neubeck, C. Residual gammaH2AX foci in head and neck squamous cell carcinomas as predictors for tumour radiosensitivity: Evaluation in pre-clinical xenograft models and clinical specimens. Radiother. Oncol. 2019, 137, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Rassamegevanon, T.; Löck, S.; Baumann, M.; Krause, M.; von Neubeck, C. Comparable radiation response of ex vivo and in vivo irradiated tumor samples determined by residual γH2AX. Radiother. Oncol. 2019, 139, 94–100. [Google Scholar] [CrossRef]
- Zech, H.B.; Berger, J.; Mansour, W.Y.; Nordquist, L.; von Bargen, C.M.; Bußmann, L.; Oetting, A.; Christiansen, S.; Möckelmann, N.; Böttcher, A.; et al. Patient derived ex vivo tissue slice cultures demonstrate a profound DNA double-strand break repair defect in HPV-positive oropharyngeal head and neck cancer. Radiother. Oncol. 2022, 168, 138–146. [Google Scholar] [CrossRef]





| Pat. No. | Age | Sex | Subsite | cTNM Ed. 7 | ECS | Smoking Status | Pack Years | Alcohol Status | Treatment | Follow-Up Time (Month)/Recurrence | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | N | M | ||||||||||
| OC14 | 74 | F | B.o.t./tongue | 4a | 2c | 0 | no | smoker | 40 | moderate | OP +RT | 44/no |
| OC17 | 76 | M | tongue | 3 | 0 | 0 | no | ex-smoker | 25 | abuse | OP | 24/no (other cause of death) |
| OC20 | 64 | F | tongue | 3 | 2b | 0 | yes | ex-smoker | 50 | no | OP + RT | 5/regional and lung metastasis |
| OC21 | 69 | F | retromolar trigone | 4a | 1 | 0 | yes | ex-smoker | 7 | light | OP + RT | 5/lung metastasis |
| OC22 | 66 | M | f.o.m. | 2 | 2c | 0 | yes | ex-smoker | 36 | moderate | OP + CRT | 25/no (other cause of death) |
| OC23 | 48 | M | f.o.m./tongue | 3 | 2b | 0 | yes | smoker | unknown | abuse | OP + CRT | 34/no |
| OC25 | 68 | M | cheek | 2 | 0 | 0 | no | smoker | 30 | abuse | OP | 27/local |
| OC27 | 75 | M | tongue | 3 | 2b | 0 | no | ex-smoker | unknown | moderate | OP + RT | 30/no (other cause of death) |
| OC28 | 73 | F | f.o.m. | 2 | 1 | 0 | no | smoker | unknown | light | OP | <1/no (other cause of death) |
| OC30 | 43 | M | tongue | 2 | 0 | 0 | no | smoker | unknown | abuse | OP | 29/no |
| OC32 | 76 | M | tongue/f.o.m. | 3 | 0 | 0 | no | non-smoker | 0 | light | OP + RT | 3/regional |
| OC33 | 71 | M | f.o.m. | 4a | 0 | 0 | no | smoker | 19 | heavy | OP + RT | 23/no |
| OC42 | 71 | M | tongue | 4a | 0 | 0 | no | ex-smoker | unknown | light | OP + RT | 6/local and regional |
| OC43 | 78 | M | tongue | 3 | 0 | 0 | no | ex-smoker | 30 | no | OP + RT | 2/no (other cause of death) |
| OC46 | 67 | M | tongue | 2 | 0 | 0 | no | non-smoker | 0 | ex-heavy | OP | 13/no |
| OC48 | 76 | M | alveolar ridge maxilla | 4a | 3b | 0 | yes | ex-smoker | unknown | light | OP + RT | 10/no |
| OC51 | 48 | F | tongue | 3 | 3b | 0 | yes | non-smoker | 0 | light | OP + CRT | 10/no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capala, M.E.; Pachler, K.S.; Lauwers, I.; de Korte, M.A.; Verkaik, N.S.; Mast, H.; Jonker, B.P.; Sewnaik, A.; Hardillo, J.A.; Keereweer, S.; et al. Ex Vivo Functional Assay for Evaluating Treatment Response in Tumor Tissue of Head and Neck Squamous Cell Carcinoma. Cancers 2023, 15, 478. https://doi.org/10.3390/cancers15020478
Capala ME, Pachler KS, Lauwers I, de Korte MA, Verkaik NS, Mast H, Jonker BP, Sewnaik A, Hardillo JA, Keereweer S, et al. Ex Vivo Functional Assay for Evaluating Treatment Response in Tumor Tissue of Head and Neck Squamous Cell Carcinoma. Cancers. 2023; 15(2):478. https://doi.org/10.3390/cancers15020478
Chicago/Turabian StyleCapala, Marta E., Katrin S. Pachler, Iris Lauwers, Maarten A. de Korte, Nicole S. Verkaik, Hetty Mast, Brend P. Jonker, Aniel Sewnaik, Jose A. Hardillo, Stijn Keereweer, and et al. 2023. "Ex Vivo Functional Assay for Evaluating Treatment Response in Tumor Tissue of Head and Neck Squamous Cell Carcinoma" Cancers 15, no. 2: 478. https://doi.org/10.3390/cancers15020478





