The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond
Abstract
:Simple Summary
Abstract
1. Introduction
2. ALDH1A3 Is a Member of the ALDH Superfamily
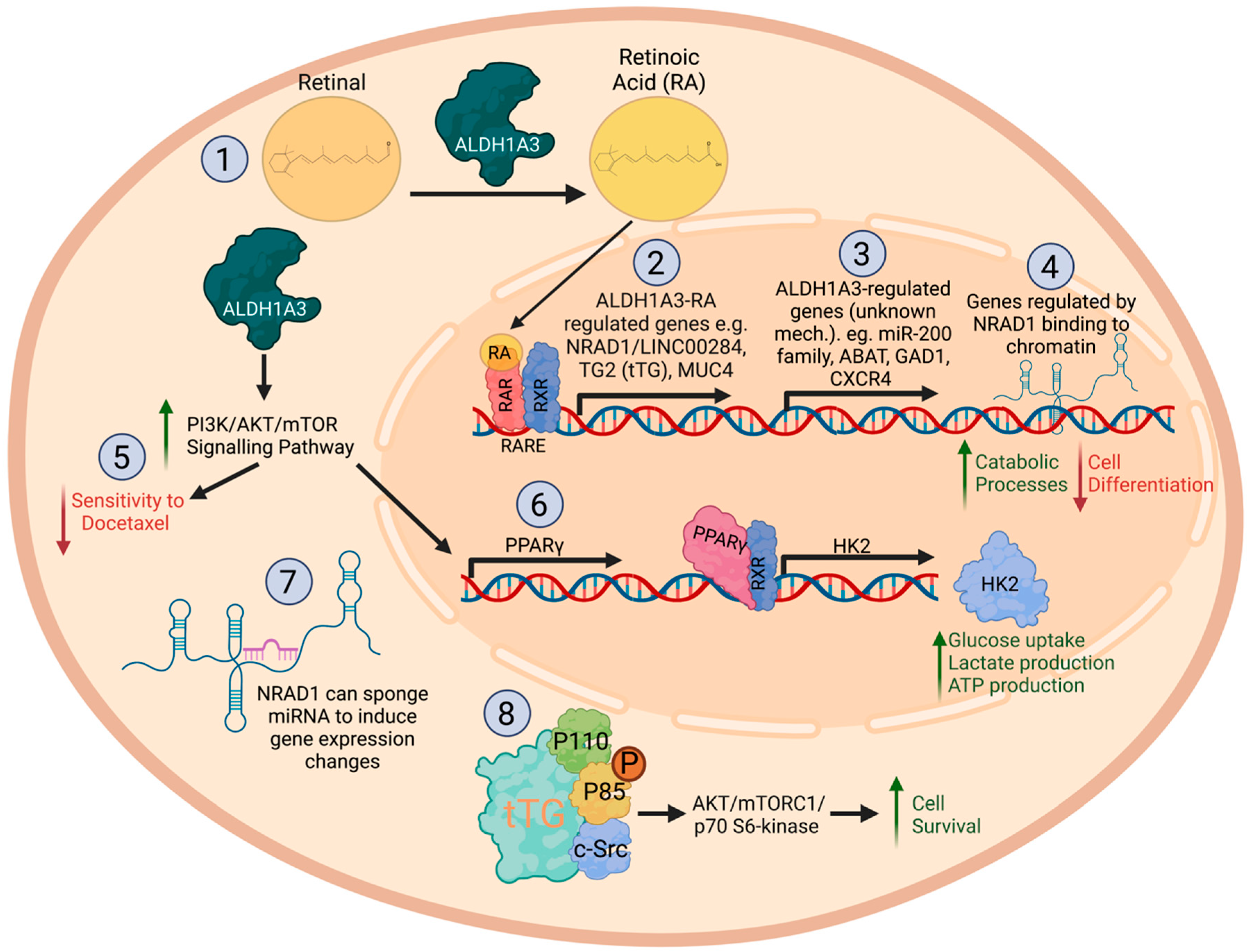
3. Retinoic Acid Signaling Is a Key Function of ALDH1A Enzymes
4. ALDH1A3 Is a Cancer Stem Cell Marker
5. ALDH1A3 Is Associated with Worse Prognosis in Cancer
6. ALDH1A3 Promotes Tumor Progression
7. The Role of ALDH1A3 Multiple Drug Resistance
8. Regulation of ALDH1A3 in Cancer

9. Mechanisms of ALDH1A3 in Cancer: Effects on Gene Expression
10. Effects of ALDH1A3 on Glycometabolism and Other Metabolic Pathways in Cancer
11. Role of ALDH1A3 in Type 2 Diabetes
12. The Role of ALDH1A3 in Cardiac Function and Pathology
13. Targeting CSCs and ALDH1A3
14. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marcato, P.; Dean, C.A.; Da, P.; Araslanova, R.; Gillis, M.; Joshi, M.; Helyer, L.; Pan, L.; Leidal, A.; Gujar, S.; et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells Is Primarily Due to Isoform ALDH1A3 and Its Expression Is Predictive of Metastasis. Stem Cells 2011, 29, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.C.; Chang, W.C.; Hiraoka, L.; Hsieh, C.L. Molecular Cloning, Genomic Organization, and Chromosomal Localization of an Additional Human Aldehyde Dehydrogenase Gene, ALDH6. Genomics 1994, 24, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Koppaka, V.; Thompson, D.C.; Chen, Y.; Ellermann, M.; Nicolaou, K.C.; Juvonen, R.O.; Petersen, D.; Deitrich, R.A.; Hurley, T.D.; Vasiliou Dr., V. Aldehyde Dehydrogenase Inhibitors: A Comprehensive Review of the Pharmacology, Mechanism of Action, Substrate Specificity, and Clinical Application. Pharmacol. Rev. 2012, 64, 520–539. [Google Scholar] [CrossRef] [Green Version]
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 Aldehyde Oxidizing Enzymes: The Aldehyde Dehydrogenase Superfamily. Expert Opin. Drug Metab. Toxicol. 2008, 4, 697–720. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wagner, E.; McCaffery, P.; Smith, D.; Andreadis, A.; Dräger, U.C. A Retinoic Acid Synthesizing Enzyme in Ventral Retina and Telencephalon of the Embryonic Mouse. Mech. Dev. 2000, 95, 283–289. [Google Scholar] [CrossRef]
- Mic, F.A.; Molotkov, A.; Fan, X.; Cuenca, A.E.; Duester, G. RALDH3, a Retinaldehyde Dehydrogenase That Generates Retinoic Acid, Is Expressed in the Ventral Retina, Otic Vesicle and Olfactory Pit during Mouse Development. Mech. Dev. 2000, 97, 227–230. [Google Scholar] [CrossRef]
- Yahyavi, M.; Abouzeid, H.; Gawdat, G.; de Preux, A.S.; Xiao, T.; Bardakjian, T.; Schneider, A.; Choi, A.; Jorgenson, E.; Baier, H.; et al. ALDH1A3 Loss of Function Causes Bilateral Anophthalmia/Microphthalmia and Hypoplasia of the Optic Nerve and Optic Chiasm. Hum. Mol. Genet. 2013, 22, 3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molotkova, N.; Molotkov, A.; Duester, G. Role of Retinoic Acid during Forebrain Development Begins Late When Raldh3 Generates Retinoic Acid in the Ventral Subventricular Zone. Dev. Biol. 2007, 303, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sima, A.; Parisotto, M.; Mader, S.; Bhat, P.V. Kinetic Characterization of Recombinant Mouse Retinal Dehydrogenase Types 3 and 4 for Retinal Substrates. Biochim. Biophys. Acta 2009, 1790, 1660–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; McCaffery, P.; Ivins, K.J.; Neve, R.L.; Hogan, P.; Chin, W.W.; Dräger, U.C. Molecular Identification of a Major Retinoic-Acid-Synthesizing Enzyme, a Retinaldehyde-Specific Dehydrogenase. J. Biol. Inorg. Chem. 1996, 240, 15–22. [Google Scholar] [CrossRef]
- Black, W.; Vasiliou, V. The Aldehyde Dehydrogenase Gene Superfamily Resource Center. Hum. Genom. 2009, 4, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyle, K.M.; Sultan, M.; Thomas, M.L.; Vaghar-Kashani, A.; Marcato, P.; Mohammad Sultan, K.M.C. Retinoid Signaling in Cancer and Its Promise for Therapy. J. Carcinog. Mutagen. 2013, 7, 16–18. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, T.J.; Duester, G. Mechanisms of Retinoic Acid Signalling and Its Roles in Organ and Limb Development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [Green Version]
- Lalevee, S.; Anno, Y.N.; Chatagnon, A.; Samarut, E.; Poch, O.; Laudet, V.; Benoit, G.; Lecompte, O.; Rochette-Egly, C. Genome-Wide in Silico Identification of New Conserved and Functional Retinoic Acid Receptor Response Elements (Direct Repeats Separated by 5 Bp). J. Biol. Chem. 2011, 286, 33322–33334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arteaga, M.F.; Mikesch, J.-H.; Fung, T.-K.; So, C.W.E. Epigenetics in Acute Promyelocytic Leukaemia Pathogenesis and Treatment Response: A TRAnsition to Targeted Therapies. Br. J. Cancer 2014, 112, 413–418. [Google Scholar] [CrossRef]
- Yoshida, H.; Kitamura, K.; Tanaka, K.; Omura, S.; Miyazaki, T.; Hachiya, T.; Ohno, R.; Naoe, T. Accelerated Degradation of PML-Retinoic Acid Receptor Alpha (PML-RARA) Oncoprotein by All-Trans-Retinoic Acid in Acute Promyelocytic Leukemia: Possible Role of the Proteasome Pathway. Cancer Res. 1996, 56, 2945–2948. [Google Scholar]
- Ablain, J.; de Thé, H. Retinoic Acid Signaling in Cancer: The Parable of Acute Promyelocytic Leukemia. Int. J. Cancer 2014, 135, 2262–2272. [Google Scholar] [CrossRef]
- Chen, F.; Shao, F.; Hinds, A.; Yao, S.; Ram-Mohan, S.; Norman, T.A.; Krishnan, R.; Fine, A. Retinoic Acid Signaling Is Essential for Airway Smooth Muscle Homeostasis. J. Clin. Investig. 2018, 3, e120398. [Google Scholar] [CrossRef] [Green Version]
- Defnet, A.E.; Shah, S.D.; Huang, W.; Shapiro, P.; Deshpande, D.A.; Kane, M.A. Dysregulated Retinoic Acid Signaling in Airway Smooth Muscle Cells in Asthma. FASEB J. 2021, 35, e22016. [Google Scholar] [CrossRef]
- Chen, F.; Marquez, H.; Kim, Y.K.; Qian, J.; Shao, F.; Fine, A.; Cruikshank, W.W.; Quadro, L.; Cardoso, W.V. Prenatal Retinoid Deficiency Leads to Airway Hyperresponsiveness in Adult Mice. J. Clin. Investg. 2014, 124, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Clagett-Dame, M.; Knutson, D. Vitamin A in Reproduction and Development. Nutrients 2011, 3, 385–428. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zan, J. The Identification of Childhood Asthma Progression-Related LncRNAs and MRNAs Suitable as Biomarkers Using Weighted Gene Coexpression Network Analysis. Genet. Res. 2021, 2021, 5511507. [Google Scholar] [CrossRef]
- Murar, M.; Vaidya, A. Cancer Stem Cell Markers: Premises and Prospects. Biomarkers Med. 2015, 9, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W.K. Aldehyde Dehydrogenase Its Role as a Cancer Stem Cell Marker Comes down to the Specific Isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, A.M.; Wan, T.S.; Leung, J.C.; Chan, L.Y.; Huang, H.; Kwong, Y.L.; Liang, R.; Leung, A.Y. Aldehyde Dehydrogenase Activity in Leukemic Blasts Defines a Subgroup of Acute Myeloid Leukemia with Adverse Prognosis and Superior NOD/SCID Engrafting Potential. Leukemia 2007, 21, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, K.W.; Lee, T.K.; Tang, K.H.; Wo, J.Y.; Zheng, B.J.; Guan, X.Y. Aldehyde Dehydrogenase Discriminates the CD133 Liver Cancer Stem Cell Populations. Mol. Cancer Res. 2008, 6, 1146–1153. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.C.; Chen, Y.W.; Hsu, H.S.; Tseng, L.M.; Huang, P.I.; Lu, K.H.; Chen, D.T.; Tai, L.K.; Yung, M.C.; Chang, S.C.; et al. Aldehyde Dehydrogenase 1 Is a Putative Marker for Cancer Stem Cells in Head and Neck Squamous Cancer. Biochem. Biophys. Res. Commun. 2009, 385, 307–313. [Google Scholar] [CrossRef]
- Jiang, F.; Qiu, Q.; Khanna, A.; Todd, N.W.; Deepak, J.; Xing, L.; Wang, H.; Liu, Z.; Su, Y.; Stass, S.A.; et al. Aldehyde Dehydrogenase 1 Is a Tumor Stem Cell-Associated Marker in Lung Cancer. Mol. Cancer Res. 2009, 7, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.P.; Fleming, J.B.; Wang, H.; Abbruzzese, J.L.; Choi, W.; Kopetz, S.; McConkey, D.J.; Evans, D.B.; Gallick, G.E. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PLoS ONE 2011, 6, e20636. [Google Scholar] [CrossRef] [Green Version]
- Rao, Q.X.; Yao, T.T.; Zhang, B.Z.; Lin, R.C.; Chen, Z.L.; Zhou, H.; Wang, L.J.; Lu, H.W.; Chen, Q.; Di, N.; et al. Expression and Functional Role of ALDH1 in Cervical Carcinoma Cells. Asian Pac. J. Cancer Prev. 2012, 13, 1325–1331. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Iovino, F.; Eterno, V.; Cammareri, P.; Gambara, G.; Espina, V.; Gulotta, G.; Dieli, F.; Giordano, S.; De, M.R.; et al. Tumorigenic and Metastatic Activity of Human Thyroid Cancer Stem Cells. Cancer Res. 2010, 70, 8874–8885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Hoogen, C.; van der Horst, G.; Cheung, H.; Buijs, J.T.; Lippitt, J.M.; Guzman-Ramirez, N.; Hamdy, F.C.; Eaton, C.L.; Thalmann, G.N.; Cecchini, M.G.; et al. High Aldehyde Dehydrogenase Activity Identifies Tumor-Initiating and Metastasis-Initiating Cells in Human Prostate Cancer. Cancer Res. 2010, 70, 5163–5173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde Dehydrogenase 1 Is a Marker for Normal and Malignant Human Colonic Stem Cells (SC) and Tracks SC Overpopulation during Colon Tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Qiu, Q.; Zhang, X.; Jiang, Z.; Leng, Q.; Liu, Z.; Stass, S.A.; Jiang, F. Aldehyde Dehydrogenase 1 A1-Positive Cell Population Is Enriched in Tumor-Initiating Cells and Associated with Progression of Bladder Cancer. Cancer Epidemiol. Biomark. Prev. 2010, 19, 327–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of Primitive Human Hematopoietic Progenitors on the Basis of Aldehyde Dehydrogenase Activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Dallaglio, K.; Chen, Y.; Robinson, W.A.; Robinson, S.E.; McCarter, M.D.; Wang, J.; Gonzalez, R.; Thompson, D.C.; Norris, D.A.; et al. ALDH1A Isozymes Are Markers of Human Melanoma Stem Cells and Potential Therapeutic Targets. Stem Cells 2012, 30, 2100–2113. [Google Scholar] [CrossRef] [Green Version]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal Glioma Stem Cells Are Maintained by Activated Glycolytic Metabolism Involving Aldehyde Dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA 2013, 110, 8644–8649. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Sullivan, J.P.; Girard, L.; Augustyn, A.; Yenerall, P.; Rodriguez-Canales, J.; Liu, H.; Behrens, C.; Shay, J.W.; Wistuba, I.I.; et al. Essential Role of Aldehyde Dehydrogenase 1A3 for the Maintenance of Non-Small Cell Lung Cancer Stem Cells Is Associated with the STAT3 Pathway. Clin. Cancer Res. 2014, 20, 4154–4166. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Liu, Y.Y.Y.; Bian, X.; Zhou, F.; Liu, Y.Y.Y. ALDH1A3 Affects Colon Cancer in Vitro Proliferation and Invasion Depending on CXCR4 Status. Br. J. Cancer 2017, 118, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Weng, J.-J.; Cheng, C.-T.; Wu, R.-C.; Huang, S.-C.; Wu, C.-E.; Chung, Y.-H.; Liu, C.-Y.; Chang, M.-H.; Chiang, K.-C.; et al. ALDH1A3, the Major Aldehyde Dehydrogenase Isoform in Human Cholangiocarcinoma Cells, Affects Prognosis and Gemcitabine Resistance in Cholangiocarcinoma Patients. Clin. Cancer Res. 2016, 22, 4225–4235. [Google Scholar] [CrossRef] [PubMed]
- Kurth, I.; Hein, L.; Mäbert, K.; Peitzsch, C.; Koi, L.; Cojoc, M.; Kunz-Schughart, L.; Baumann, M.; Dubrovska, A.; Kurth, I.; et al. Cancer Stem Cell Related Markers of Radioresistance in Head and Neck Squamous Cell Carcinoma. Oncotarget 2015, 6, 34494–34509. [Google Scholar] [CrossRef] [Green Version]
- Lang, T.; Xu, J.; Zhou, L.; Zhang, Z.; Ma, X.; Gu, J.; Liu, J.; Li, Y.; Ding, D.; Qiu, J. Disruption of KDM4C-ALDH1A3 Feed-Forward Loop Inhibits Stemness, Tumorigenesis and Chemoresistance of Gastric Cancer Stem Cells. Signal Transduct. Target. Ther. 2021, 6, 336. [Google Scholar] [CrossRef] [PubMed]
- Greve, B.; Kelsch, R.; Spaniol, K.; Eich, H.T.; Götte, M. Flow Cytometry in Cancer Stem Cell Analysis and Separation. Cytom. A 2012, 81, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Marcato, P.; Dean, C.A.; Liu, R.-Z.; Coyle, K.M.; Bydoun, M.; Wallace, M.; Clements, D.; Turner, C.; Mathenge, E.G.; Gujar, S.A.; et al. Aldehyde Dehydrogenase 1A3 Influences Breast Cancer Progression via Differential Retinoic Acid Signaling. Mol. Oncol. 2014, 9, 17–31. [Google Scholar] [CrossRef]
- Wang, S.; Liang, C.; Bao, M.; Li, X.; Zhang, L.; Li, S.; Qin, C.; Shao, P.; Li, J.; Hua, L.; et al. ALDH1A3 Correlates with Luminal Phenotype in Prostate Cancer. Tumour Biol. 2017, 39, 1010428317703652. [Google Scholar] [CrossRef] [Green Version]
- Gan, C.; Pierscianek, D.; El Hindy, N.; Ahmadipour, Y.; Keyvani, K.; Sure, U.; Zhu, Y. The Predominant Expression of Cancer Stem Cell Marker ALDH1A3 in Tumor Infiltrative Area Is Associated with Shorter Overall Survival of Human Glioblastoma. BMC Cancer 2020, 20, 672. [Google Scholar] [CrossRef]
- Flahaut, M.; Jauquier, N.; Chevalier, N.; Nardou, K.; Balmas Bourloud, K.; Joseph, J.-M.; Barras, D.; Widmann, C.; Gross, N.; Renella, R.; et al. Aldehyde Dehydrogenase Activity Plays a Key Role in the Aggressive Phenotype of Neuroblastoma. BMC Cancer 2016, 16, 781. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.L.; Yang, L.; Zou, Q.; Yuan, Y.; Li, J.; Liang, L.; Zeng, G.; Chen, S. Positive ALDH1A3 and Negative GPX3 Expressions Are Biomarkers for Poor Prognosis of Gallbladder Cancer. Dis. Markers 2013, 35, 163–172. [Google Scholar] [CrossRef]
- Duan, J.J.; Wang, D.; Cai, J.; Chen, J.J.; Zheng, X.X.; Chen, T.Q.; Wang, J.; Zhang, X.; Yang, Q.K.; Yu, S.C. An Aldehyde Dehydrogenase 1A3 Inhibitor Attenuates the Metastasis of Human Colorectal Cancer. Cancer Lett. 2022, 536, 215662. [Google Scholar] [CrossRef]
- Kawakami, R.; Mashima, T.; Kawata, N.; Kumagai, K.; Migita, T.; Sano, T.; Mizunuma, N.; Yamaguchi, K.; Seimiya, H. ALDH1A3-MTOR Axis as a Therapeutic Target for Anticancer Drug-Tolerant Persister Cells in Gastric Cancer. Cancer Sci. 2020, 111, 962–973. [Google Scholar] [CrossRef]
- Nie, S.; Qian, X.; Shi, M.; Li, H.; Peng, C.; Ding, X.; Zhang, S.; Zhang, B.; Xu, G.; Lv, Y.; et al. ALDH1A3 Accelerates Pancreatic Cancer Metastasis by Promoting Glucose Metabolism. Front. Oncol. 2020, 10, 915. [Google Scholar] [CrossRef]
- Kim, Y.J.; Yoon, H.Y.; Kim, J.S.; Kang, H.W.; Min, B.D.; Kim, S.K.; Ha, Y.S.; Kim, I.Y.; Ryu, K.H.; Lee, S.C.; et al. HOXA9, ISL1 and ALDH1A3 Methylation Patterns as Prognostic Markers for Nonmuscle Invasive Bladder Cancer: Array-Based DNA Methylation and Expression Profiling. Int. J. Cancer 2013, 133, 1135–1142. [Google Scholar] [CrossRef]
- Ma, Y.M.; Zhao, S. Prognostic Values of Aldehyde Dehydrogenase 1 Isoenzymes in Ovarian Cancer. Onco Targets Ther. 2016, 9, 1981–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samson, J.M.; Ravindran Menon, D.; Smith, D.E.; Baird, E.; Kitano, T.; Gao, D.; Tan, A.C.; Fujita, M. Clinical Implications of ALDH1A1 and ALDH1A3 MRNA Expression in Melanoma Subtypes. Chem. Biol. Interact. 2019, 314, 108822. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Duan, P.G.; Gao, Z.Z.; Dai, M. MicroRNA-487b-3p Inhibits Osteosarcoma Chemoresistance and Metastasis by Targeting ALDH1A3. Oncol. Rep. 2020, 44, 2691–2700. [Google Scholar] [CrossRef]
- Pérez-Alea, M.; Mcgrail, K.; Sánchez-Redondo, S.; Ferrer, B.; Fournet, G.; Cortés, J.; Muñoz, E.; Hernandez-Losa, J.; Tenbaum, S.; Martin, G.; et al. ALDH1A3 Is Epigenetically Regulated during Melanocyte Transformation and Is a Target for Melanoma Treatment. Nat. Publ. Group 2017, 36, 5695–5708. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.N.M.; Namkung, J.; Phan, A.N.H.; Vo, V.T.A.; Kim, M.-K.; Jeong, Y.; Choi, J.-W. PPARgamma-Mediated ALDH1A3 Suppression Exerts Anti-Proliferative Effects in Lung Cancer by Inducing Lipid Peroxidation. J. Recept. Signal Transduct. 2018, 38, 191–197. [Google Scholar] [CrossRef]
- Croker, A.K.; Rodriguez-Torres, M.; Xia, Y.; Pardhan, S.; Sing Leong, H.; Lewis, J.D.; Allan, A.L. Differential Functional Roles of ALDH1A1 and ALDH1A3 in Mediating Metastatic Behavior and Therapy Resistance of Human Breast Cancer Cells. Int. J. Mol. Sci. 2017, 18, 2039. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic Assay of Cells in Vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Thomas, M.L.; de Antueno, R.; Coyle, K.M.; Sultan, M.; Cruickshank, B.M.; Giacomantonio, M.A.; Giacomantonio, C.A.; Duncan, R.; Marcato, P. Citral Reduces Breast Tumor Growth by Inhibiting the Cancer Stem Cell Marker ALDH1A3. Mol. Oncol. 2016, 10, 1485–1496. [Google Scholar] [CrossRef]
- Huang, X.; Hou, Y.; Weng, X.; Pang, W.; Hou, L.; Liang, Y.; Wang, Y.; Du, L.; Wu, T.; Yao, M.; et al. Oncogenesis Diethyldithiocarbamate-Copper Complex (CuET) Inhibits Colorectal Cancer Progression via MiR-16-5p and 15b-5p/ALDH1A3/PKM2 Axis-Mediated Aerobic Glycolysis Pathway. Oncogenesis 2021, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Liu, Y.; Gao, Z.; Huang, W. Recent Advances in Drug Delivery Systems for Targeting Cancer Stem Cells. Acta Pharm. Sin. B 2021, 11, 55–70. [Google Scholar] [CrossRef]
- Durinikova, E.; Kozovska, Z.; Poturnajova, M.; Plava, J.; Cierna, Z.; Babelova, A.; Bohovic, R.; Schmidtova, S.; Tomas, M.; Kucerova, L.; et al. ALDH1A3 Upregulation and Spontaneous Metastasis Formation Is Associated with Acquired Chemoresistance in Colorectal Cancer Cells. BMC Cancer 2018, 18, 848. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.Z.; He, Y.F.; Wen, J.; Niu, Y.N.; Gao, Y.; Liu, L.H.; Zhang, X.P.; Wang, Y.; Zhang, X.L.; Zhang, H.F.; et al. Epithelial Cell Adhesion Molecule Promotes Breast Cancer Resistance Protein-Mediated Multidrug Resistance in Breast Cancer by Inducing Partial Epithelial-Mesenchymal Transition. Cell Biol. Int. 2021, 45, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Jung, K.H.; Byun, Y.; Lee, J.H.; Moon, S.H.; Cho, Y.S.; Lee, K.H. ATP-Binding Cassette Transporters Substantially Reduce Estimates of ALDH-Positive Cancer Cells Based on Aldefluor and AldeRed588 Assays. Sci. Rep. 2019, 9, 6462. [Google Scholar] [CrossRef] [Green Version]
- Bauzone, M.; Souidi, M.; Dessein, A.F.; Wisztorski, M.; Vincent, A.; Gimeno, J.P.; Monte, D.; van Seuningen, I.; Gespach, C.; Huet, G. Cross-Talk between YAP and RAR-RXR Drives Expression of Stemness Genes to Promote 5-FU Resistance and Self-Renewal in Colorectal Cancer Cells. Mol. Cancer Res. 2021, 19, 612–622. [Google Scholar] [CrossRef]
- Yun, X.; Zhang, K.; Wang, J.J.; Pangeni, R.P.; Yang, L.; Bonner, M.; Wu, J.; Wang, J.J.; Nardi, I.K.; Gao, M.; et al. Targeting USP22 Suppresses Tumorigenicity and Enhances Cisplatin Sensitivity through ALDH1A3 Downregulation in Cancer-Initiating Cells from Lung Adenocarcinoma. Mol. Cancer Res. 2018, 16, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Canino, C.; Luo, Y.Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. A STAT3-NFkB/DDIT3/CEBPβ Axis Modulates ALDH1A3 Expression in Chemoresistant Cell Subpopulations. Oncotarget 2015, 6, 12637. [Google Scholar] [CrossRef] [Green Version]
- Cioce, M.; Sacconi, A.; Pass, H.I.; Canino, C.; Strano, S.; Blandino, G.; Fazio, V.M. Insights into Intra-Tumoral Heterogeneity: Transcriptional Profiling of Chemoresistant MPM Cell Subpopulations Reveals Involvement of NFkB and DNA Repair Pathways and Contributes a Prognostic Signature. Int. J. Mol. Sci. 2021, 22, 12071. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Qin, H.; Yang, L.; Chen, M.; Yang, Y.; Zhang, W.; Hao, J.; Lu, Q.; Shi, J.; Zhuang, J.; et al. CircCYP24A1 Promotes Docetaxel Resistance in Prostate Cancer by Upregulating ALDH1A3. Biomark. Res. 2022, 10, 48. [Google Scholar] [CrossRef]
- Yao, Y.; Li, N. MIR600HG Suppresses Metastasis and Enhances Oxaliplatin Chemosensitivity by Targeting ALDH1A3 in Colorectal Cancer. Biosci. Rep. 2020, 40, 20200390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidtova, S.; Kalavska, K.; Gercakova, K.; Cierna, Z.; Miklikova, S.; Smolkova, B.; Buocikova, V.; Miskovska, V.; Durinikova, E.; Burikova, M.; et al. Disulfiram Overcomes Cisplatin Resistance in Human Embryonal Carcinoma Cells. Cancers 2019, 11, 1224. [Google Scholar] [CrossRef] [Green Version]
- Moreb, J.S.; Muhoczy, D.; Ostmark, B.; Zucali, J.R. RNAi-Mediated Knockdown of Aldehyde Dehydrogenase Class-1A1 and Class-3A1 Is Specific and Reveals That Each Contributes Equally to the Resistance against 4-Hydroperoxycyclophosphamide. Cancer Chemother. Pharmacol. 2007, 59, 127–136. [Google Scholar] [CrossRef]
- Ekhart, C.; Doodeman, V.D.; Rodenhuis, S.; Smits, P.H.M.; Beijnen, J.H.; Huitema, A.D.R. Influence of Polymorphisms of Drug Metabolizing Enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the Pharmacokinetics of Cyclophosphamide and 4-Hydroxycyclophosphamide. Pharmacogenet. Genom. 2008, 18, 515–523. [Google Scholar] [CrossRef]
- Sládek, N.E.; Kollander, R.; Sreerama, L.; Kiang, D.T. Cellular Levels of Aldehyde Dehydrogenases (ALDH1A1 and ALDH3A1) as Predictors of Therapeutic Responses to Cyclophosphamide-Based Chemotherapy of Breast Cancer: A Retrospective Study. Rational Individualization of Oxazaphosphorine-Based Cancer Chemotherapeutic Regimens. Cancer Chemother. Pharmacol. 2002, 49, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-Regulated Long Non-Coding RNA NRAD1 Is a Potential Novel Target for Triple-Negative Breast Tumors and Cancer Stem Cells. Cell Death Differ. 2020, 27, 363–378. [Google Scholar] [CrossRef]
- Sullivan, K.E.; Rojas, K.; Cerione, R.A.; Nakano, I.; Wilson, K.F. The Stem Cell/Cancer Stem Cell Marker ALDH1A3 Regulates the Expression of the Survival Factor Tissue Transglutaminase, in Mesenchymal Glioma Stem Cells. Oncotarget 2017, 8, 22325. [Google Scholar] [CrossRef] [Green Version]
- Thamrongwaranggoon, U.; Detarya, M.; Seubwai, W.; Saengboonmee, C.; Hino, S.; Koga, T.; Nakao, M.; Wongkham, S. Lactic Acidosis Promotes Aggressive Features of Cholangiocarcinoma Cells via Upregulating ALDH1A3 Expression through EGFR Axis. Life Sci. 2022, 302, 120648. [Google Scholar] [CrossRef]
- Corominas-Faja, B.; Oliveras-Ferraros, C.; Cuyàs, E.; Segura-Carretero, A.; Joven, J.; Martin-Castillo, B.; Barrajón-Catalán, E.; Micol, V.; Bosch-Barrera, J.; Menendez, J.A. Stem Cell-like ALDH(Bright) Cellular States in EGFR-Mutant Non-Small Cell Lung Cancer: A Novel Mechanism of Acquired Resistance to Erlotinib Targetable with the Natural Polyphenol Silibinin. Cell Cycle 2013, 12, 3390–3404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, P.; Wang, J.; Waghmare, I.; Sartini, S.; Coviello, V.; Zhang, Z.; Kim, S.H.; Mohyeldin, A.; Pavlyukov, M.S.; Minata, M.; et al. FOXD1-ALDH1A3 Signaling Is a Determinant for the Self-Renewal and Tumorigenicity of Mesenchymal Glioma Stem Cells. Cancer Res. 2016, 76, 7219–7230. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.W.; Wang, S.; Fan, L.; Feng, S.; Cai, X.; Peng, C.; Wu, X.; Lu, J.; Chen, D.; et al. USP9X Deubiquitinates ALDH1A3 and Maintains Mesenchymal Identity in Glioblastoma Stem Cells. J. Clin. Investig. 2019, 129, 2043–2055. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Yan, W.; You, G.; Bao, Z.; Wang, Y.; Liu, Y.; You, Y.; Jiang, T. Genome-Wide DNA Methylation Profiling Identifies ALDH1A3 Promoter Methylation as a Prognostic Predictor in G-CIMP- Primary Glioblastoma. Cancer Lett. 2013, 328, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Luo, L.; Ping, Z.; Yuan, H.P.; Zhao, X.D.; Xu, W. Prognostic Value of ALDH1A3 Promoter Methylation in Gliob;Astoma: A Single Center Experience in Western China. Asian Pac. J. Cancer Prev. 2015, 16, 591–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Wasson, M.C.D.; Marcato, P. The Missing Lnc: The Potential of Targeting Triple-Negative Breast Cancer and Cancer Stem Cells by Inhibiting Long Non-Coding RNAs. Cells 2020, 9, 763. [Google Scholar] [CrossRef] [Green Version]
- Kwak, P.B.; Iwasaki, S.; Tomari, Y. The MicroRNA Pathway and Cancer. Cancer Sci. 2010, 101, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Wasson, M.C.D.; Brown, J.M.; Fernando, W.; Marcato, P. LncRNA-MiRNA Axes in Breast Cancer: Novel Points of Interaction for Strategic Attack. Cancer Lett. 2021, 509, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Li, M.; You, C.; Zhao, F.; Guo, M.; Xu, H.; Li, L.; Wang, L.; Dou, J. Inhibition of Breast Cancer Growth via MiR-7 Suppressing ALDH1A3 Activity Concomitant with Decreasing Breast Cancer Stem Cell Subpopulation. J. Cell Physiol. 2020, 235, 1405–1416. [Google Scholar] [CrossRef] [Green Version]
- Coyle, K.M.; Maxwell, S.; Thomas, M.L.; Marcato, P. Profiling of the Transcriptional Response to All-Trans Retinoic Acid in Breast Cancer Cells Reveals RARE-Independent Mechanisms of Gene Expression. Sci. Rep. 2017, 7, 16684. [Google Scholar] [CrossRef] [Green Version]
- Ke, C.; Shen, M.; Wang, P.; Chen, Z.; Lin, S.; Dong, F. ALDH1A3-Linc00284 Axis Mediates the Invasion of Colorectal Cancer by Targeting TGF β Signaling via Sponging MiR-361-5p. Int. J. Genom. 2022, 2022, 6561047. [Google Scholar] [CrossRef]
- Yan, D.; Wu, F.; Peng, C.; Wang, M. Silencing of LINC00284 Inhibits Cell Proliferation and Migration in Oral Squamous Cell Carcinoma by the MiR-211-3p/MAFG Axis and FUS/KAZN Axis. Cancer Biol. Ther. 2021, 22, 149–163. [Google Scholar] [CrossRef]
- Zhu, M.; Yan, X.; Zhao, Y.; Xue, H.; Wang, Z.; Wu, B.; Li, X.; Shen, Y. LncRNA LINC00284 Promotes Nucleus Pulposus Cell Proliferation and ECM Synthesis via Regulation of the MiR-205-3p/Wnt/β-Catenin Axis. Mol. Med. Rep. 2022, 25, 179. [Google Scholar] [CrossRef]
- Zhou, B.; Ge, Y.; Shao, Q.; Yang, L.; Chen, X.; Jiang, G. ARTICLE Long Noncoding RNA LINC00284 Facilitates Cell Proliferation in Papillary Thyroid Cancer via Impairing MiR-3127-5p Targeted E2F7 Suppression. Cell Death Discov. 2021, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Li, J.; Ke, C.; Xiao, Y.; Lu, C.; Huang, F.; Mi, Y.; Xia, R.; Li, Q. Oncogenic Long Intervening Noncoding RNA Linc00284 Promotes C-Met Expression by Sponging MiR-27a in Colorectal Cancer. Oncogene 2021, 40, 4151–4166. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Y.; Liu, X.; Wang, Z.; Zhang, C.; Wu, F.; Jiang, H.; Zhang, W.; Bao, Z.; Wang, Y.; et al. ALDH1A3 Induces Mesenchymal Differentiation and Serves as a Predictor for Survival in Glioblastoma. Cell Death Dis. 2018, 9, 1190. [Google Scholar] [CrossRef] [Green Version]
- Dahn, M.L.; Walsh, H.R.; Dean, C.A.; Giacomantonio, M.A.; Fernando, W.; Murphy, J.P.; Walker, O.L.; Wasson, M.C.D.; Gujar, S.; Pinto, D.M.; et al. Metabolite Profiling Reveals a Connection between Aldehyde Dehydrogenase 1A3 and GABA Metabolism in Breast Cancer Metastasis. Metabolomics 2022, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Kim-Muller, J.Y.; Fan, J.; Kim, Y.J.R.; Lee, S.A.; Ishida, E.; Blaner, W.S.; Accili, D. Aldehyde Dehydrogenase 1a3 Defines a Subset of Failing Pancreatic β Cells in Diabetic Mice. Nat. Commun. 2016, 7, 12631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Mohan, R.; Chen, X.; Matson, K.; Waugh, J.; Mao, Y.; Zhang, S.; Li, W.; Tang, X.X.; Satin, L.S.; et al. MicroRNA-483 Protects Pancreatic β-Cells by Targeting ALDH1A3. Endocrinology 2021, 162, bqab031. [Google Scholar] [CrossRef]
- Walsh, H.R.; Cruickshank, B.M.; Brown, J.M.; Marcato, P. The Flick of a Switch: Conferring Survival Advantage to Breast Cancer Stem Cells Through Metabolic Plasticity. Front. Oncol. 2019, 9, 753. [Google Scholar] [CrossRef]
- Cinti, F.; Bouchi, R.; Kim-Muller, J.Y.; Ohmura, Y.; Sandoval, P.R.; Masini, M.; Marselli, L.; Suleiman, M.; Ratner, L.E.; Marchetti, P.; et al. Evidence of β-Cell Dedifferentiation in Human Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 1044–1054. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.Y.; Chen, S.H.; Lin, Y.S.; Wu, H.P.; Chao, P.M. An Antidiabetic Nutraceutical Combination of Red Yeast Rice (Monascus Purpureus), Bitter Gourd (Momordica Charantia), and Chromium Alleviates Dedifferentiation of Pancreatic β Cells in Db/Db Mice. Food Sci. Nutr. 2020, 8, 6718–6726. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, A.; Norlin, S.; Steneberg, P.; Remeseiro, S.; Edlund, H.; Hörnblad, A. Pan-AMPK Activator O304 Prevents Gene Expression Changes and Remobilisation of Histone Marks in Islets of Diet-Induced Obese Mice. Sci. Rep. 2021, 11, 24410. [Google Scholar] [CrossRef]
- Puttini, S.; Plaisance, I.; Barile, L.; Cervio, E.; Milano, G.; Marcato, P.; Pedrazzini, T.; Vassalli, G. ALDH1A3 Is the Key Isoform That Contributes to Aldehyde Dehydrogenase Activity and Affects in Vitro Proliferation in Cardiac Atrial Appendage Progenitor Cells. Front. Cardiovasc. Med. 2018, 5, 90. [Google Scholar] [CrossRef] [Green Version]
- Roehrich, M.E.; Spicher, A.; Milano, G.; Vassalli, G. Characterization of Cardiac-Resident Progenitor Cells Expressing High Aldehyde Dehydrogenase Activity. Biomed. Res. Int. 2013, 2013, 503047. [Google Scholar] [CrossRef] [Green Version]
- Koninckx, R.; Daniëls, A.; Windmolders, S.; Mees, U.; MacIanskiene, R.; Mubagwa, K.; Steels, P.; Jamaer, L.; Dubois, J.; Robic, B.; et al. The Cardiac Atrial Appendage Stem Cell: A New and Promising Candidate for Myocardial Repair. Cardiovasc. Res. 2013, 97, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Shao, N.Y.; Moonen, J.R.M.D.; Zhao, Z.; Shi, M.; Otsuki, S.; Wang, L.; Elaine Yan, T.N.; Marciano, D.P.; Contrepois, K.; et al. ALDH1A3 Coordinates Metabolism With Gene Regulation in Pulmonary Arterial Hypertension. Circulation 2021, 143, 2074–2090. [Google Scholar] [CrossRef] [PubMed]
- Hosono, H.; Ohishi, T.; Takei, J.; Asano, T.; Sayama, Y.; Kawada, M.; Kaneko, M.K.; Kato, Y. The Anti-epithelial Cell Adhesion Molecule (EpCAM) Monoclonal Antibody EpMab-16 Exerts Antitumor Activity in a Mouse Model of Colorectal Adenocarcinoma. Oncol. Lett. 2020, 20, 383. [Google Scholar] [CrossRef]
- Desai, A.; Yan, Y.; Gerson, S.L. Concise Reviews: Cancer Stem Cell Targeted Therapies: Toward Clinical Success. Stem Cells Transl. Med. 2019, 8, 75–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanbari-Movahed, M.; Ghanbari-Movahed, Z.; Momtaz, S.; Kilpatrick, K.L.; Farzaei, M.H.; Bishayee, A. Unlocking the Secrets of Cancer Stem Cells with γ-Secretase Inhibitors: A Novel Anticancer Strategy. Molecules 2021, 26, 972. [Google Scholar] [CrossRef]
- Pal, A.; Pattanayak, R.D.; Sagar, R. Tracing the Journey of Disulfiram: From an Unintended Discovery to a Treatment Option for Alcoholism. J. Ment. Health Hum. Behav. 2015, 20, 41. [Google Scholar] [CrossRef]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent Advances in Repurposing Disulfiram and Disulfiram Derivatives as Copper-Dependent Anticancer Agents. Front. Mol. Biosci. 2021, 8, 741316. [Google Scholar] [CrossRef]
- Lin, J.; Haffner, M.C.; Zhang, Y.; Lee, B.H.; Brennen, W.N.; Britton, J.; Kachhap, S.K.; Shim, J.S.; Liu, J.O.; Nelson, W.G.; et al. Disulfiram Is a DNA Demethylating Agent and Inhibits Prostate Cancer Cell Growth. Prostate 2011, 71, 333–343. [Google Scholar] [CrossRef] [Green Version]
- Kona, F.R.; Buac, D.; Burger, A.M. Disulfiram, and Disulfiram Derivatives as Novel Potential Anticancer Drugs Targeting the Ubiquitin-Proteasome System in Both Preclinical and Clinical Studies. Curr. Cancer Drug Targets 2011, 11, 338–346. [Google Scholar] [CrossRef]
- Zirjacks, L.; Stransky, N.; Klumpp, L.; Prause, L.; Eckert, F.; Zips, D.; Schleicher, S.; Handgretinger, R.; Huber, S.M.; Ganser, K. Repurposing Disulfiram for Targeting of Glioblastoma Stem Cells: An In Vitro Study. Biomolecules 2021, 11, 1561. [Google Scholar] [CrossRef]
- Yamashita, D.; Minata, M.; Ibrahim, A.N.; Yamaguchi, S.; Coviello, V.; Bernstock, J.D.; Harada, S.; Cerione, R.A.; Tannous, B.A.; la Motta, C.; et al. Identification of ALDH1A3 as a Viable Therapeutic Target in Breast Cancer Metastasis-Initiating Cells. Mol. Cancer Ther. 2020, 19, 1134–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelardi, E.L.M.; Colombo, G.; Picarazzi, F.; Ferraris, D.M.; Mangione, A.; Petrarolo, G.; Aronica, E.; Rizzi, M.; Mori, M.; la Motta, C.; et al. A Selective Competitive Inhibitor of Aldehyde Dehydrogenase 1A3 Hinders Cancer Cell Growth, Invasiveness and Stemness In Vitro. Cancers 2021, 13, 356. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, H.; Miyano, M.; Ito, D.; Kimura, T.; Hagiwara, K.; Kogai, H.; Kaburagi, Y.; Kotake, Y.; Takase, Y. Identification of a Novel ALDH1A3-Selective Inhibitor by a Chemical Probe with Unrelated Bioactivity: An Approach to Affinity-Based Drug Target Discovery. Chem. Biol. Drug Des. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Li, J.; Donini, S.; Sobol, R.W.; Rizzi, M.; Garavaglia, S. Crystal Structure of Human Aldehyde Dehydrogenase 1A3 Complexed with NAD+ and Retinoic Acid. Sci. Rep. 2016, 6, 35710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Garavaglia, S.; Ye, Z.; Moretti, A.; Belyaeva, O.V.; Beiser, A.; Ibrahim, M.; Wilk, A.; McClellan, S.; Klyuyeva, A.V.; et al. A Specific Inhibitor of ALDH1A3 Regulates Retinoic Acid Biosynthesis in Glioma Stem Cells. Commun. Biol. 2021, 4, 1420. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, J.; Zhang, Z.; Qian, Y.; Wang, G.; Duan, M.; Zhao, H.; Yang, Z.; Jiang, X. Mesenchymal Stem Cell-Derived Exosome: A Tumor Regulator and Carrier for Targeted Tumor Therapy. Cancer Lett. 2022, 526, 29–40. [Google Scholar] [CrossRef]
| Cancer Dataset | Patient Number | Amplification | Missense Mutations | Deep Deletion | Truncation Mutation |
|---|---|---|---|---|---|
| Prostate (TCGA, Firehose Legacy) | 489 | 0.2% | 0.2% | 0.2% | 0.0% |
| Prostate Metastasis (SU2C/PCF Dream Team, PNAS 2019) | 429 | 1.4% | 0.2% | 0.9% | 0.0% |
| Colorectal (TCGA, Firehose Legacy) | 220 | 0.45% | 1.8% | 0.0% | 0.45% |
| Breast (TCGA, Cell 2015) | 816 | 3.2% | 0.5% | 0.4% | 0.1% |
| Triple Negative Breast Cancer (TCGA, Cell 2015) | 82 | 9% | 0.0% | 0.0% | 0.0% |
| Breast Metastasis (Provisional, December 2021) | 301 | 9.3% | 0.0% | 2.3% | 0.0% |
| Pancreatic (TCGA, Firehose Legacy) | 149 | 2.7% | 1.3% | 0.0% | 0.0% |
| Glioblastoma (CPTAC, Cell 2021) | 96 | 2.1% | 0.0% | 0.0% | 0.0% |
| Melanoma (TCGA, Cell 2015) | 344 | 0.0% | 0.6% | 0.0% | 0.0% |
| Melanoma Metastasis (DFCI, Science 2015) | 110 | 6.3% | 1.8% | 0.9% | 0.0% |
| Lung (TCGA, Firehose Legacy) | 230 | 1.7% | 0.0% | 0.4% | 0.0% |
| Lung Never Smokers (NCI, Nature Genetics 2021) | 232 | 0.0% | 0.4% | 0.0% | 0.0% |
| Non-Small Cell Lung (TRACERx, NEJM & Nature 2017) | 100 | 0.0% | 1.0% | 0.0% | 1.0% |
| Bile Duct (TCGA, Firehose Legacy) | 35 | 0.0% | 0.0% | 0.0% | 0.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McLean, M.E.; MacLean, M.R.; Cahill, H.F.; Arun, R.P.; Walker, O.L.; Wasson, M.-C.D.; Fernando, W.; Venkatesh, J.; Marcato, P. The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond. Cancers 2023, 15, 492. https://doi.org/10.3390/cancers15020492
McLean ME, MacLean MR, Cahill HF, Arun RP, Walker OL, Wasson M-CD, Fernando W, Venkatesh J, Marcato P. The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond. Cancers. 2023; 15(2):492. https://doi.org/10.3390/cancers15020492
Chicago/Turabian StyleMcLean, Meghan E., Maya R. MacLean, Hannah F. Cahill, Raj Pranap Arun, Olivia L. Walker, Marie-Claire D. Wasson, Wasundara Fernando, Jaganathan Venkatesh, and Paola Marcato. 2023. "The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond" Cancers 15, no. 2: 492. https://doi.org/10.3390/cancers15020492
APA StyleMcLean, M. E., MacLean, M. R., Cahill, H. F., Arun, R. P., Walker, O. L., Wasson, M.-C. D., Fernando, W., Venkatesh, J., & Marcato, P. (2023). The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond. Cancers, 15(2), 492. https://doi.org/10.3390/cancers15020492






