The Effect of Cannabis Plant Extracts on Head and Neck Squamous Cell Carcinoma and the Quest for Cannabis-Based Personalized Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phytocannabinoid Extraction and Sample Preparation
2.2. Phytocannabinoid Identification and Quantification
2.3. Fractionation of the Whole Extract
2.4. Reagents
2.5. Cell Cultures
2.6. Cell Viability Assays
2.7. Cell Apoptosis Assay (Annexin V-APC/PI Assay)
2.8. Cell lysis and Western Blot Analyses
2.9. RNA Extraction
2.10. Real-Time Quantitative PCR
2.11. Statistical Analysis
2.12. SynergyFinder
3. Results
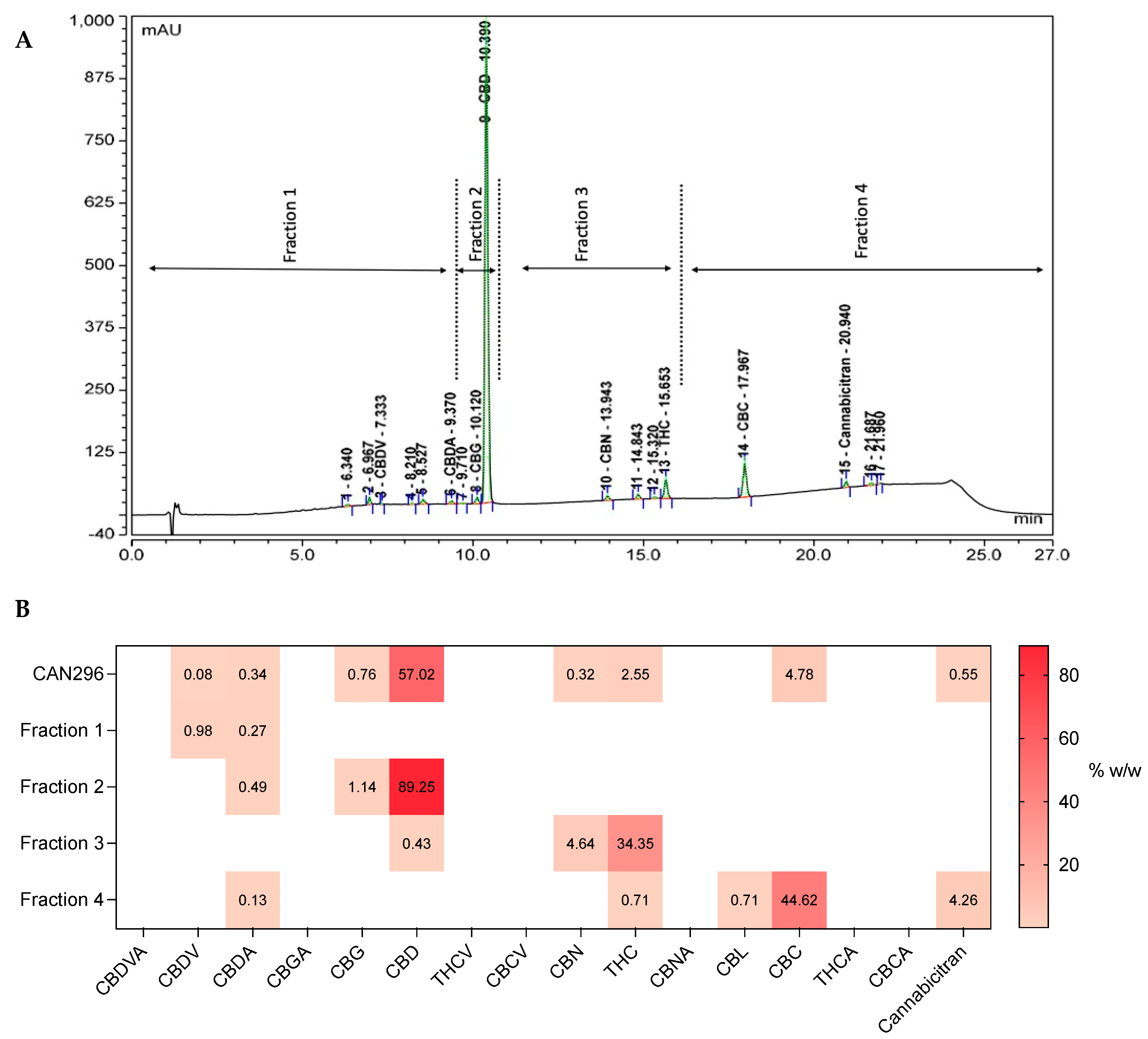
3.1. The Heterogeneous Composition of Cannabis Extracts
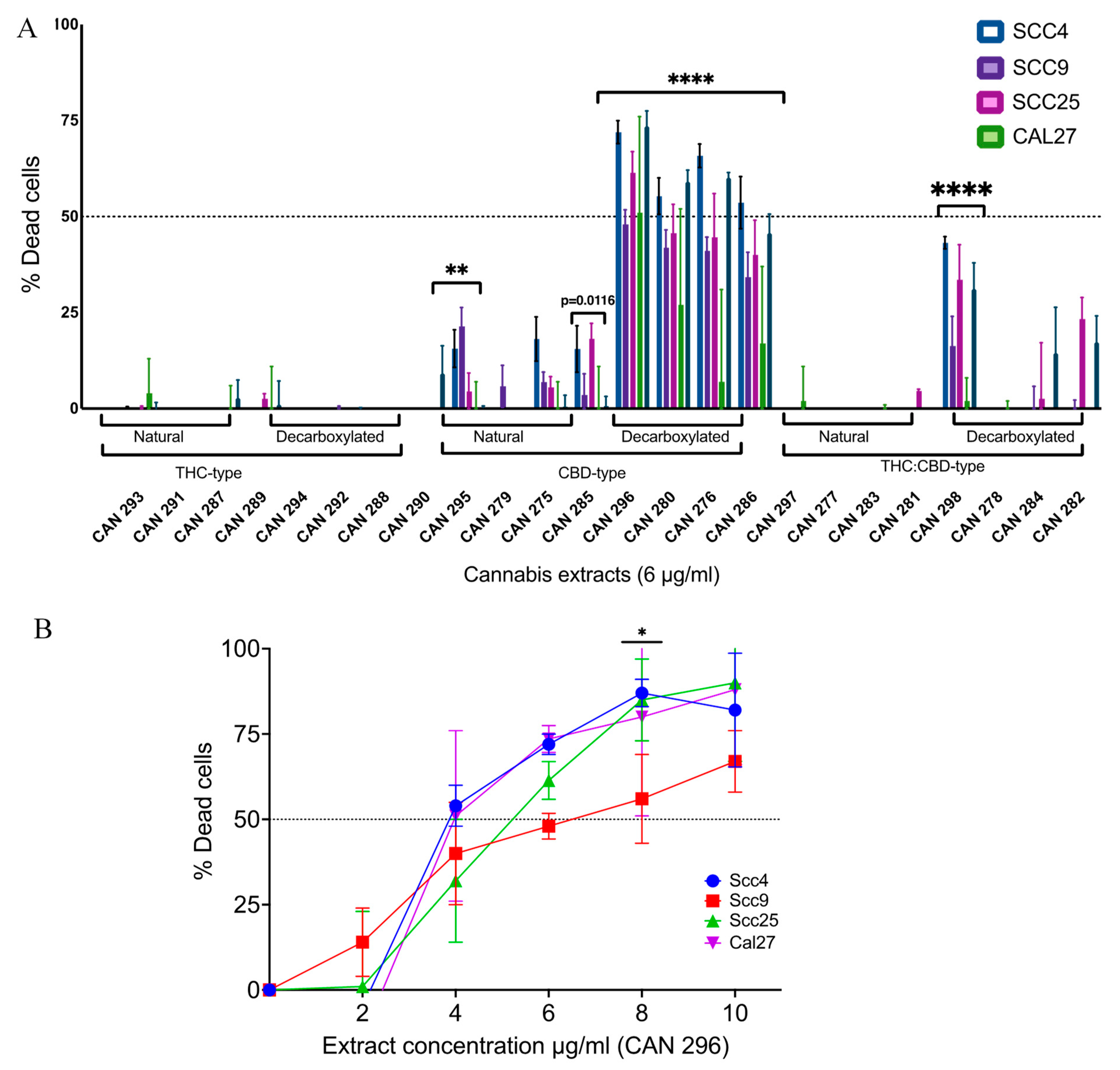
3.2. Decarboxylated High-CBD Extracts Are the Most Effective in Inducing HNSCC Cell Death
3.3. Cannabis Extract CAN296 Shows Apoptotic Effects in HNSCC
3.4. Cannabis Extract CAN296 Is More Selective Towards HNSCC Cells Compared to Non-Cancer Fibroblastic Cells
3.5. Endocannabinoid Receptors Expression in HNSCC Cells
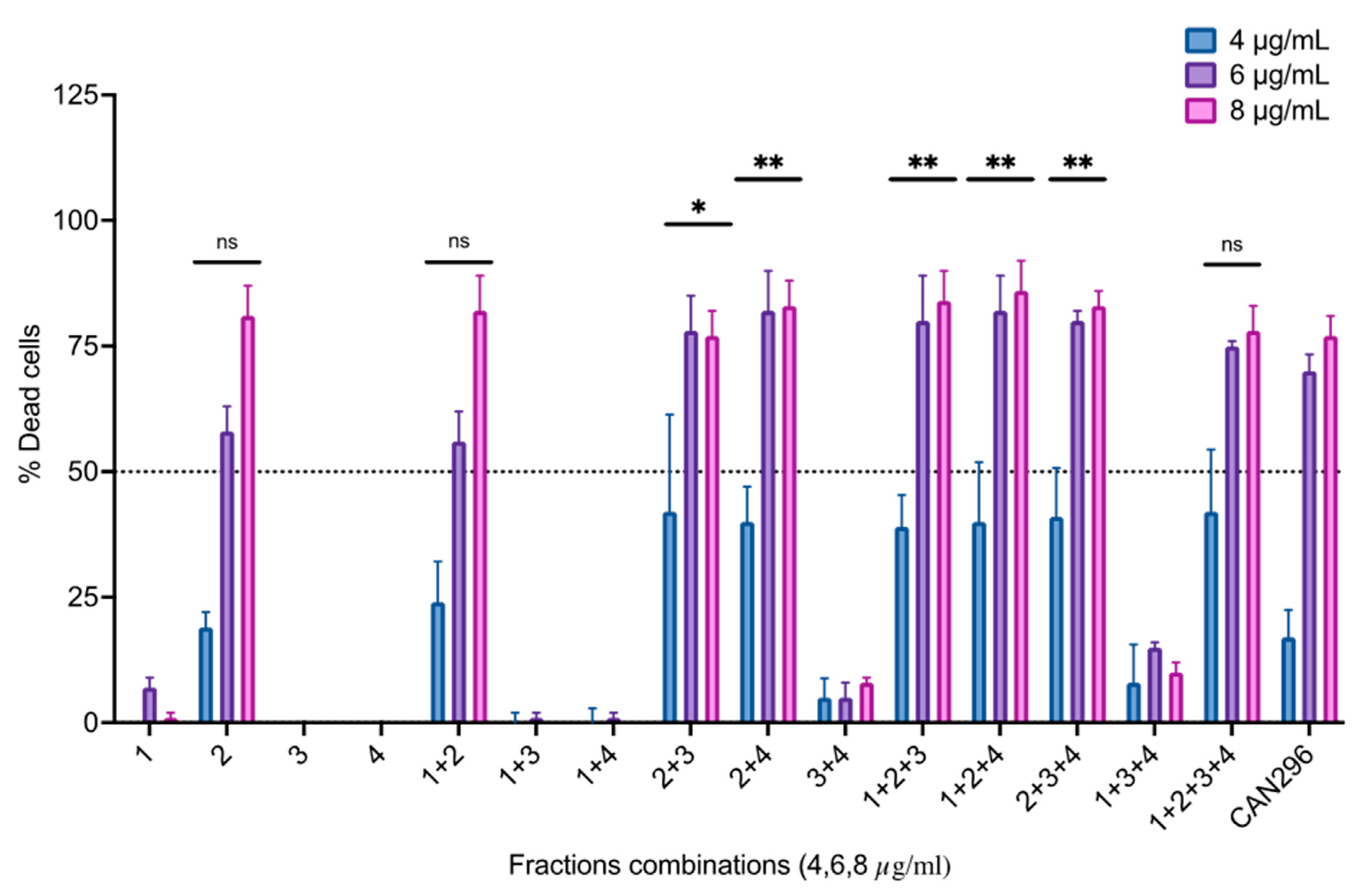
3.6. Identifying the Active Molecules in CAN296 Responsible for the Apoptotic Effect
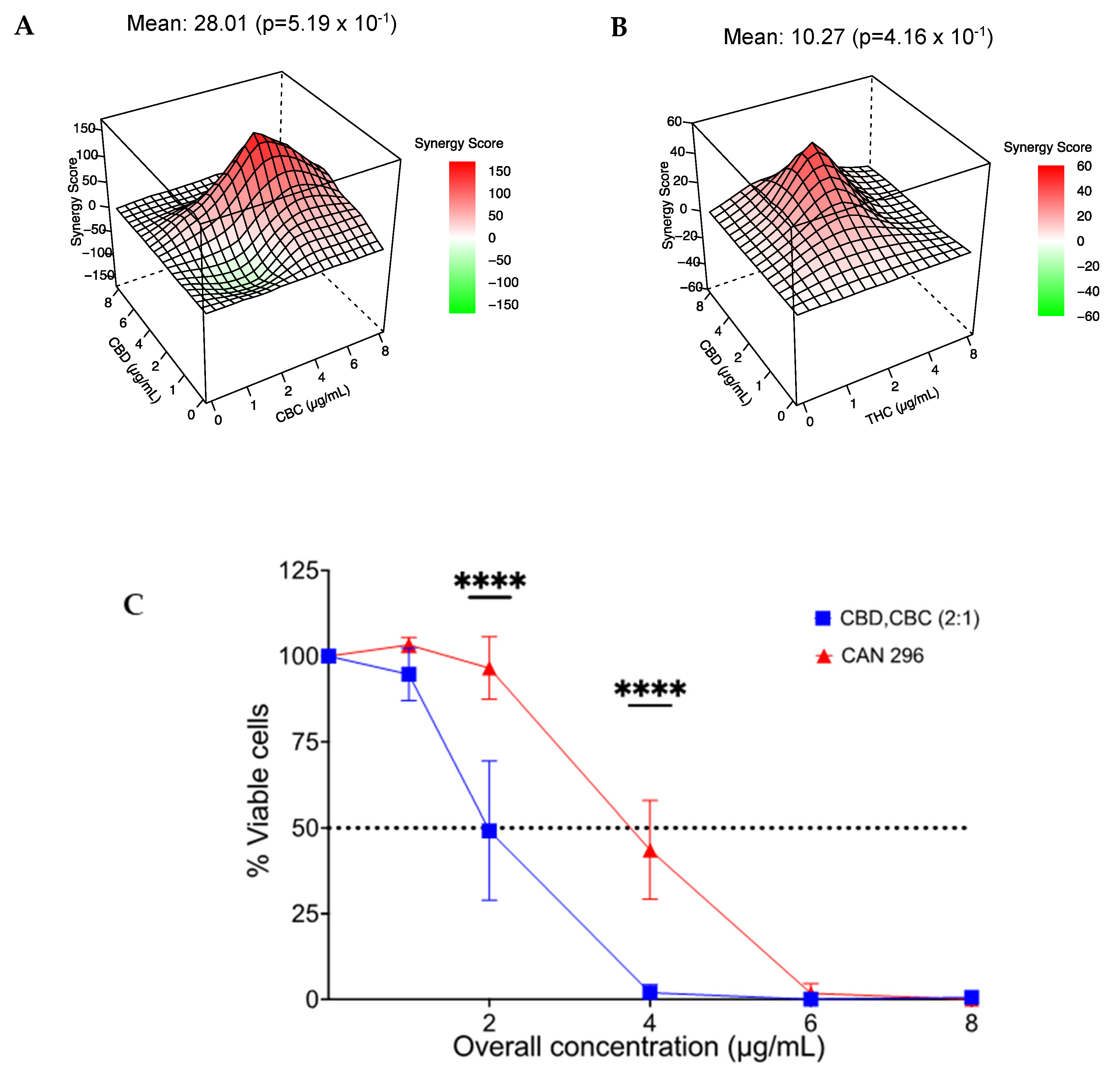
3.7. Combination of CBD with CBC or THC Results in a Synergistic Effect on HNSCC Cell Viability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CB1 | Cannabinoid receptor 1 |
| CB2 | Cannabinoid receptor 2 |
| CBD | Cannabidiol |
| CBC | Cannabichromene |
| DMSO | Dimethyl Sulfoxide |
| EBV | Epstein-Barr Virus |
| ECS | Endocannabinoid System |
| ESI | Electrospray Ionization |
| FACS | Flow Cytometry Cell Sorting |
| GPCRs | G-protein coupled receptors |
| HNSCC | Head and Neck Squamous Cell Carcinoma |
| HPLC | High Performance Liquid Chromatography |
| HPV | Human Papilloma Virus |
| LC | Liquid Chromatography |
| MS | Mass Spectrometry |
| PARP | poly ADP ribose polymerase |
| PPARγ | Peroxisome Proliferator Activated Receptor gamma |
| THC | (−)-trans-Δ⁹-tetrahydrocannabinol |
| TRPV | Transient Receptor Potential cation channel subfamily V |
| UHPLC | Ultra-HPLC |
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global cancer ob-servatory: Cancer today. Lyon Fr. Int. Agency Res. Cancer 2018, 3, 2019. [Google Scholar]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Kumar, M.; Nanavati, R.; Modi, T.G.; Dobariya, C. Oral cancer: Etiology and risk factors: A review. J. Cancer Res. Ther. 2016, 12, 458–463. [Google Scholar] [CrossRef]
- Pulte, D.; Brenner, H. Changes in Survival in Head and Neck Cancers in the Late 20th and Early 21st Century: A Period Analysis. Oncologist 2010, 15, 994–1001. [Google Scholar] [CrossRef]
- Baram, L.; Peled, E.; Berman, P.; Yellin, B.; Besser, E.; Benami, M.; Louria-Hayon, I.; Lewitus, G.M.; Meiri, D. The heterogeneity and complexity of Cannabis extracts as antitumor agents. Oncotarget 2019, 10, 4091–4106. [Google Scholar] [CrossRef]
- Velasco, G.; Sánchez, C.; Guzmán, M. Anticancer Mechanisms of Cannabinoids. Curr. Oncol. 2016, 23, S23–S32. [Google Scholar] [CrossRef]
- Andre, C.M.; Hausman, J.-F.; Guerriero, G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef]
- van Velzen, R.; Schranz, M.E. Origin and Evolution of the Cannabinoid Oxidocyclase Gene Family. Genome Biol. Evol. 2021, 13, evab130. [Google Scholar] [CrossRef]
- Mechoulam, R.; Gaoni, Y. Hashish—IV. The isolation and structure of cannabinolic cannabidiolic and cannabigerolic acids. Tetrahedron 1965, 21, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.C.; Perry, E.; MacDougall, L.; Ammerman, Y.; Cooper, T.; Wu, Y.-T.; Braley, G.; Gueorguieva, R.; Krystal, J.H. The Psychotomimetic Effects of Intravenous Delta-9-Tetrahydrocannabinol in Healthy Individuals: Implications for Psychosis. Neuropsychopharmacology 2004, 29, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Grof, C.P.L. Cannabis, from plant to pill. Br. J. Clin. Pharmacol. 2018, 84, 2463–2467. [Google Scholar] [CrossRef]
- Izzo, A.A.; Borrelli, F.; Capasso, R.; Di Marzo, V.; Mechoulam, R. Non-psychotropic plant cannabinoids: New therapeutic opportunities from an ancient herb. Trends Pharmacol. Sci. 2009, 30, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shabat, S.; Fride, E.; Sheskin, T.; Tamiri, T.; Rhee, M.-H.; Vogel, Z.; Bisogno, T.; De Petrocellis, L.; Di Marzo, V.; Mechoulam, R. An entourage effect: Inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur. J. Pharmacol. 1998, 353, 23–31. [Google Scholar] [CrossRef]
- Donvito, G.; Nass, S.; Wilkerson, J.; Curry, Z.; Schurman, L.; Kinsey, S.G.; Lichtman, A.H. The Endogenous Cannabinoid System: A Budding Source of Targets for Treating Inflammatory and Neuropathic Pain. Neuropsychopharmacology 2017, 43, 52–79. [Google Scholar] [CrossRef]
- Pandey, R.; Mousawy, K.; Nagarkatti, M.; Nagarkatti, P. Endocannabinoids and immune regulation. Pharmacol. Res. 2009, 60, 85–92. [Google Scholar] [CrossRef]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.H.; Di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid Receptors and Their Ligands: Beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef]
- Pacher, P.; Bátkai, S.; Kunos, G. The Endocannabinoid System as an Emerging Target of Pharmacotherapy. Pharmacol. Rev. 2006, 58, 389–462. [Google Scholar] [CrossRef]
- McPartland, J.M. Phylogenomic and chemotaxonomic analysis of the endocannabinoid system. Brain Res. Rev. 2004, 45, 18–29. [Google Scholar] [CrossRef]
- Pertwee, R.G. Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacol. Ther. 1997, 74, 129–180. [Google Scholar] [CrossRef]
- Pertwee, R.G. GPR55: A new member of the cannabinoid receptor clan? Br. J. Pharmacol. 2007, 152, 984–986. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Ligresti, A.; Moriello, A.S.; Allarà, M.; Bisogno, T.; Petrosino, S.; Stott, C.G.; Di Marzo, V. Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br. J. Pharmacol. 2011, 163, 1479–1494. [Google Scholar] [CrossRef]
- Moreno, E.; Cavic, M.; Krivokuca, A.; Casadó, V.; Canela, E. The Endocannabinoid System as a Target in Cancer Diseases: Are We There Yet? Front. Pharmacol. 2019, 10, 339. [Google Scholar] [CrossRef]
- Śledziński, P.; Zeyland, J.; Słomski, R.; Nowak, A. The current state and future perspectives of cannabinoids in cancer biology. Cancer Med. 2018, 7, 765–775. [Google Scholar] [CrossRef]
- Mangal, N.; Erridge, S.; Habib, N.; Sadanandam, A.; Reebye, V.; Sodergren, M.H. Cannabinoids in the landscape of cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 2507–2534. [Google Scholar] [CrossRef]
- Tapasak, B.; Edelmayer, L.; Seidman, M.D. Endocannabinoid System and the Otolaryngologist. Otolaryngol. Clin. North Am. 2022, 55, 1101–1110. [Google Scholar] [CrossRef]
- Wang, D.; Wang, H.; Ning, W.; Backlund, M.G.; Dey, S.K.; DuBois, R.N. Loss of Cannabinoid Receptor 1 Accelerates Intestinal Tumor Growth. Cancer Res. 2008, 68, 6468–6476. [Google Scholar] [CrossRef]
- Orellana-Serradell, O.; Poblete, C.E.; Sanchez, C.; Castellón, E.A.; Gallegos, I.; Huidobro, C.; Llanos, M.N.; Contreras, H.R. Proapoptotic effect of endocannabinoids in prostate cancer cells. Oncol. Rep. 2015, 33, 1599–1608. [Google Scholar] [CrossRef]
- Perez-Gomez, E.; Andradas, C.; Blasco-Benito, S.; Caffarel, M.M.; García-Taboada, E.; Villa-Morales, M.C.; Moreno, E.; Hamann, S.; Martin-Villar, E.; Flores, J.M.; et al. Role of Cannabinoid Receptor CB2 in HER2 Pro-oncogenic Signaling in Breast Cancer. J. Natl. Cancer Inst. 2015, 107, djv077. [Google Scholar] [CrossRef]
- Hasenoehrl, C.; Feuersinger, D.; Sturm, E.M.; Bärnthaler, T.; Heitzer, E.; Graf, R.; Grill, M.; Pichler, M.; Beck, S.; Butcher, L.; et al. G protein-coupled receptor GPR55 promotes colorectal cancer and has opposing effects to cannabinoid receptor 1. Int. J. Cancer 2018, 142, 121–132. [Google Scholar] [CrossRef]
- Maccarrone, M.; Lorenzon, T.; Bari, M.; Melino, G.; Finazzi-Agrò, A. Anandamide induces apoptosis in human cells via vanilloid receptors. Evidence for a protective role of cannabinoid receptors. J. Biol. Chem. 2000, 275, 31938–31945. [Google Scholar] [CrossRef]
- Fonseca, B.M.; Correia-Da-Silva, G.; Teixeira, N.A. Cannabinoid-induced cell death in endometrial cancer cells: Involvement of TRPV1 receptors in apoptosis. J. Physiol. Biochem. 2018, 74, 261–272. [Google Scholar] [CrossRef]
- Pagano, E.; Borrelli, F. Targeting cannabinoid receptors in gastrointestinal cancers for therapeutic uses: Current status and future perspectives. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 871–873. [Google Scholar] [CrossRef]
- Seltzer, E.S.; Watters, A.K.; MacKenzie, J.D., Jr.; Granat, L.M.; Zhang, D. Cannabidiol (CBD) as a Promising Anti-Cancer Drug. Cancers 2020, 12, 3203. [Google Scholar] [CrossRef]
- Laezza, C.; Pagano, C.; Navarra, G.; Pastorino, O.; Proto, M.C.; Fiore, D.; Piscopo, C.; Gazzerro, P.; Bifulco, M. The Endocannabinoid System: A Target for Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 747. [Google Scholar] [CrossRef]
- Velasco, G.; Sánchez, C.; Guzmán, M. Endocannabinoids and Cancer. Endocannabinoids 2015, 231, 449–472. [Google Scholar] [CrossRef]
- Armstrong, J.L.; Hill, D.S.; McKee, C.S.; Hernandez-Tiedra, S.; Lorente, M.; Lopez-Valero, I.; Anagnostou, M.E.; Babatunde, F.; Corazzari, M.; Redfern, C.P.F.; et al. Exploiting Cannabinoid-Induced Cytotoxic Autophagy to Drive Melanoma Cell Death. J. Investig. Dermatol. 2015, 135, 1629–1637. [Google Scholar] [CrossRef]
- Berman, P.; Futoran, K.; Lewitus, G.M.; Mukha, D.; Benami, M.; Shlomi, T.; Meiri, D. A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in Cannabis. Sci. Rep. 2018, 8, 14280. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genom. Proteom. Bioinform. 2022, 20, 587–596. [Google Scholar] [CrossRef]
- Procaccia, S.; Lewitus, G.M.; Feder, C.L.; Shapira, A.; Berman, P.; Meiri, D. Cannabis for Medical Use: Versatile Plant Rather Than a Single Drug. Front. Pharmacol. 2022, 13, 894960. [Google Scholar] [CrossRef]
- Galve-Roperh, I.; Sánchez, C.; Cortés, M.L.; Del Pulgar, T.G.; Izquierdo, M.; Guzmán, M. Anti-tumoral action of cannabinoids: Involvement of sustained ceramide accumulation and extracellular signal-regulated kinase activation. Nat. Med. 2000, 6, 313–319. [Google Scholar] [CrossRef]
- Blázquez, C.; Carracedo, A.; Barrado, L.; Real, P.J.; Fernández-Luna, J.L.; Velasco, G.; Malumbres, M.; Guzmán, M. Cannabinoid receptors as novel targets for the treatment of melanoma. FASEB J. 2006, 20, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Romano, B.; Borrelli, F.; Pagano, E.; Cascio, M.G.; Pertwee, R.G.; Izzo, A.A. Inhibition of colon carcinogenesis by a standardized Cannabis sativa extract with high content of cannabidiol. Phytomedicine 2013, 21, 631–639. [Google Scholar] [CrossRef]
- Ligresti, A.; Moriello, A.S.; Starowicz, K.; Matias, I.; Pisanti, S.; De Petrocellis, L.; Laezza, C.; Portella, G.; Bifulco, M.; Di Marzo, V. Antitumor Activity of Plant Cannabinoids with Emphasis on the Effect of Cannabidiol on Human Breast Carcinoma. J. Pharmacol. Exp. Ther. 2006, 318, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Velasco, G.; Sánchez, C.; Guzmán, M. Towards the use of cannabinoids as antitumour agents. Nat. Rev. Cancer 2012, 12, 436–444. [Google Scholar] [CrossRef]
- Nabissi, M.; Morelli, M.B.; Offidani, M.; Amantini, C.; Gentili, S.; Soriani, A.; Cardinali, C.; Leoni, P.; Santoni, G. Cannabinoids synergize with carfilzomib, reducing multiple myeloma cells viability and migration. Oncotarget 2016, 7, 77543–77557. [Google Scholar] [CrossRef]
- Benito, S.B.; Seijo-Vila, M.; Caro-Villalobos, M.; Tundidor, I.; Andradas, C.; García-Taboada, E.; Wade, J.; Smith, S.; Guzmán, M.; Pérez-Gómez, E.; et al. Appraising the “entourage effect”: Antitumor action of a pure cannabinoid versus a botanical drug preparation in preclinical models of breast cancer. Biochem. Pharmacol. 2018, 157, 285–293. [Google Scholar] [CrossRef]
- Zuardi, A.W.; Shirakawa, I.; Finkelfarb, E.; Karniol, I.G. Action of cannabidiol on the anxiety and other effects produced by delta 9-THC in normal subjects. Psychopharmacology 1982, 76, 245–250. [Google Scholar] [CrossRef]
- Russo, E.; Guy, G.W. A tale of two cannabinoids: The therapeutic rationale for combining tetrahydrocannabinol and cannabidiol. Med. Hypotheses 2006, 66, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Anis, O.; Vinayaka, A.; Shalev, N.; Namdar, D.; Nadarajan, S.; Anil, S.; Cohen, O.; Belausov, E.; Ramon, J.; Gati, E.M.; et al. Cannabis-Derived Compounds Cannabichromene and Δ9-Tetrahydrocannabinol Interact and Exhibit Cytotoxic Activity against Urothelial Cell Carcinoma Correlated with Inhibition of Cell Migration and Cytoskeleton Organization. Molecules 2021, 26, 465. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.B. Taming THC: Potential cannabis synergy and phytocannabinoid-terpenoid entourage effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef]
- Russo, E.B. The Case for the Entourage Effect and Conventional Breeding of Clinical Cannabis: No “Strain,” No Gain. Front. Plant Sci. 2019, 9, 1969. [Google Scholar] [CrossRef]
- Koltai, H.; Namdar, D. Cannabis Phytomolecule ‘Entourage’: From Domestication to Medical Use. Trends Plant Sci. 2020, 25, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qin, Y.; Pan, Z.; Li, M.; Liu, X.; Chen, X.; Qu, G.; Zhou, L.; Xu, M.; Zheng, Q.; et al. Cannabidiol Induces Cell Cycle Arrest and Cell Apoptosis in Human Gastric Cancer SGC-7901 Cells. Biomolecules 2019, 9, 302. [Google Scholar] [CrossRef]
- Schoeman, R.; Beukes, N.; Frost, C. Cannabinoid Combination Induces Cytoplasmic Vacuolation in MCF-7 Breast Cancer Cells. Molecules 2020, 25, 4682. [Google Scholar] [CrossRef]
- Muller, C.; Morales, P.; Reggio, P.H. Cannabinoid Ligands Targeting TRP Channels. Front. Mol. Neurosci. 2019, 11, 487. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Vellani, V.; Schiano-Moriello, A.; Marini, P.; Magherini, P.C.; Orlando, P.; Di Marzo, V. Plant-Derived Cannabinoids Modulate the Activity of Transient Receptor Potential Channels of Ankyrin Type-1 and Melastatin Type-8. J. Pharmacol. Exp. Ther. 2008, 325, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, F.; Pagano, E.; Romano, B.; Panzera, S.; Maiello, F.; Coppola, D.; De Petrocellis, L.; Buono, L.; Orlando, P.; Izzo, A.A. Colon carcinogenesis is inhibited by the TRPM8 antagonist cannabigerol, a Cannabis-derived non-psychotropic cannabinoid. Carcinogenesis 2014, 35, 2787–2797. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blal, K.; Besser, E.; Procaccia, S.; Schwob, O.; Lerenthal, Y.; Abu Tair, J.; Meiri, D.; Benny, O. The Effect of Cannabis Plant Extracts on Head and Neck Squamous Cell Carcinoma and the Quest for Cannabis-Based Personalized Therapy. Cancers 2023, 15, 497. https://doi.org/10.3390/cancers15020497
Blal K, Besser E, Procaccia S, Schwob O, Lerenthal Y, Abu Tair J, Meiri D, Benny O. The Effect of Cannabis Plant Extracts on Head and Neck Squamous Cell Carcinoma and the Quest for Cannabis-Based Personalized Therapy. Cancers. 2023; 15(2):497. https://doi.org/10.3390/cancers15020497
Chicago/Turabian StyleBlal, Kifah, Elazar Besser, Shiri Procaccia, Ouri Schwob, Yaniv Lerenthal, Jawad Abu Tair, David Meiri, and Ofra Benny. 2023. "The Effect of Cannabis Plant Extracts on Head and Neck Squamous Cell Carcinoma and the Quest for Cannabis-Based Personalized Therapy" Cancers 15, no. 2: 497. https://doi.org/10.3390/cancers15020497
APA StyleBlal, K., Besser, E., Procaccia, S., Schwob, O., Lerenthal, Y., Abu Tair, J., Meiri, D., & Benny, O. (2023). The Effect of Cannabis Plant Extracts on Head and Neck Squamous Cell Carcinoma and the Quest for Cannabis-Based Personalized Therapy. Cancers, 15(2), 497. https://doi.org/10.3390/cancers15020497





