The Role of Intratumor Microbiomes in Cervical Cancer Metastasis
Abstract
:Simple Summary
Abstract
1. Introduction
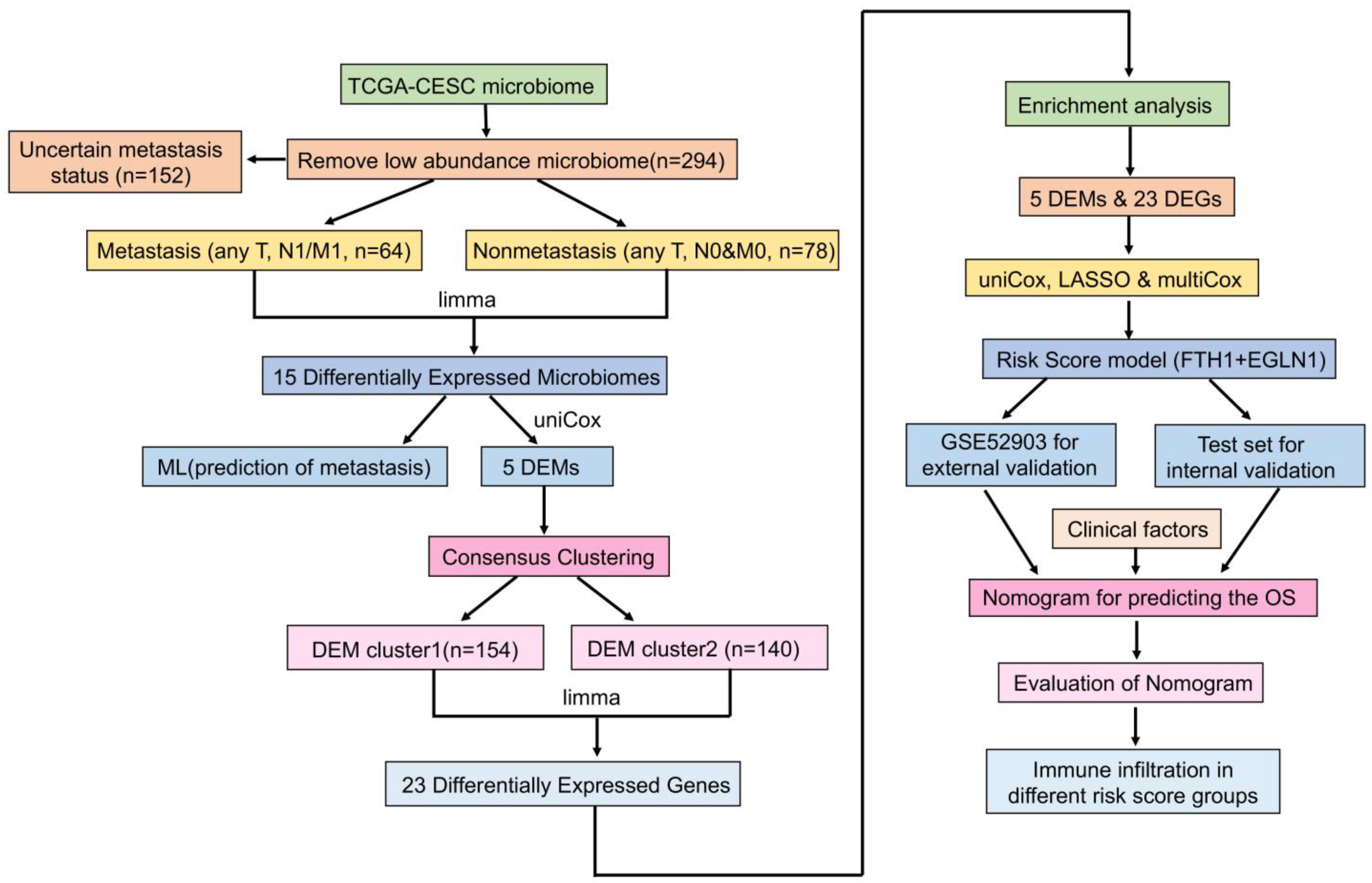
2. Materials and Methods
2.1. Preparation of Data
2.2. Identification of Tumor Metastasis-Associated Microbiomes and Evaluation of Machine Learning Classification Models
2.3. Construction and Evaluation of Nomogram
2.4. Tumor Typing Based on Metastasis-Associated Microbiomes
2.5. Immune Infiltration of Tumors with Different DEM Clusters
2.6. Gene Expression Analysis and Functional Enrichment of Tumors with Different DEM Clusters
2.7. Development and Validation of a Risk Scoring System Based on DEM and DEG
2.8. Validation and Development of a Prognostic Nomogram for Cervical Cancer
2.9. The Relationship between Risk Score and Immune Infiltration
2.10. Statistical Analysis
3. Results
3.1. Identification of Microbiomes Associated with Cervical Cancer Metastasis
3.2. Model Construction and Feature Selection in Machine Learning
3.3. The Nomogram Model for Prediction of Cervical Cancer Metastasis
3.4. DEM-Based Tumor Typing and Prognosis, Immune Infiltration
3.5. The DEGs and Functional Enrichment Derived from the DEM Cluster
3.6. The Development and Validation of a Cervical Cancer Prognostic Risk Score Model
3.7. Nomogram Model for Cervical Cancer Prognosis and Validation
3.8. The Relationship between Prognostic Risk Score and Immune Cell Infiltration
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; de Sanjosé, S.; Fakhry, C.; Monk, B.J.; Stanley, M.A.; Franceschi, S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Prim. 2016, 2, 16086. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; MacIntyre, D.A.; Marchesi, J.R.; Lee, Y.S.; Bennett, P.R.; Kyrgiou, M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: What do we know and where are we going next? Microbiome 2016, 4, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Luo, M.; Zhang, Y.; Cao, G.; Wang, S. Association of high-risk human papillomavirus infection duration and cervical lesions with vaginal microbiota composition. Ann. Transl. Med. 2020, 8, 1161. [Google Scholar] [CrossRef]
- So, K.A.; Yang, E.J.; Kim, N.R.; Hong, S.R.; Lee, J.H.; Hwang, C.S.; Shim, S.H.; Lee, S.J.; Kim, T.J. Changes of vaginal microbiota during cervical carcinogenesis in women with human papillomavirus infection. PLoS ONE 2020, 15, e0238705. [Google Scholar] [CrossRef]
- Qian, X.B.; Chen, T.; Xu, Y.P.; Chen, L.; Sun, F.X.; Lu, M.P.; Liu, Y.X. A guide to human microbiome research: Study design, sample collection, and bioinformatics analysis. Chin. Med. J. 2020, 133, 1844–1855. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Livyatan, I.; Nejman, D.; Shental, N.; Straussman, R. Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. Oncoimmunology 2020, 9, 1800957. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e26. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Olawaiye, A.B.; Baker, T.P.; Washington, M.K.; Mutch, D.G. The new (Version 9) American Joint Committee on Cancer tumor, node, metastasis staging for cervical cancer. CA Cancer J. Clin. 2021, 71, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Medina-Martinez, I.; Barrón, V.; Roman-Bassaure, E.; Juárez-Torres, E.; Guardado-Estrada, M.; Espinosa, A.M.; Bermudez, M.; Fernández, F.; Venegas-Vega, C.; Orozco, L.; et al. Impact of gene dosage on gene expression, biological processes and survival in cervical cancer: A genome-wide follow-up study. PLoS ONE 2014, 9, e97842. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, J.; Ge, L.; Hu, Q. Identification of Immune-Related Biomarkers for Sciatica in Peripheral Blood. Front. Genet. 2021, 12, 781945. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.-X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Rokos, T.; Holubekova, V.; Kolkova, Z.; Hornakova, A.; Pribulova, T.; Kozubik, E.; Biringer, K.; Kudela, E. Is the Physiological Composition of the Vaginal Microbiome Altered in High-Risk HPV Infection of the Uterine Cervix? Viruses 2022, 14, 2130. [Google Scholar] [CrossRef] [PubMed]
- Frąszczak, K.; Barczyński, B.; Kondracka, A. Does Lactobacillus Exert a Protective Effect on the Development of Cervical and Endometrial Cancer in Women? Cancers 2022, 14, 4909. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Wu, Y.; Li, M.; An, F.; Yao, L.; Wang, M.; Wang, X.; Yuan, J.; Jiang, K.; Li, W.; et al. Lactobacillus spp. create a protective micro-ecological environment through regulating the core fucosylation of vaginal epithelial cells against cervical cancer. Cell Death Dis. 2021, 12, 1094. [Google Scholar] [CrossRef] [PubMed]
- Wong-Rolle, A.; Wei, H.K.; Zhao, C.; Jin, C. Unexpected guests in the tumor microenvironment: Microbiome in cancer. Protein Cell 2021, 12, 426–435. [Google Scholar] [CrossRef]
- Hermida, L.C.; Gertz, E.M.; Ruppin, E. Predicting cancer prognosis and drug response from the tumor microbiome. Nat. Commun. 2022, 13, 2896. [Google Scholar] [CrossRef]
- Cheng, T.; Huang, S. Roles of Non-Coding RNAs in Cervical Cancer Metastasis. Front. Oncol. 2021, 11, 646192. [Google Scholar] [CrossRef]
- Leone, L.; Mazzetta, F.; Martinelli, D.; Valente, S.; Alimandi, M.; Raffa, S.; Santino, I. Klebsiella pneumoniae Is Able to Trigger Epithelial-Mesenchymal Transition Process in Cultured Airway Epithelial Cells. PLoS ONE 2016, 11, e0146365. [Google Scholar] [CrossRef] [Green Version]
- Weniger, M.; Hank, T.; Qadan, M.; Ciprani, D.; Michelakos, T.; Niess, H.; Heiliger, C.; Ilmer, M.; D’Haese, J.G.; Ferrone, C.R.; et al. Influence of Klebsiella pneumoniae and quinolone treatment on prognosis in patients with pancreatic cancer. Br. J. Surg. 2021, 108, 709–716. [Google Scholar]
- Hu, M.; Bai, W.; Zhao, C.; Wang, J. Distribution of esophagus flora in esophageal squamous cell carcinoma and its correlation with clinicopathological characteristics. Transl. Cancer Res. 2020, 9, 3973–3985. [Google Scholar] [CrossRef]
- Kaur, C.P.; Vadivelu, J.; Chandramathi, S. Impact of Klebsiella pneumoniae in lower gastrointestinal tract diseases. J. Dig. Dis. 2018, 19, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Mansour, B.; Monyók, Á.; Makra, N.; Gajdács, M.; Vadnay, I.; Ligeti, B.; Juhász, J.; Szabó, D.; Ostorházi, E. Bladder cancer-related microbiota: Examining differences in urine and tissue samples. Sci. Rep. 2020, 10, 11042. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- López-Filloy, M.; Cortez, F.J.; Gheit, T.; Cruz, Y.C.O.; Cruz-Talonia, F.; Chávez-Torres, M.; Arteaga-Gómez, C.; Mancilla-Herrera, I.; Montesinos, J.J.; Cortés-Morales, V.A.; et al. Altered Vaginal Microbiota Composition Correlates With Human Papillomavirus and Mucosal Immune Responses in Women With Symptomatic Cervical Ectopy. Front. Cell. Infect. Microbiol. 2022, 12, 884272. [Google Scholar] [CrossRef]
- Tian, Y.; Lu, J.; Hao, X.; Li, H.; Zhang, G.; Liu, X.; Li, X.; Zhao, C.; Kuang, W.; Chen, D.; et al. FTH1 Inhibits Ferroptosis Through Ferritinophagy in the 6-OHDA Model of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1796–1812. [Google Scholar] [CrossRef]
- Ma, R.; Fang, L.; Chen, L.; Wang, X.; Jiang, J.; Gao, L. Ferroptotic stress promotes macrophages against intracellular bacteria. Theranostics 2022, 12, 2266–2289. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]








| Characteristic | Non-Metastasis | Metastasis | All * |
|---|---|---|---|
| n | 78 | 64 | 294 |
| M, n (%) | |||
| M0 | 78 (100%) | 25 (39.1%) | 111 (37.8%) |
| M1 | 0 (0%) | 9 (14.1%) | 9 (3.1%) |
| MX | 0 (0%) | 30 (46.8%) | 174 (59.1%) |
| N, n (%) | |||
| N0 | 78 (100%) | 2 (3.1%) | 129 (43.9%) |
| N1 | 0 (0%) | 57 (89.1%) | 57 (19.4%) |
| NX | 0 (0%) | 5 (7.8%) | 108 (36.7%) |
| T, n (%) | |||
| T1 | 56 (71.8%) | 30 (46.9%) | 134 (45.6%) |
| T2 | 20 (25.6%) | 22 (34.4%) | 71 (24.2%) |
| T3 | 2 (2.6%) | 7 (10.9%) | 20 (6.8%) |
| T4 | 0 (0%) | 3 (4.7%) | 8 (2.7%) |
| Tis | 0 (0%) | 1 (1.6%) | 1 (0.3%) |
| TX | 0 (0%) | 1 (1.6%) | 60 (20.4%) |
| age, mean ± SD | 47.26 ± 12.88 | 45.36 ± 11.93 | 48.22 ± 13.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Duan, B.; Jia, P.; Zhang, Y.; Yan, X. The Role of Intratumor Microbiomes in Cervical Cancer Metastasis. Cancers 2023, 15, 509. https://doi.org/10.3390/cancers15020509
Jiang L, Duan B, Jia P, Zhang Y, Yan X. The Role of Intratumor Microbiomes in Cervical Cancer Metastasis. Cancers. 2023; 15(2):509. https://doi.org/10.3390/cancers15020509
Chicago/Turabian StyleJiang, Lu, Baofeng Duan, Peng Jia, Yan Zhang, and Xin Yan. 2023. "The Role of Intratumor Microbiomes in Cervical Cancer Metastasis" Cancers 15, no. 2: 509. https://doi.org/10.3390/cancers15020509






