Breast Cancer Molecular Subtypes Differentially Express Gluconeogenic Rate-Limiting Enzymes—Obesity as a Crucial Player
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Ethical Approval
2.2. Sample Characteristics
2.3. Hematoxylin–Eosin Staining
2.4. Immunohistochemistry Assays
2.5. Statistical Analysis
3. Results
3.1. Demographic and Clinicopathological Data
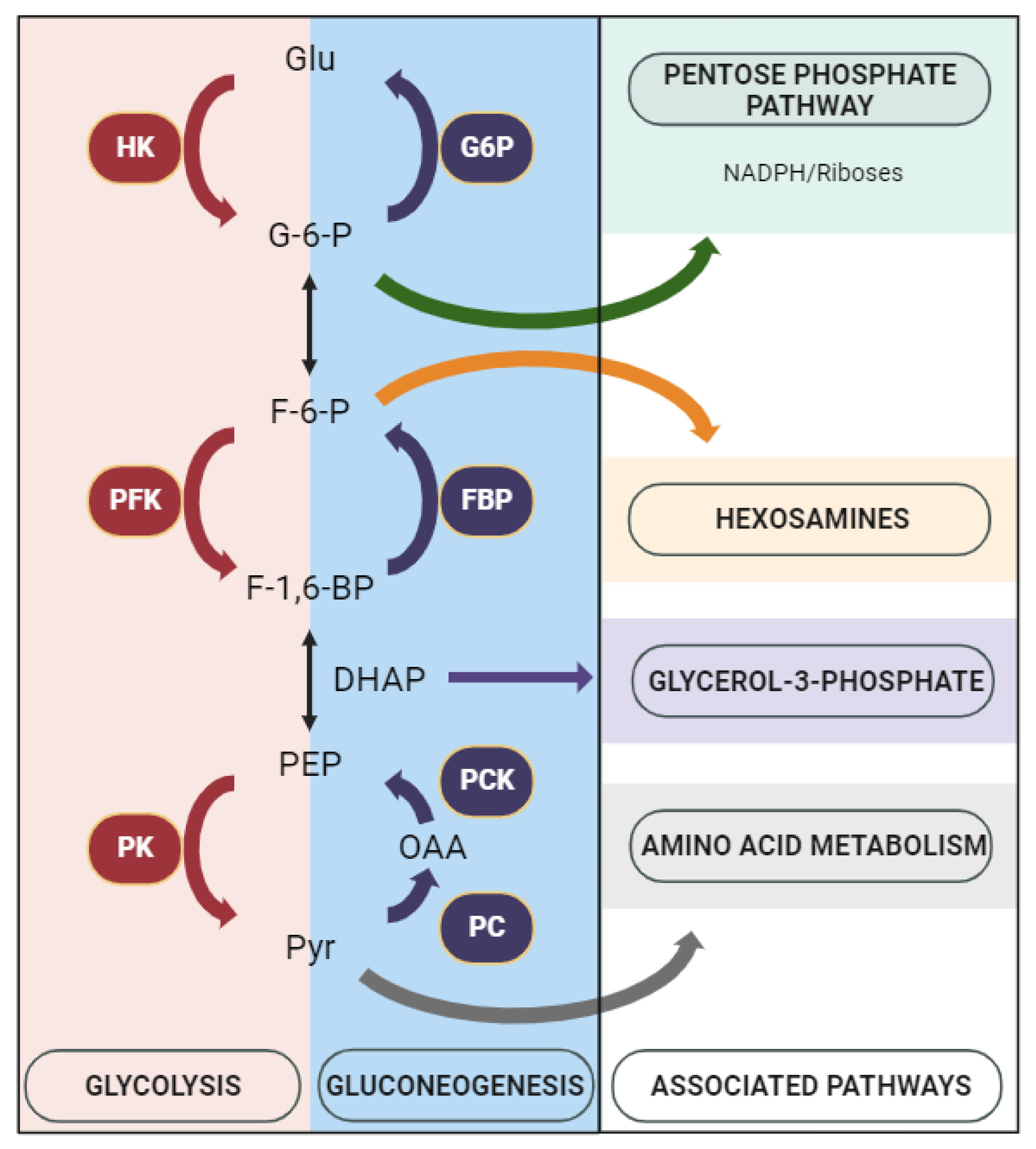
3.2. Expression of Glycolysis and Gluconeogenesis Rate-Limiting Enzymes
3.3. Correlation between Rate-Limiting Enzymes Expression and Clinicopathological Features
3.4. Intratumor Adipose Tissue Deposition
3.5. Inflammatory Infiltration
3.6. Correlation between Tumor Cell Enzymatic Expression and Obesity-Associated Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- James, W.P.T. WHO recognition of the global obesity epidemic. Int. J. Obes. 2008, 32, S120–S126. [Google Scholar] [CrossRef] [PubMed]
- Tzenios, N. Obesity as a risk factor for different types of cancer. EPRA Int. J. Multidiscip. Res. (IJMR) 2023, 8, 97–100. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 3. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Health Statistics 2022: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Argolo, D.F.; Hudis, C.A.; Iyengar, N.M. The impact of obesity on breast cancer. Curr. Oncol. Rep. 2018, 20, 47. [Google Scholar] [CrossRef]
- Yager, J.D.; Davidson, N.E. Estrogen Carcinogenesis in Breast Cancer. N. Engl. J. Med. 2006, 354, 270–282. [Google Scholar] [CrossRef]
- Cao, H. Adipocytokines in obesity and metabolic disease. J. Endocrinol. 2014, 220, 2. [Google Scholar] [CrossRef] [PubMed]
- Eliyatkin, N.; Yalcin, E.; Zengel, B.; Aktaş, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Warburg, O. The metabolism of carcinoma cells. J Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Semenza, G.L. Tumor metabolism: Cancer cells give and take lactate. J. Clin. Investig. 2008, 118, 3835–3837. [Google Scholar] [CrossRef]
- Pavlides, S.; Whitaker-Menezes, D.; Castello-Cros, R.; Flomenberg, N.; Witkiewicz, A.K.; Frank, P.G.; Casimiro, M.C.; Wang, C.; Fortina, P.; Addya, S.; et al. The reverse Warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009, 8, 3984–4001. [Google Scholar] [CrossRef] [PubMed]
- Luis, C.; Duarte, F.; Faria, I.; Jarak, I.; Oliveira, P.F.; Alves, M.G.; Soares, R.; Fernandes, R. Warburg Effect Inversion: Adiposity shifts central primary metabolism in MCF-7 breast cancer cells. Life Sci. 2019, 223, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Jelliffe, D.B.; Jelliffe, E.F. Underappreciated pioneers. Quetelet: Man and index. Am. J. Clin. Nutr. 1979, 32, 2519–2521. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Edge, S.B.; Giuliano, A. New and Important Changes in the TNM Staging System for Breast Cancer. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 457–467. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412. [Google Scholar] [CrossRef]
- Rybinska, I.; Agresti, R.; Trapani, A.; Tagliabue, E.; Triulzi, T. Adipocytes in breast cancer, the thick and the thin. Cells 2020, 9, 560. [Google Scholar] [CrossRef]
- Jimenez, R.E.; Wallis, T.; Visscher, D.W. Centrally necrotizing carcinomas of the breast: A distinct histologic subtype with aggressive clinical behavior. Am. J. Surg. Pathol. 2001, 25, 331–337. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, J.; Zhao, Q.; Fu, S.; Jin, J. Emerging roles of aerobic glycolysis in breast cancer. Clin. Transl. Oncol. 2020, 22, 631–646. [Google Scholar] [CrossRef]
- Shi, L.; Zhao, C.; Pu, H.; Zhang, Q. FBP1 expression is associated with basal-like breast carcinoma. Oncol. Lett. 2017, 13, 3046–3056. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.S.; Kim, N.H.; Yun, J.S.; Cho, S.B.; Kim, H.S.; Yook, J.I. Breast Cancer Subtypes Underlying EMT-Mediated Catabolic Metabolism. Cells 2020, 9, 2064. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, V.; Iorio, E.; Miodini, P.; Silvestri, M.; Dugo, M.; Daidone, M.G. Metabolic Footprints and Molecular Subtypes in Breast Cancer. Dis. Markers 2017, 2017, 7687851. [Google Scholar] [CrossRef] [PubMed]
- Phannasil, P.; Israr-ul, H.A.; El Azzouny, M.; Longacre, M.J.; Rattanapornsompong, K.; Burant, C.F.; MacDonald, M.J.; Jitrapakdee, S. Mass spectrometry analysis shows the biosynthetic pathways supported by pyruvate carboxylase in highly invasive breast cancer cells. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef]
- Hsu, H.P.; Chu, P.Y.; Chang, T.M.; Huang, K.W.; Hung, W.C.; Jiang, S.S.; Lin, H.Y.; Tsai, H.J. Mitochondrial phosphoenolpyruvate carboxykinase promotes tumor growth in estrogen receptor-positive breast cancer via regulation of the mTOR pathway. Cancer Med. 2023, 12, 1588–1601. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Meng, S.; Xiang, M.; Ma, H. Phosphoenolpyruvate carboxykinase in cell metabolism: Roles and mechanisms beyond gluconeogenesis. Mol. Metab. 2021, 53, 101257. [Google Scholar] [CrossRef]
- Ferrante, A.W. Obesity-induced inflammation: A metabolic dialogue in the language of inflammation. J. Intern. Med. 2007, 262, 408–414. [Google Scholar] [CrossRef]
- Rybinska, I.; Mangano, N.; Tagliabue, E.; Triulzi, T. Cancer-associated adipocytes in breast cancer: Causes and consequences. Int. J. Mol. Sci. 2021, 22, 3775. [Google Scholar] [CrossRef]
- Crespi, E.; Bottai, G.; Santarpia, L. Role of inflammation in obesity-related breast cancer. Curr. Opin. Pharmacol. 2016, 31, 114–122. [Google Scholar] [CrossRef]
- Blücher, C.; Stadler, S.C. Obesity and breast cancer: Current insights on the role of fatty acids and lipid metabolism in promoting breast cancer growth and progression. Front. Endocrinol. 2017, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- García-Estévez, L.; Cortés, J.; Pérez, S.; Calvo, I.; Gallegos, I.; Moreno-Bueno, G. Obesity and Breast Cancer: A Paradoxical and Controversial Relationship Influenced by Menopausal Status. Front. Oncol. 2021, 11, 705911. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.H.; Domingo-Vidal, M.; Roche, M.; Lin, Z.; Whitaker-Menezes, D.; Seifert, E.; Capparelli, C.; Tuluc, M.; Birbe, R.C.; Tassone, P.; et al. TP53-inducible glycolysis and apoptosis regulator (TIGAR) metabolically reprograms carcinoma and stromal cells in breast cancer. J. Biol. Chem. 2016, 291, 26291–26303. [Google Scholar] [CrossRef] [PubMed]
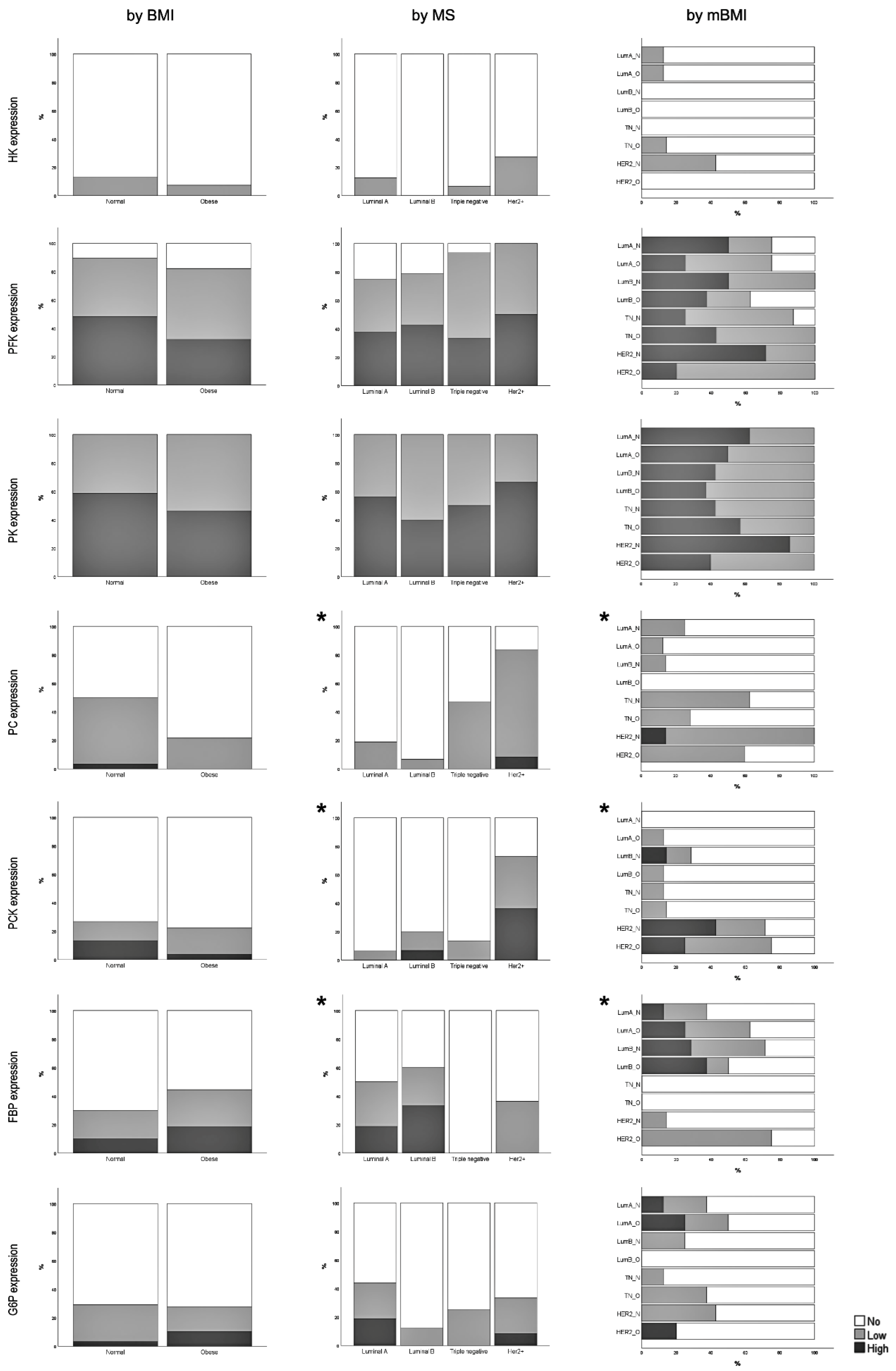

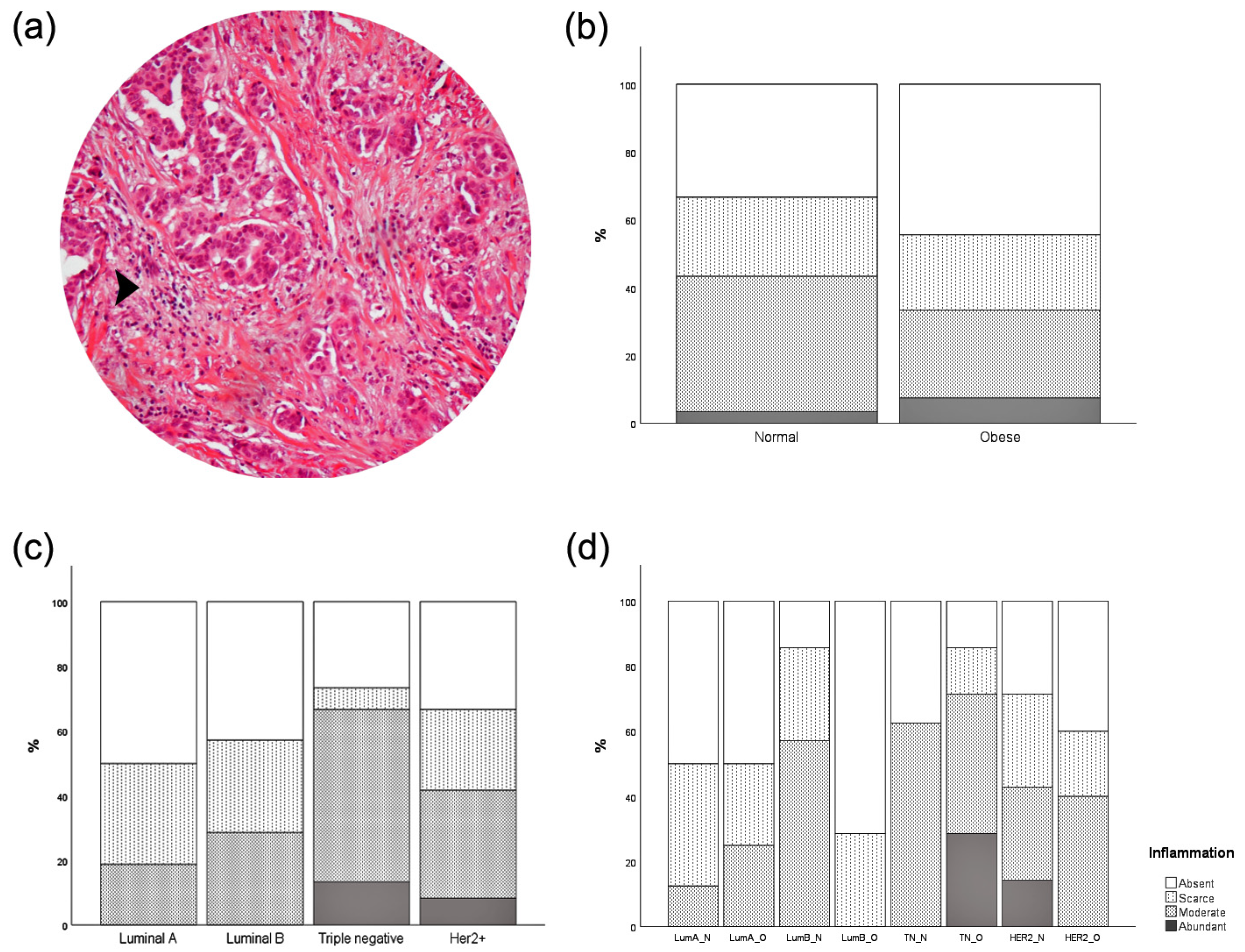
| n (%) | Total | p-Value BMI Analysis | p-Value MS Analysis | p-Value mBMI Analysis |
|---|---|---|---|---|
| 62 (100.0%) | ||||
| Age at diagnosis | ||||
| (Mean ± SD) | 57.48 ± 10.53 | 0.480 | 0.218 | 0.604 |
| BMI categories | ||||
| Normal weight | 32 (51.6%) | - | 0.974 | - |
| Obese | 30 (48.4%) | |||
| Molecular subtypes | ||||
| Luminal A | 16 (25.8%) | 0.974 | - | - |
| Luminal B | 16 (25.8%) | |||
| Triple negative | 16 (25.8%) | |||
| HER2+ | 14 (22.6%) | |||
| Topographic localization | ||||
| CQ | 3 (4.8%) | 0.680 | 0.052 | 0.075 |
| IOQ | 4 (6.5%) | |||
| SOQ | 21 (33.9%) | |||
| SIQ | 4 (6.5%) | |||
| Multiple | 27 (43.5%) | |||
| Other | 3 (4.8%) | |||
| Laterality | ||||
| Right | 32 (51.6%) | 0.806 | 0.632 | 0.075 |
| Left | 30 (48.4%) | |||
| Pathological stage | ||||
| Stage I | 36 (59.0%) | 0.321 | 0.037 * | 0.051 |
| Stage II | 22 (36.1%) | |||
| Stage III | 3 (4.9%) | |||
| Histological grade | ||||
| Grade I Grade II | 0 (0%) 37 (59.7%) | 0.227 | <0.001 * | <0.001 * |
| Grade III | 25 (40.3%) | |||
| Carcinoma in situ | ||||
| Presence | 24 (38.7%) | 0.469 | 0.282 | 0.357 |
| Size. cm (mean ± SD) | 1.35 ± 2.47 | 0.649 | 0.798 | 0.463 |
| Extensive | 20 (32.3%) | 0.472 | 0.367 | 0.253 |
| Microcalcifications | 28 (45.2%) | 0.818 | 0.236 | 0.333 |
| Necrosis | 39 (62.9%) | 0.946 | 0.042 * | 0.126 |
| Invasive carcinoma | ||||
| Size. cm (mean ± SD) | 1.76 ± 0.75 | 0.465 | 0.197 | 0.210 |
| Multifocal | 14 (22.6%) | 0.891 | 0.564 | 0.656 |
| Invasion | 19 (30.6%) | 0.511 | 0.661 | 0.783 |
| Metastatic nodes | ||||
| 0 | 41 (66.2%) | 0.364 | 0.144 | 0.082 |
| 1–3 | 18 (29.0%) | |||
| 4–9 | 1 (1.6%) | |||
| >10 | 2 (3.2%) | |||
| Size cm (mean ± SD) (Larger metastasis) | 11.48 ± 31.49 | 0.764 | 0.056 | 0.340 |
| HK | PFK | PK | PC | PCK | FBP | G6P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | |
| BMI | −0.147 | 0.275 | −0.162 | 0.229 | −0.059 | 0.650 | −0.303 | 0.021 * | −0.138 | 0.305 | 0.138 | 0.305 | 0.047 | 0.720 |
| ER | −0.151 | 0.268 | −0.177 | 0.192 | −0.168 | 0.196 | −0.514 | <0.001 * | −0.284 | 0.034 * | 0.463 | <0.001 * | 0.020 | 0.881 |
| PR | −0.151 | 0.268 | −0.177 | 0.192 | −0.168 | 0.196 | −0.514 | <0.001 * | −0.284 | 0.034 * | 0.463 | <0.001 * | 0.020 | 0.881 |
| HER2 | 0.277 | 0.039 * | 0.259 | 0.054 | 0.139 | 0.285 | 0.503 | <0.001 * | 0.569 | <0.001 * | −0.075 | 0.580 | 0.059 | 0.658 |
| Ki67 | −0.045 | 0.799 | 0.122 | 0.486 | 0.123 | 0.463 | 0.089 | 0.604 | 0.288 | 0.093 | −0.038 | 0.829 | 0.009 | 0.956 |
| TL | 0.043 | 0.752 | 0.185 | 0.169 | 0.132 | 0.306 | 0.105 | 0.433 | −0.141 | 0.296 | −0.209 | 0.119 | −0.209 | 0.110 |
| LAT | −0.086 | 0.524 | 0.113 | 0.401 | 0.131 | 0.312 | −0.084 | 0.529 | −0.028 | 0.834 | 0.156 | 0.246 | −0.046 | 0.726 |
| pST | 0.053 | 0.698 | 0.138 | 0.310 | 0.263 | 0.041 * | 0.437 | <0.001 * | 0.185 | 0.172 | −0.215 | 0.111 | 0.154 | 0.246 |
| HG | 0.078 | 0.566 | 0.106 | 0.434 | 0.120 | 0.353 | 0.452 | <0.001 * | 0.238 | 0.075 | −0.396 | 0.002 * | 0.014 | 0.918 |
| HGnc | 0.117 | 0.384 | 0.198 | 0.139 | 0.137 | 0.287 | 0.249 | 0.060 | 0.193 | 0.149 | −0.304 | 0.021 * | 0.059 | 0.656 |
| HGm | 0.050 | 0.711 | 0.032 | 0.812 | 0.034 | 0.796 | 0.392 | 0.002 * | 0.272 | 0.041 * | −0.320 | 0.015 * | −0.007 | 0.955 |
| HGtf | −0.083 | 0.541 | −0.021 | 0.875 | 0.090 | 0.489 | 0.243 | 0.066 | 0.036 | 0.793 | −0.144 | 0.286 | −0.020 | 0.881 |
| ISs | −0.016 | 0.906 | −0.120 | 0.373 | −0.094 | 0.469 | −0.071 | 0.595 | −0.151 | 0.263 | 0.088 | 0.514 | 0.230 | 0.077 |
| ISext | 0.026 | 0.846 | −0.027 | 0.840 | −0.063 | 0.624 | −0.166 | 0.212 | −0.173 | 0.198 | 0.142 | 0.293 | 0.212 | 0.194 |
| ISmc | 0.122 | 0.365 | −0.082 | 0.546 | −0.187 | 0.146 | −0.148 | 0.267 | −0.143 | 0.289 | 0.183 | 0.172 | 0.063 | 0.631 |
| ISnec | 0.154 | 0.254 | 0.098 | 0.468 | −0.091 | 0.483 | 0.196 | 0.139 | 0.025 | 0.854 | 0.032 | 0.813 | 0.026 | 0.842 |
| INVs | −0.016 | 0.907 | 0.043 | 0.753 | 0.155 | 0.229 | 0.242 | 0.067 | 0.055 | 0.683 | −0.237 | 0.075 | 0.210 | 0.108 |
| INVm | 0.234 | 0.080 | −0.137 | 0.309 | −0.053 | 0.680 | −0.031 | 0.816 | 0.014 | 0.918 | −0.014 | 0.920 | 0.109 | 0.407 |
| INVvi | 0.269 | 0.043 * | 0.160 | 0.234 | 0.094 | 0.467 | 0.077 | 0.567 | −0.027 | 0.840 | −0.012 | 0.930 | 0.059 | 0.656 |
| METs | −0.090 | 0.504 | 0.089 | 0.509 | −0.093 | 0.474 | 0.048 | 0.720 | −0.154 | 0.253 | 0.009 | 0.947 | 0.076 | 0.564 |
| Enzymes | χ2 Test | Spearman Correlation | |
|---|---|---|---|
| p-Value | R | p-Value | |
| HK | 0.635 | −0.038 | 0.784 |
| PFK | 0.509 | −0.054 | 0.690 |
| PK | 0.318 | −0.113 | 0.403 |
| PC | 0.580 | 0.035 | 0.796 |
| PCK | 0.590 | −0.111 | 0.415 |
| FBP | 0.222 | −0.190 | 0.160 |
| G6P | 0.569 | −0.049 | 0.718 |
| Enzymes | χ2 Test | Spearman Correlation | |
|---|---|---|---|
| p-Value | r | p-Value | |
| HK | 0.411 | 0.209 | 0.126 |
| PFK | 0.739 | 0.117 | 0.391 |
| PK | 0.264 | −0.017 | 0.900 |
| PC | <0.001 * | 0.248 | 0.062 |
| PCK | 0.411 | 0.179 | 0.186 |
| FBP | 0.465 | −0.071 | 0.604 |
| G6P | 0.938 | 0.097 | 0.471 |
| n (%) | Total | p-Value BMI Analysis | p-Value MS Analysis | p-Value mBMI Analysis |
|---|---|---|---|---|
| 62 (100.0%) | ||||
| Diabetes | ||||
| Yes | 7 (11.5%) | 0.589 | 0.883 | 0.791 |
| No | 54 (88.5%) | |||
| Hypertension | ||||
| Yes | 25 (41.0%) | 0.008 * | 0.949 | 0.088 |
| No | 36 (59.0%) | |||
| Dyslipidemia | ||||
| Yes | 39 (63.9%) | 0.175 | 0.495 | 0.549 |
| No | 22 (36.1%) |
| Enzymes | Diabetes | Hypertension | Dyslipidemia | |||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| HK | −0.131 | 0.337 | −0.065 | 0.635 | 0.079 | 0.564 |
| PFK | 0.070 | 0.607 | 0.177 | 0.187 | 0.180 | 0.180 |
| PK | −0.099 | 0.447 | 0.025 | 0.849 | 0.029 | 0.822 |
| PC | −0.035 | 0.794 | −0.142 | 0.287 | 0.040 | 0.768 |
| PCK | 0.304 | 0.022 * | 0.189 | 0.159 | 0.157 | 0.244 |
| FBP | 0.057 | 0.676 | 0.320 | 0.015 * | 0.328 | 0.013 * |
| G6P | 0.240 | 0.065 | 0.082 | 0.531 | 0.149 | 0.257 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luís, C.; Schmitt, F.; Fernandes, R.; Coimbra, N.; Rigor, J.; Dias, P.; Leitão, D.; Fernandes, R.; Soares, R. Breast Cancer Molecular Subtypes Differentially Express Gluconeogenic Rate-Limiting Enzymes—Obesity as a Crucial Player. Cancers 2023, 15, 4936. https://doi.org/10.3390/cancers15204936
Luís C, Schmitt F, Fernandes R, Coimbra N, Rigor J, Dias P, Leitão D, Fernandes R, Soares R. Breast Cancer Molecular Subtypes Differentially Express Gluconeogenic Rate-Limiting Enzymes—Obesity as a Crucial Player. Cancers. 2023; 15(20):4936. https://doi.org/10.3390/cancers15204936
Chicago/Turabian StyleLuís, Carla, Fernando Schmitt, Rute Fernandes, Nuno Coimbra, Joana Rigor, Paula Dias, Dina Leitão, Rúben Fernandes, and Raquel Soares. 2023. "Breast Cancer Molecular Subtypes Differentially Express Gluconeogenic Rate-Limiting Enzymes—Obesity as a Crucial Player" Cancers 15, no. 20: 4936. https://doi.org/10.3390/cancers15204936
APA StyleLuís, C., Schmitt, F., Fernandes, R., Coimbra, N., Rigor, J., Dias, P., Leitão, D., Fernandes, R., & Soares, R. (2023). Breast Cancer Molecular Subtypes Differentially Express Gluconeogenic Rate-Limiting Enzymes—Obesity as a Crucial Player. Cancers, 15(20), 4936. https://doi.org/10.3390/cancers15204936










