CD44 Intracellular Domain: A Long Tale of a Short Tail
Abstract
:Simple Summary
Abstract
1. Introduction
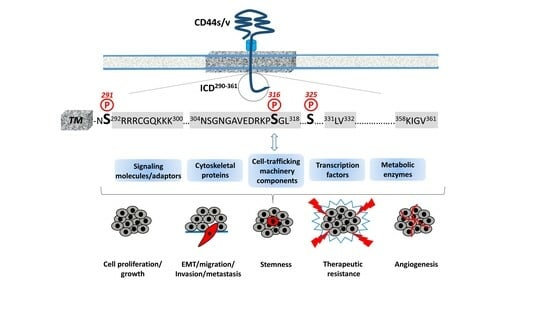
2. Structural Features of CD44 ICD
3. Hyaluronan Internalization and Interactions with the Cell-Trafficking Machinery
4. Regulation of Cytoskeletal Organization and Cell Phenotype
5. Regulation of Cell–Cell Contact Inhibition and Cell Growth
6. Cleavage and Intracellular Release of CD44 ICD: A Master Regulator of the Transcriptome
7. Regulation of Cell Metabolism
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J.I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell 1989, 56, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Isacke, C.M. The role of the cytoplasmic domain in regulating CD44 function. J. Cell Sci. 1994, 107 Pt 9, 2353–2359. [Google Scholar] [CrossRef] [PubMed]
- Thorne, R.F.; Legg, J.W.; Isacke, C.M. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 2004, 117, 373–380. [Google Scholar] [CrossRef]
- Heldin, P.; Kolliopoulos, C.; Lin, C.Y.; Heldin, C.H. Involvement of hyaluronan and CD44 in cancer and viral infections. Cell Signal. 2020, 65, 109427. [Google Scholar] [CrossRef]
- Kincade, P.W.; Zheng, Z.; Katoh, S.; Hanson, L. The importance of cellular environment to function of the CD44 matrix receptor. Curr. Opin. Cell Biol. 1997, 9, 635–642. [Google Scholar] [CrossRef]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef]
- Elkashty, O.A.; Elghanam, G.A.; Su, X.; Liu, Y.; Chauvin, P.J.; Tran, S.D. Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas. Carcinogenesis 2020, 41, 458–466. [Google Scholar] [CrossRef]
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Li, J.; Zhou, B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal. 2019, 63, 109377. [Google Scholar] [CrossRef] [PubMed]
- Lesley, J.; He, Q.; Miyake, K.; Hamann, A.; Hyman, R.; Kincade, P.W. Requirements for hyaluronic acid binding by CD44: A role for the cytoplasmic domain and activation by antibody. J. Exp. Med. 1992, 175, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Neame, S.J.; Isacke, C.M. The cytoplasmic tail of CD44 is required for basolateral localization in epithelial MDCK cells but does not mediate association with the detergent-insoluble cytoskeleton of fibroblasts. J. Cell Biol. 1993, 121, 1299–1310. [Google Scholar] [CrossRef]
- Uff, C.R.; Neame, S.J.; Isacke, C.M. Hyaluronan binding by CD44 is regulated by a phosphorylation-independent mechanism. Eur. J. Immunol. 1995, 25, 1883–1887. [Google Scholar] [CrossRef]
- Lokeshwar, V.B.; Fregien, N.; Bourguignon, L.Y. Ankyrin-binding domain of CD44(GP85) is required for the expression of hyaluronic acid-mediated adhesion function. J. Cell Biol. 1994, 126, 1099–1109. [Google Scholar] [CrossRef]
- Thomas, L.; Byers, H.R.; Vink, J.; Stamenkovic, I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J. Cell Biol. 1992, 118, 971–977. [Google Scholar] [CrossRef]
- Gal, I.; Lesley, J.; Ko, W.; Gonda, A.; Stoop, R.; Hyman, R.; Mikecz, K. Role of the extracellular and cytoplasmic domains of CD44 in the rolling interaction of lymphoid cells with hyaluronan under physiologic flow. J. Biol. Chem. 2003, 278, 11150–11158. [Google Scholar] [CrossRef]
- Legg, J.W.; Isacke, C.M. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998, 8, 705–708. [Google Scholar] [CrossRef]
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998, 140, 885–895. [Google Scholar] [CrossRef]
- Sheikh, H.; Isacke, C.M. A di-hydrophobic Leu-Val motif regulates the basolateral localization of CD44 in polarized Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 1996, 271, 12185–12190. [Google Scholar] [CrossRef] [PubMed]
- Neame, S.J.; Isacke, C.M. Phosphorylation of CD44 in vivo requires both Ser323 and Ser325, but does not regulate membrane localization or cytoskeletal interaction in epithelial cells. EMBO J. 1992, 11, 4733–4738. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.A.; Townsend, P.A.; Isacke, C.M. Ca(2+)/calmodulin-dependent protein kinase mediates the phosphorylation of CD44 required for cell migration on hyaluronan. Biochem. J. 2001, 357, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Peck, D.; Isacke, C.M. Hyaluronan-dependent cell migration can be blocked by a CD44 cytoplasmic domain peptide containing a phosphoserine at position 325. J. Cell Sci. 1998, 111 Pt 11, 1595–1601. [Google Scholar] [CrossRef]
- Peck, D.; Isacke, C.M. CD44 phosphorylation regulates melanoma cell and fibroblast migration on, but not attachment to, a hyaluronan substratum. Curr. Biol. 1996, 6, 884–890. [Google Scholar] [CrossRef]
- Galandrini, R.; De Maria, R.; Piccoli, M.; Frati, L.; Santoni, A. CD44 triggering enhances human NK cell cytotoxic functions. J. Immunol. 1994, 153, 4399–4407. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Lokeshwar, V.B.; Chen, X.; Kerrick, W.G. Hyaluronic acid-induced lymphocyte signal transduction and HA receptor (GP85/CD44)-cytoskeleton interaction. J. Immunol. 1993, 151, 6634–6644. [Google Scholar] [CrossRef]
- Racine, R.R.; Manalo, N.A.; Hall, J.M.F.; Dibas, A.; Raffel, G.D.; Mummert, M.E. CD44 induced enhancement of phosphatase activity and calcium influx: Modifications of EGR-1 expression and cell proliferation. Biochem. Biophys. Rep. 2016, 6, 172–178. [Google Scholar] [CrossRef]
- Meyer, T.; Hanson, P.I.; Stryer, L.; Schulman, H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science 1992, 256, 1199–1202. [Google Scholar] [CrossRef]
- Legg, J.W.; Lewis, C.A.; Parsons, M.; Ng, T.; Isacke, C.M. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 2002, 4, 399–407. [Google Scholar] [CrossRef]
- Tzircotis, G.; Thorne, R.F.; Isacke, C.M. Directional sensing of a phorbol ester gradient requires CD44 and is regulated by CD44 phosphorylation. Oncogene 2006, 25, 7401–7410. [Google Scholar] [CrossRef] [PubMed]
- Heilker, R.; Spiess, M.; Crottet, P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999, 21, 558–567. [Google Scholar] [CrossRef]
- Tammi, R.; Rilla, K.; Pienimaki, J.P.; MacCallum, D.K.; Hogg, M.; Luukkonen, M.; Hascall, V.C.; Tammi, M. Hyaluronan enters keratinocytes by a novel endocytic route for catabolism. J. Biol. Chem. 2001, 276, 35111–35122. [Google Scholar] [CrossRef] [PubMed]
- Knudson, W.; Chow, G.; Knudson, C.B. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002, 21, 15–23. [Google Scholar] [CrossRef]
- Thankamony, S.P.; Knudson, W. Acylation of CD44 and its association with lipid rafts are required for receptor and hyaluronan endocytosis. J. Biol. Chem. 2006, 281, 34601–34609. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, D.J.; Knudson, W.; Knudson, C.B. Internalization of the hyaluronan receptor CD44 by chondrocytes. Exp. Cell Res. 1999, 252, 292–302. [Google Scholar] [CrossRef]
- Eyster, C.A.; Cole, N.B.; Petersen, S.; Viswanathan, K.; Fruh, K.; Donaldson, J.G. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell 2011, 22, 3218–3230. [Google Scholar] [CrossRef]
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular hyaluronan: Importance for cellular functions. Semin Cancer Biol. 2020, 62, 20–30. [Google Scholar] [CrossRef]
- Maldonado-Baez, L.; Donaldson, J.G. Hook1, microtubules, and Rab22: Mediators of selective sorting of clathrin-independent endocytic cargo proteins on endosomes. Bioarchitecture 2013, 3, 141–146. [Google Scholar] [CrossRef]
- Skandalis, S.S.; Kozlova, I.; Engstrom, U.; Hellman, U.; Heldin, P. Proteomic identification of CD44 interacting proteins. IUBMB Life 2010, 62, 833–840. [Google Scholar] [CrossRef]
- Murai, T. Lipid Raft-Mediated Regulation of Hyaluronan-CD44 Interactions in Inflammation and Cancer. Front. Immunol. 2015, 6, 420. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Y.; Cao, M.; Fan, W.; Cui, Y.; Cui, Y.; Zhan, Y.; Gu, R.; Tian, F.; Zhang, S.; et al. A novel EHD1/CD44/Hippo/SP1 positive feedback loop potentiates stemness and metastasis in lung adenocarcinoma. Clin. Transl. Med. 2022, 12, e836. [Google Scholar] [CrossRef] [PubMed]
- Naslavsky, N.; Caplan, S. EHD proteins: Key conductors of endocytic transport. Trends Cell Biol. 2011, 21, 122–131. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, H.; Liu, S.; Kim, C.K.; Xu, Y.; Hurley, L.A.; Nishikawa, R.; Nagane, M.; Hu, B.; Stegh, A.H.; et al. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc. Natl. Acad. Sci. USA 2017, 114, 8366–8371. [Google Scholar] [CrossRef] [PubMed]
- Heiska, L.; Alfthan, K.; Gronholm, M.; Vilja, P.; Vaheri, A.; Carpen, O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 1998, 273, 21893–21900. [Google Scholar] [CrossRef]
- Granes, F.; Urena, J.M.; Rocamora, N.; Vilaro, S. Ezrin links syndecan-2 to the cytoskeleton. J. Cell Sci. 2000, 113 Pt 7, 1267–1276. [Google Scholar] [CrossRef]
- Ivetic, A.; Deka, J.; Ridley, A.; Ager, A. The cytoplasmic tail of L-selectin interacts with members of the Ezrin-Radixin-Moesin (ERM) family of proteins: Cell activation-dependent binding of Moesin but not Ezrin. J. Biol. Chem. 2002, 277, 2321–2329. [Google Scholar] [CrossRef]
- Obremski, V.J.; Hall, A.M.; Fernandez-Valle, C. Merlin, the neurofibromatosis type 2 gene product, and beta1 integrin associate in isolated and differentiating Schwann cells. J Neurobiol. 1998, 37, 487–501. [Google Scholar] [CrossRef]
- Curto, M.; McClatchey, A.I. Nf2/Merlin: A coordinator of receptor signalling and intercellular contact. Br. J. Cancer 2008, 98, 256–262. [Google Scholar] [CrossRef]
- Sato, T.; Sekido, Y. NF2/Merlin Inactivation and Potential Therapeutic Targets in Mesothelioma. Int. J. Mol. Sci. 2018, 19, 988. [Google Scholar] [CrossRef]
- Mranda, G.M.; Xiang, Z.P.; Liu, J.J.; Wei, T.; Ding, Y. Advances in prognostic and therapeutic targets for hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The hippo signaling pathway. Front. Oncol. 2022, 12, 937957. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Zhao, L.; Ma, Z.; An, H.; Marrink, S.J.; Sun, F. Molecular basis of PIP2-dependent conformational switching of phosphorylated CD44 in binding FERM. Biophys. J. 2023, 122, 2675–2685. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, A.; Edwards, K.; Fehon, R.G. ERM proteins and merlin: Integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 2002, 3, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Jokela, T.; Oikari, S.; Takabe, P.; Rilla, K.; Karna, R.; Tammi, M.; Tammi, R. Interleukin-1beta-induced Reduction of CD44 Ser-325 Phosphorylation in Human Epidermal Keratinocytes Promotes CD44 Homomeric Complexes, Binding to Ezrin, and Extended, Monocyte-adhesive Hyaluronan Coats. J. Biol. Chem. 2015, 290, 12379–12393. [Google Scholar] [CrossRef]
- Nunomura, W.; Takakuwa, Y.; Tokimitsu, R.; Krauss, S.W.; Kawashima, M.; Mohandas, N. Regulation of CD44-protein 4.1 interaction by Ca2+ and calmodulin. Implications for modulation of CD44-ankyrin interaction. J. Biol. Chem. 1997, 272, 30322–30328. [Google Scholar] [CrossRef]
- Donatello, S.; Babina, I.S.; Hazelwood, L.D.; Hill, A.D.; Nabi, I.R.; Hopkins, A.M. Lipid raft association restricts CD44-ezrin interaction and promotion of breast cancer cell migration. Am. J. Pathol. 2012, 181, 2172–2187. [Google Scholar] [CrossRef]
- Sherman, L.; Xu, H.M.; Geist, R.T.; Saporito-Irwin, S.; Howells, N.; Ponta, H.; Herrlich, P.; Gutmann, D.H. Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene 1997, 15, 2505–2509. [Google Scholar] [CrossRef]
- Tang, X.; Jang, S.W.; Wang, X.; Liu, Z.; Bahr, S.M.; Sun, S.Y.; Brat, D.; Gutmann, D.H.; Ye, K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat. Cell Biol. 2007, 9, 1199–1207. [Google Scholar] [CrossRef]
- Thaxton, C.; Lopera, J.; Bott, M.; Fernandez-Valle, C. Neuregulin and laminin stimulate phosphorylation of the NF2 tumor suppressor in Schwann cells by distinct protein kinase A and p21-activated kinase-dependent pathways. Oncogene 2008, 27, 2705–2715. [Google Scholar] [CrossRef]
- Ng, T.; Parsons, M.; Hughes, W.E.; Monypenny, J.; Zicha, D.; Gautreau, A.; Arpin, M.; Gschmeissner, S.; Verveer, P.J.; Bastiaens, P.I.; et al. Ezrin is a downstream effector of trafficking PKC-integrin complexes involved in the control of cell motility. EMBO J. 2001, 20, 2723–2741. [Google Scholar] [CrossRef]
- Choi, Y.; Yun, J.H.; Yoo, J.; Lee, I.; Kim, H.; Son, H.N.; Kim, I.S.; Yoon, H.S.; Zimmermann, P.; Couchman, J.R.; et al. New structural insight of C-terminal region of Syntenin-1, enhancing the molecular dimerization and inhibitory function related on Syndecan-4 signaling. Sci. Rep. 2016, 6, 36818. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, R.; Trapasso, F.; Sama, I.; Le Pera, I.; Martelli, M.L.; Lembo, F.; Santoro, M.; Viglietto, G.; Chiariotti, L.; Fusco, A. Rat protein tyrosine phosphatase eta physically interacts with the PDZ domains of syntenin. FEBS Lett. 2001, 500, 41–44. [Google Scholar] [CrossRef]
- Bass, M.D.; Humphries, M.J. Cytoplasmic interactions of syndecan-4 orchestrate adhesion receptor and growth factor receptor signalling. Biochem. J. 2002, 368, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.W.; Sacks, D.B. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003, 4, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Noritake, J.; Watanabe, T.; Sato, K.; Wang, S.; Kaibuchi, K. IQGAP1: A key regulator of adhesion and migration. J. Cell Sci. 2005, 118, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.D.; Sacks, D.B. IQGAP1 in cellular signaling: Bridging the GAP. Trends Cell Biol. 2006, 16, 242–249. [Google Scholar] [CrossRef]
- Kozlova, I.; Ruusala, A.; Voytyuk, O.; Skandalis, S.S.; Heldin, P. IQGAP1 regulates hyaluronan-mediated fibroblast motility and proliferation. Cell Signal. 2012, 24, 1856–1862. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Gilad, E.; Rothman, K.; Peyrollier, K. Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J. Biol. Chem. 2005, 280, 11961–11972. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Zhu, H.; Shao, L.; Zhu, D.; Chen, Y.W. Rho-kinase (ROK) promotes CD44v(3,8-10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil. Cytoskelet. 1999, 43, 269–287. [Google Scholar] [CrossRef]
- Oliferenko, S.; Paiha, K.; Harder, T.; Gerke, V.; Schwarzler, C.; Schwarz, H.; Beug, H.; Gunthert, U.; Huber, L.A. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 1999, 146, 843–854. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Singleton, P.A.; Diedrich, F.; Stern, R.; Gilad, E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 2004, 279, 26991–27007. [Google Scholar] [CrossRef]
- Triantafilou, M.; Miyake, K.; Golenbock, D.T.; Triantafilou, K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J. Cell Sci. 2002, 115, 2603–2611. [Google Scholar] [CrossRef]
- Grass, G.D.; Dai, L.; Qin, Z.; Parsons, C.; Toole, B.P. CD147: Regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv. Cancer Res. 2014, 123, 351–373. [Google Scholar] [PubMed]
- Wu, A.; Wojtowicz, K.; Savary, S.; Hamon, Y.; Trombik, T. Do ABC transporters regulate plasma membrane organization? Cell. Mol. Biol. Lett. 2020, 25, 37. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Q. E-cadherin negatively regulates CD44-hyaluronan interaction and CD44-mediated tumor invasion and branching morphogenesis. J. Biol. Chem. 2003, 278, 8661–8668. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Lin, S.C.; Wang, J.H.; Bigby, M.; Sy, M.S. Palmitoylation of CD44 interferes with CD3-mediated signaling in human T lymphocytes. Int. Immunol. 1994, 6, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P.; Woods, E.C.; Bertozzi, C.R.; Tammi, M.I.; Lidke, D.S.; Johnson, P.; et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e10. [Google Scholar] [CrossRef]
- Ahandoust, S.; Li, K.; Sun, X.; Li, B.Y.; Yokota, H.; Na, S. Intracellular and extracellular moesins differentially regulate Src activity and beta-catenin translocation to the nucleus in breast cancer cells. Biochem. Biophys. Res. Commun. 2023, 639, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Shi, X.; Liu, Y.; He, Y.; Du, Y.; Zhang, G.; Yang, C.; Gao, F. CD44 cross-linking increases malignancy of breast cancer via upregulation of p-Moesin. Cancer Cell Int. 2020, 20, 563. [Google Scholar] [CrossRef]
- Brown, K.L.; Birkenhead, D.; Lai, J.C.; Li, L.; Li, R.; Johnson, P. Regulation of hyaluronan binding by F-actin and colocalization of CD44 and phosphorylated ezrin/radixin/moesin (ERM) proteins in myeloid cells. Exp. Cell Res. 2005, 303, 400–414. [Google Scholar] [CrossRef]
- Liu, D.; Liu, T.; Sy, M.S. Identification of two regions in the cytoplasmic domain of CD44 through which PMA, calcium, and foskolin differentially regulate the binding of CD44 to hyaluronic acid. Cell Immunol. 1998, 190, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Lesley, J.; Hascall, V.C.; Tammi, M.; Hyman, R. Hyaluronan binding by cell surface CD44. J. Biol. Chem. 2000, 275, 26967–26975. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.; Rudy, W.; Hofmann, M.; Moll, J.; Herrlich, P.; Ponta, H. Regulated clustering of variant CD44 proteins increases their hyaluronate binding capacity. J. Cell Biol. 1996, 135, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yago, T.; Zhang, N.; Abdisalaam, S.; Alexandrakis, G.; Rodgers, W.; McEver, R.P. Cytoskeletal regulation of CD44 membrane organization and interactions with E-selectin. J. Biol. Chem. 2014, 289, 35159–35171. [Google Scholar] [CrossRef] [PubMed]
- Seluanov, A.; Hine, C.; Azpurua, J.; Feigenson, M.; Bozzella, M.; Mao, Z.; Catania, K.C.; Gorbunova, V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA 2009, 106, 19352–19357. [Google Scholar] [CrossRef]
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349. [Google Scholar] [CrossRef]
- Morrison, H.; Sherman, L.S.; Legg, J.; Banine, F.; Isacke, C.; Haipek, C.A.; Gutmann, D.H.; Ponta, H.; Herrlich, P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001, 15, 968–980. [Google Scholar] [CrossRef]
- Jin, H.; Sperka, T.; Herrlich, P.; Morrison, H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 2006, 442, 576–579. [Google Scholar] [CrossRef]
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Bohme, S.; Li, Y.; Morrison, H.; Herrlich, A.; Herrlich, P. Tumor Suppressor NF2 Blocks Cellular Migration by Inhibiting Ectodomain Cleavage of CD44. Mol. Cancer Res. 2015, 13, 879–890. [Google Scholar] [CrossRef]
- Hamaratoglu, F.; Willecke, M.; Kango-Singh, M.; Nolo, R.; Hyun, E.; Tao, C.; Jafar-Nejad, H.; Halder, G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006, 8, 27–36. [Google Scholar] [CrossRef]
- Zhang, N.; Bai, H.; David, K.K.; Dong, J.; Zheng, Y.; Cai, J.; Giovannini, M.; Liu, P.; Anders, R.A.; Pan, D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 2010, 19, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M.; Kim, N.G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 2014, 127, 709–717. [Google Scholar] [CrossRef]
- Yin, F.; Yu, J.; Zheng, Y.; Chen, Q.; Zhang, N.; Pan, D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 2013, 154, 1342–1355. [Google Scholar] [CrossRef] [PubMed]
- Kissil, J.L.; Johnson, K.C.; Eckman, M.S.; Jacks, T. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. J. Biol. Chem. 2002, 277, 10394–10399. [Google Scholar] [CrossRef] [PubMed]
- Surace, E.I.; Haipek, C.A.; Gutmann, D.H. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene 2004, 23, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V.; Chen, L.; Sleeman, J.P.; Herrlich, P.; Ponta, H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002, 16, 3074–3086. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Morrison, H.; Matzke, A.; Kastilan, T.; Pace, G.; Herrlich, P.; Ponta, H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol. Biol. Cell 2007, 18, 76–83. [Google Scholar] [CrossRef]
- Ooki, T.; Murata-Kamiya, N.; Takahashi-Kanemitsu, A.; Wu, W.; Hatakeyama, M. High-Molecular-Weight Hyaluronan Is a Hippo Pathway Ligand Directing Cell Density-Dependent Growth Inhibition via PAR1b. Dev. Cell 2019, 49, 590–604.e9. [Google Scholar] [CrossRef]
- Bergamaschi, D.; Samuels, Y.; O’Neil, N.J.; Trigiante, G.; Crook, T.; Hsieh, J.K.; O’Connor, D.J.; Zhong, S.; Campargue, I.; Tomlinson, M.L.; et al. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat. Genet. 2003, 33, 162–167. [Google Scholar] [CrossRef]
- Sullivan, A.; Lu, X. ASPP: A new family of oncogenes and tumour suppressor genes. Br. J. Cancer 2007, 96, 196–200. [Google Scholar] [CrossRef]
- Yang, J.P.; Hori, M.; Sanda, T.; Okamoto, T. Identification of a novel inhibitor of nuclear factor-kappaB, RelA-associated inhibitor. J. Biol. Chem. 1999, 274, 15662–15670. [Google Scholar] [CrossRef] [PubMed]
- Trigiante, G.; Lu, X. ASPP [corrected] and cancer. Nat. Rev. Cancer 2006, 6, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Basu, K.; Ruusala, A.; Kozlova, I.; Li, Y.S.; Skandalis, S.S.; Heldin, C.H.; Heldin, P. Hyaluronan-Induced CD44-iASPP Interaction Affects Fibroblast Migration and Survival. Cancers 2023, 15, 1082. [Google Scholar] [CrossRef] [PubMed]
- Godar, S.; Ince, T.A.; Bell, G.W.; Feldser, D.; Donaher, J.L.; Bergh, J.; Liu, A.; Miu, K.; Watnick, R.S.; Reinhardt, F.; et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008, 134, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456. [Google Scholar] [CrossRef]
- Hartmann, M.; Parra, L.M.; Ruschel, A.; Lindner, C.; Morrison, H.; Herrlich, A.; Herrlich, P. Inside-out Regulation of Ectodomain Cleavage of Cluster-of-Differentiation-44 (CD44) and of Neuregulin-1 Requires Substrate Dimerization. J. Biol. Chem. 2015, 290, 17041–17054. [Google Scholar] [CrossRef]
- Parra, L.M.; Hartmann, M.; Schubach, S.; Li, Y.; Herrlich, P.; Herrlich, A. Distinct Intracellular Domain Substrate Modifications Selectively Regulate Ectodomain Cleavage of NRG1 or CD44. Mol. Cell Biol. 2015, 35, 3381–3395. [Google Scholar] [CrossRef]
- Suenaga, N.; Mori, H.; Itoh, Y.; Seiki, M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene 2005, 24, 859–868. [Google Scholar] [CrossRef]
- Terawaki, S.; Kitano, K.; Aoyama, M.; Mori, T.; Hakoshima, T. MT1-MMP recognition by ERM proteins and its implication in CD44 shedding. Genes Cells 2015, 20, 847–859. [Google Scholar] [CrossRef]
- Okamoto, I.; Tsuiki, H.; Kenyon, L.C.; Godwin, A.K.; Emlet, D.R.; Holgado-Madruga, M.; Lanham, I.S.; Joynes, C.J.; Vo, K.T.; Guha, A.; et al. Proteolytic cleavage of the CD44 adhesion molecule in multiple human tumors. Am. J. Pathol. 2002, 160, 441–447. [Google Scholar] [CrossRef]
- Wöhner, B.; Li, W.; Hey, S.; Drobny, A.; Werny, L.; Becker-Pauly, C.; Lucius, R.; Zunke, F.; Linder, S.; Arnold, P. Proteolysis of CD44 at the cell surface controls a downstream protease network. Front. Mol. Biosci. 2023, 10, 1026810. [Google Scholar] [CrossRef] [PubMed]
- Kolliopoulos, C.; Chatzopoulos, A.; Skandalis, S.S.; Heldin, C.H.; Heldin, P. TRAF4/6 Is Needed for CD44 Cleavage and Migration via RAC1 Activation. Cancers 2021, 13, 1021. [Google Scholar] [CrossRef] [PubMed]
- Lammich, S.; Okochi, M.; Takeda, M.; Kaether, C.; Capell, A.; Zimmer, A.K.; Edbauer, D.; Walter, J.; Steiner, H.; Haass, C. Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Abeta-like peptide. J. Biol. Chem. 2002, 277, 44754–44759. [Google Scholar] [CrossRef]
- Murakami, D.; Okamoto, I.; Nagano, O.; Kawano, Y.; Tomita, T.; Iwatsubo, T.; De Strooper, B.; Yumoto, E.; Saya, H. Presenilin-dependent gamma-secretase activity mediates the intramembranous cleavage of CD44. Oncogene 2003, 22, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.L.; Wang, M.J.; Chen, J.Y. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol. 2009, 185, 949–957. [Google Scholar] [CrossRef]
- Miletti-Gonzalez, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007. [Google Scholar] [CrossRef] [PubMed]
- De Falco, V.; Tamburrino, A.; Ventre, S.; Castellone, M.D.; Malek, M.; Manie, S.N.; Santoro, M. CD44 proteolysis increases CREB phosphorylation and sustains proliferation of thyroid cancer cells. Cancer Res. 2012, 72, 1449–1458. [Google Scholar] [CrossRef]
- Johansson, E.; Grassi, E.S.; Pantazopoulou, V.; Tong, B.; Lindgren, D.; Berg, T.J.; Pietras, E.J.; Axelson, H.; Pietras, A. CD44 Interacts with HIF-2alpha to Modulate the Hypoxic Phenotype of Perinecrotic and Perivascular Glioma Cells. Cell Rep. 2017, 20, 1641–1653. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Wong, G.; Earle, C.; Chen, L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J. Biol. Chem. 2012, 287, 32800–32824. [Google Scholar] [CrossRef]
- Cho, Y.; Lee, H.W.; Kang, H.G.; Kim, H.Y.; Kim, S.J.; Chun, K.H. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget 2015, 6, 8709–8721. [Google Scholar] [CrossRef]
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Matoba, S.; Kang, J.G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 regulates mitochondrial respiration. Science 2006, 312, 1650–1653. [Google Scholar] [CrossRef]
- Anastasiou, D.; Poulogiannis, G.; Asara, J.M.; Boxer, M.B.; Jiang, J.K.; Shen, M.; Bellinger, G.; Sasaki, A.T.; Locasale, J.W.; Auld, D.S.; et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 334, 1278–1283. [Google Scholar] [CrossRef]
- Gruning, N.M.; Ralser, M. Cancer: Sacrifice for survival. Nature 2011, 480, 190–191. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Cell biology. Warburg effect and redox balance. Science 2011, 334, 1219–1220. [Google Scholar] [CrossRef]
- Nam, K.; Oh, S.; Shin, I. Ablation of CD44 induces glycolysis-to-oxidative phosphorylation transition via modulation of the c-Src-Akt-LKB1-AMPKalpha pathway. Biochem. J. 2016, 473, 3013–3030. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Li, D.; Xun, J.; Zhou, W.; Li, J.; Wang, J.; Liu, C.; Li, X.; Shen, W.; Qiao, H.; et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics 2018, 8, 6248–6262. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, A.; Telang, S.; Clem, B.; Chesney, J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp. Mol. Pathol. 2009, 86, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Huang, H.; Choi, K.; Lee, D.H.; Shi, J.; Liu, T.; Chun, K.H.; Seo, J.A.; Lima, I.S.; Zabolotny, J.M.; et al. ROCK1 isoform-specific deletion reveals a role for diet-induced insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E332–E343. [Google Scholar] [CrossRef]
- Ahn, M.; Oh, E.; McCown, E.M.; Wang, X.; Veluthakal, R.; Thurmond, D.C. A requirement for PAK1 to support mitochondrial function and maintain cellular redox balance via electron transport chain proteins to prevent beta-cell apoptosis. Metabolism 2021, 115, 154431. [Google Scholar] [CrossRef]
- Ahn, M.; Yoder, S.M.; Wang, Z.; Oh, E.; Ramalingam, L.; Tunduguru, R.; Thurmond, D.C. The p21-activated kinase (PAK1) is involved in diet-induced beta cell mass expansion and survival in mice and human islets. Diabetologia 2016, 59, 2145–2155. [Google Scholar] [CrossRef]
- Lopez, J.P.; Turner, J.R.; Philipson, L.H. Glucose-induced ERM protein activation and translocation regulates insulin secretion. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E772–E785. [Google Scholar] [CrossRef]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef]
- Bourguignon, L.Y. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 2008, 18, 251–259. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Maxwell-Warburton, S.; Hasib, A.; Ma, L.; Kang, L. The membrane receptor CD44: Novel insights into metabolism. Trends Endocrinol. Metab. 2022, 33, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Hasenauer, S.; Malinger, D.; Koschut, D.; Pace, G.; Matzke, A.; von Au, A.; Orian-Rousseau, V. Internalization of Met requires the co-receptor CD44v6 and its link to ERM proteins. PLoS ONE 2013, 8, e62357. [Google Scholar] [CrossRef]
- Distel, J.S.; Ortiz Flores, R.M.; Bienvenu, A.; Aguilera, M.O.; Bonazzi, M.; Beron, W. Ezrin and CD44 participate in the internalization process of Coxiella burnetii into non-phagocytic cells. Biol. Cell 2022, 114, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Kolliopoulos, C.; Huang, C.H.; Tenhunen, J.; Heldin, C.H.; Chen, Y.H.; Heldin, P. High levels of serum hyaluronan is an early predictor of dengue warning signs and perturbs vascular integrity. EBioMedicine 2019, 48, 425–441. [Google Scholar] [CrossRef] [PubMed]


| Cellular Pathway | Cellular Function | CD44 ICD Interaction(s) | Ref(s) |
|---|---|---|---|
| Cell trafficking | HA endocytosis, pericellular matrix retention, and organization | Absence of ICD | [36] |
| CD44–HA endocytosis and degradation | MARCH-VIII ubiquitin ligase | [37] | |
| CD44 endocytic recycling and stabilization, stemness, and EMT | Hook1, EHD1, and EHD2 | [39,40,42] | |
| Sustained EGFR signaling and survival | Rab7A | [44] | |
| Cytoskeletal organization | Growth, anchorage-independent growth, migration, adhesion, chemotaxis, and leukocyte binding | ERM, merlin, and ankyrin | [4,30,31,52,53,54,55,56,57,60,69,97] |
| Migration and proliferation | IQGAP1 | [40,67,68] | |
| Contact inhibition of growth, migration, apoptosis, and proliferation | Merlin | [85,86,87,89,90,91,93] | |
| Contact inhibition of growth | PAR1b | [98] | |
| Transcription | Apoptosis, survival, migration, and adhesion | iASPP | [40,103] |
| CD44 expression | CBP/p300 transcriptional activator | [110] | |
| Cell cycle progression | CREB | [117] | |
| Stemness and therapeutic resistance | HIF-2 alpha | [118] | |
| Stemness and therapeutic resistance | Oct4, Sox2, and Nanog | [119,120] | |
| Metabolism | Survival, stemness, and therapeutic resistance | LDH, PFKFB4, ALDOC, and PDK1 | [116,130,132] |
| Survival and therapeutic resistance | PKM2 | [40,121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skandalis, S.S. CD44 Intracellular Domain: A Long Tale of a Short Tail. Cancers 2023, 15, 5041. https://doi.org/10.3390/cancers15205041
Skandalis SS. CD44 Intracellular Domain: A Long Tale of a Short Tail. Cancers. 2023; 15(20):5041. https://doi.org/10.3390/cancers15205041
Chicago/Turabian StyleSkandalis, Spyros S. 2023. "CD44 Intracellular Domain: A Long Tale of a Short Tail" Cancers 15, no. 20: 5041. https://doi.org/10.3390/cancers15205041
APA StyleSkandalis, S. S. (2023). CD44 Intracellular Domain: A Long Tale of a Short Tail. Cancers, 15(20), 5041. https://doi.org/10.3390/cancers15205041