Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies
Abstract
:Simple Summary
Abstract
1. Introduction
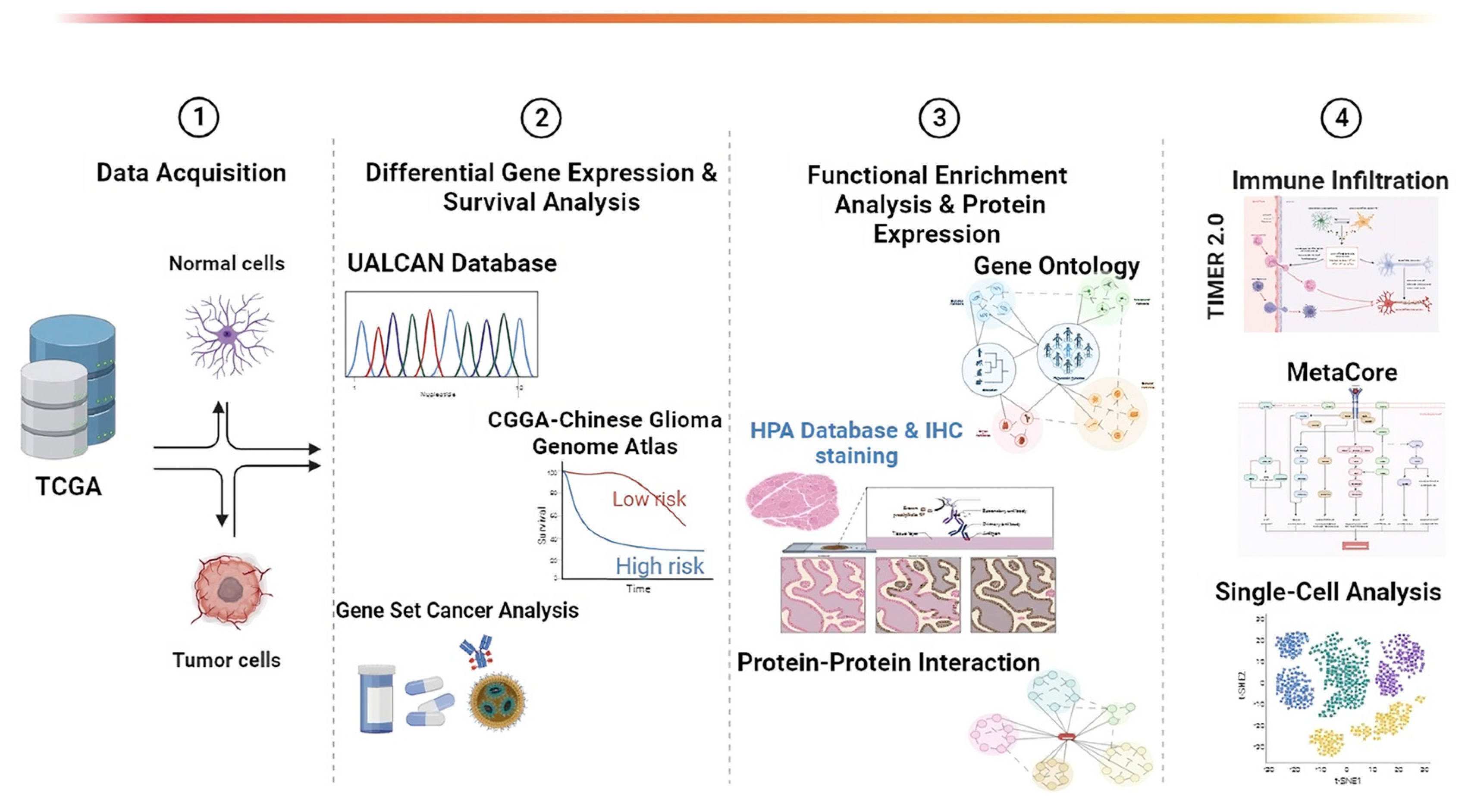
2. Materials and Methods
2.1. Use of UALCAN for Pan-Cancer and Gene Expression Analyses
2.2. Chinese Glioma Genome Atlas (CGGA) Dataset for Survival Analysis
2.3. Protein–Protein Interactions (PPIs)
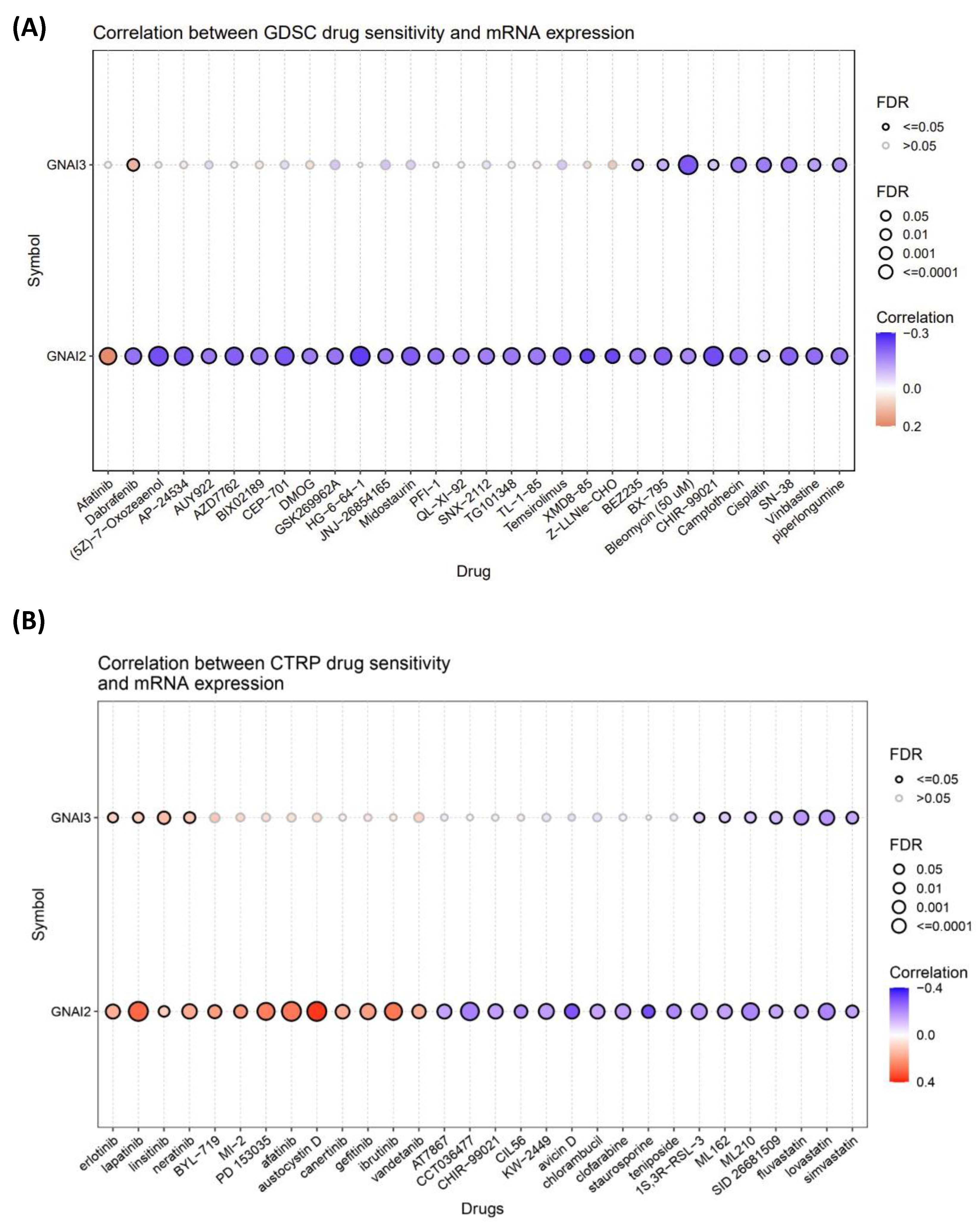
2.4. Drug Sensitivity
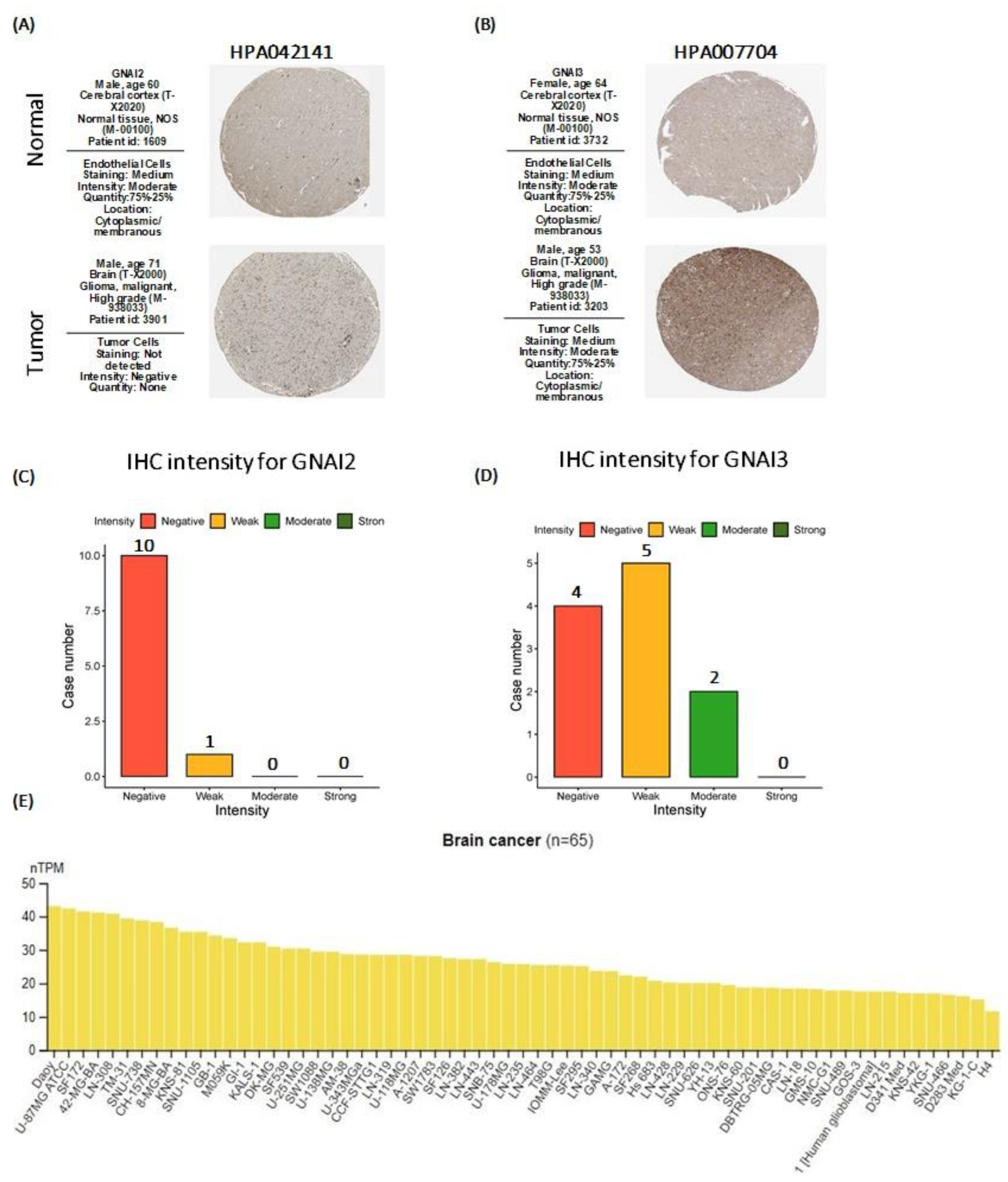
2.5. Human Protein Atlas
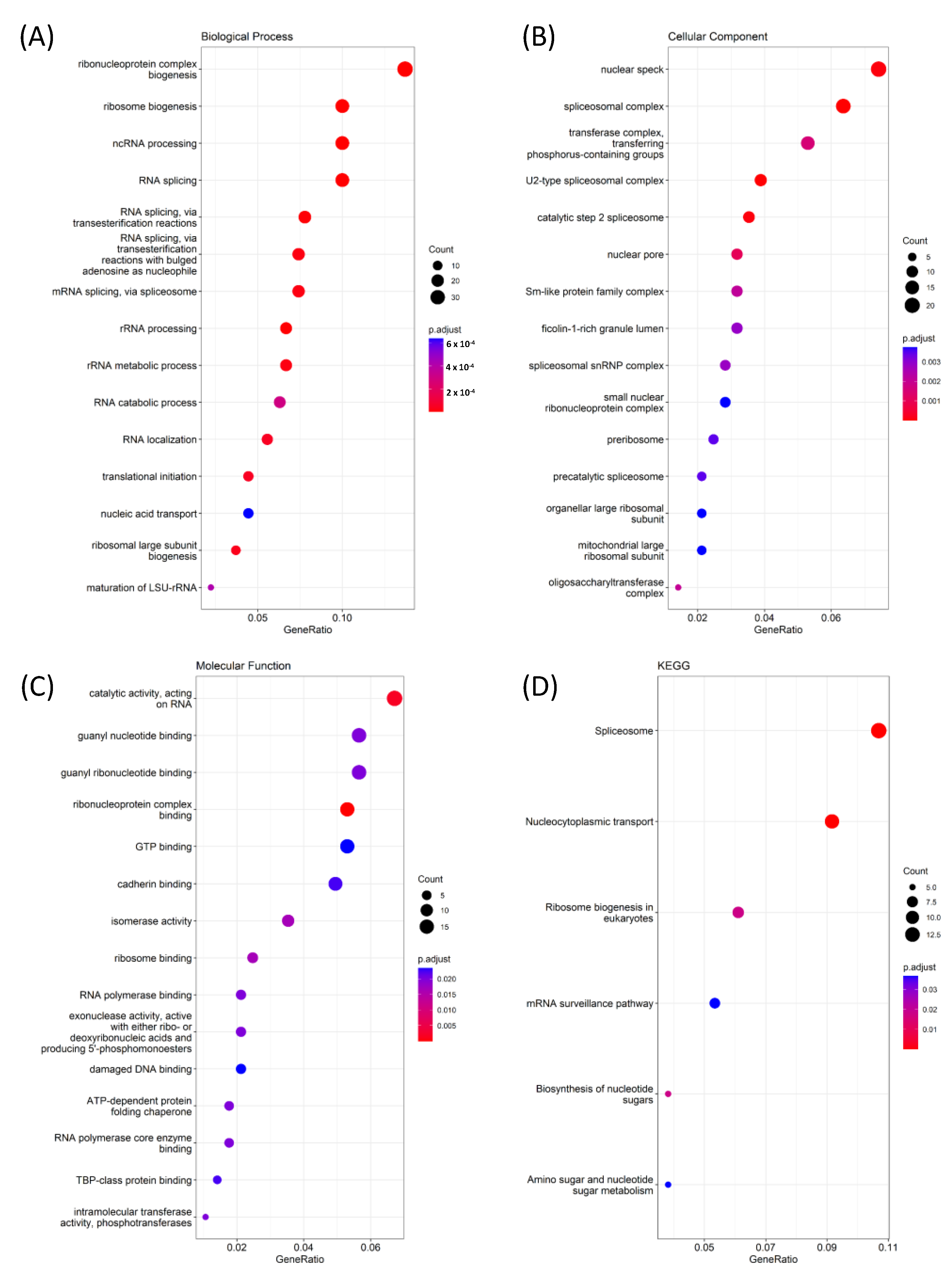
2.6. Functional Enrichment GO (Gene Ontology) Analysis
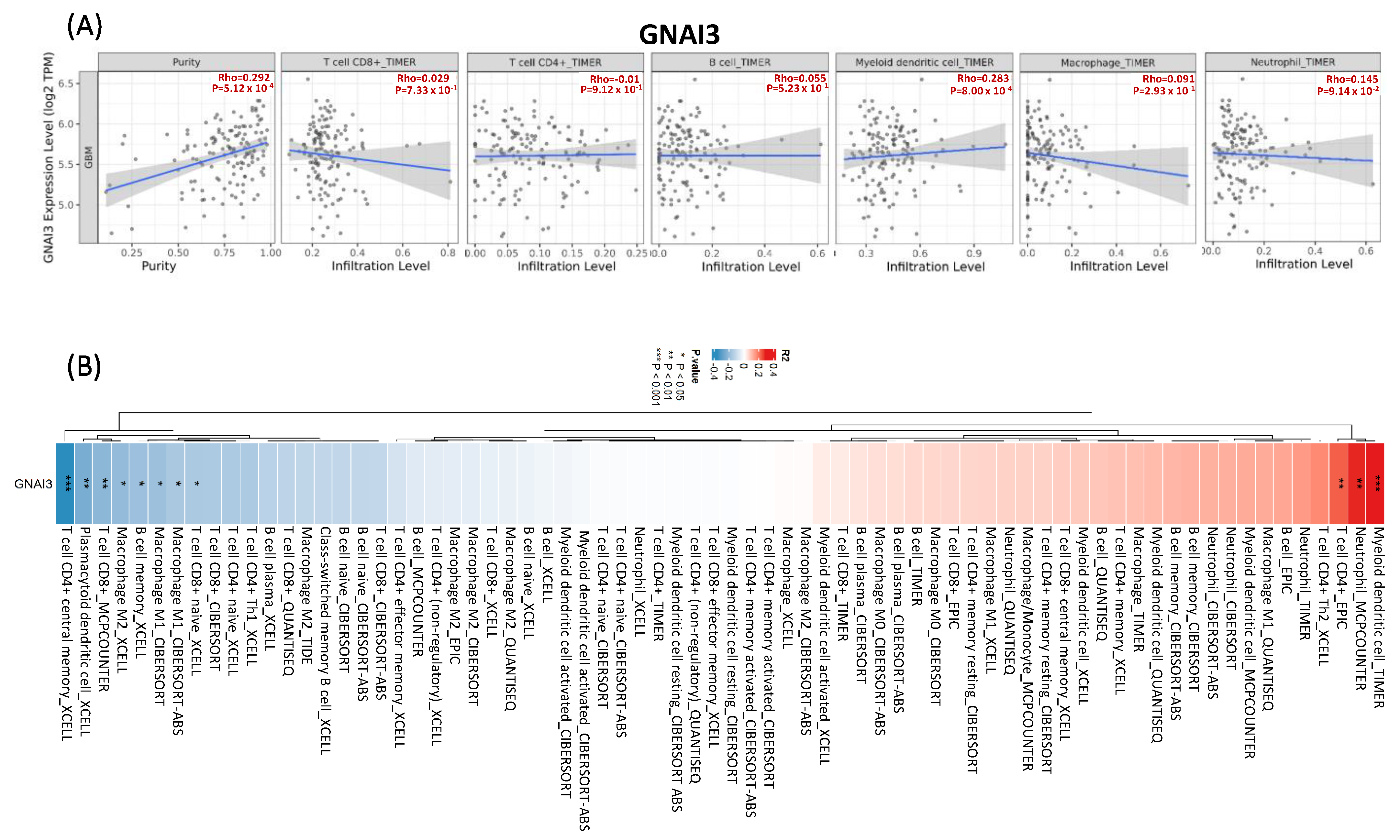
2.7. TIMER 2.0 for Immune Infiltration
2.8. Single-Cell Analysis
2.9. Statistical Analysis
3. Results
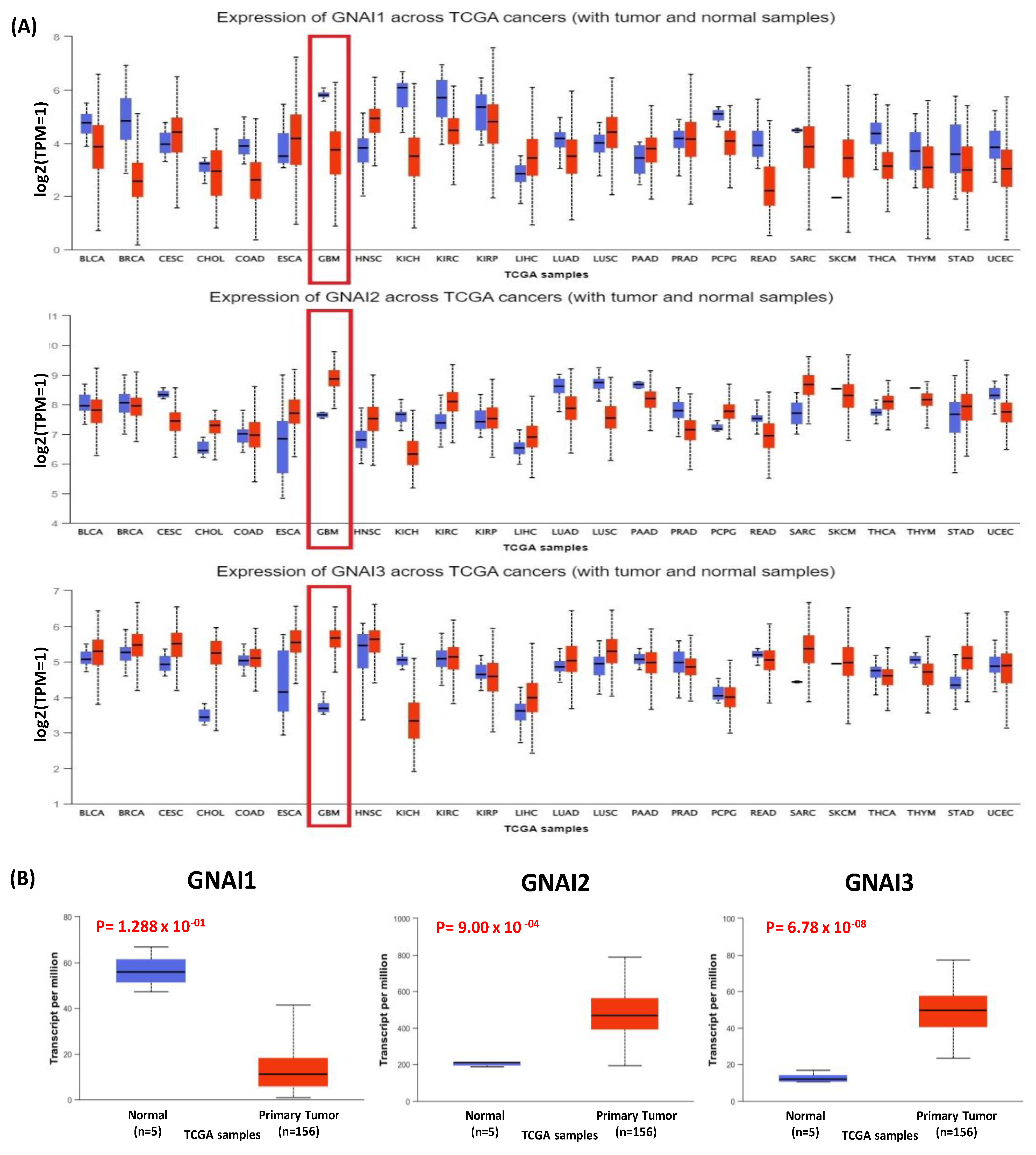
3.1. Expression of GNAI3 in a Pan-Cancer Analysis of DEGs Using TCGA Data
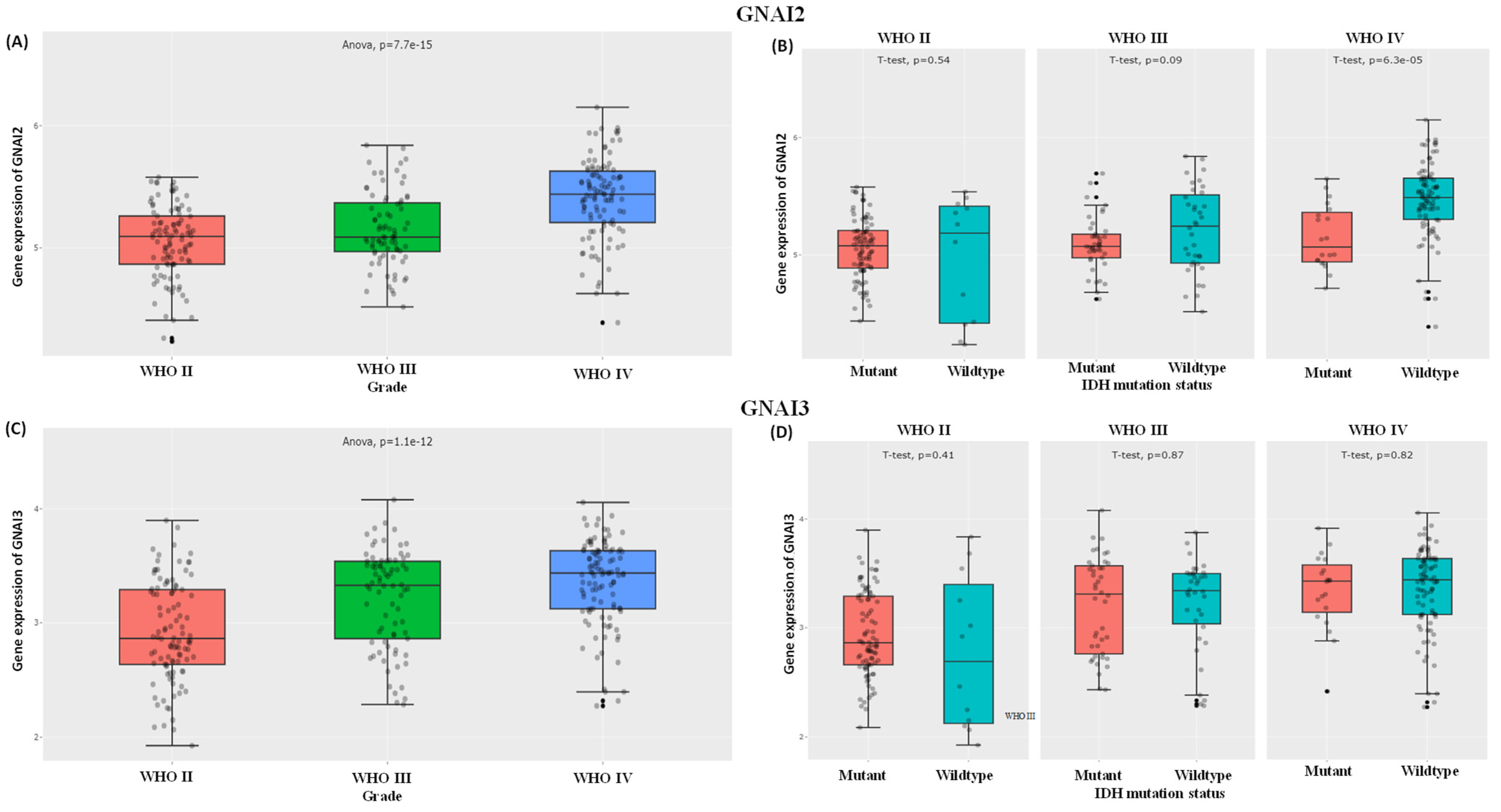
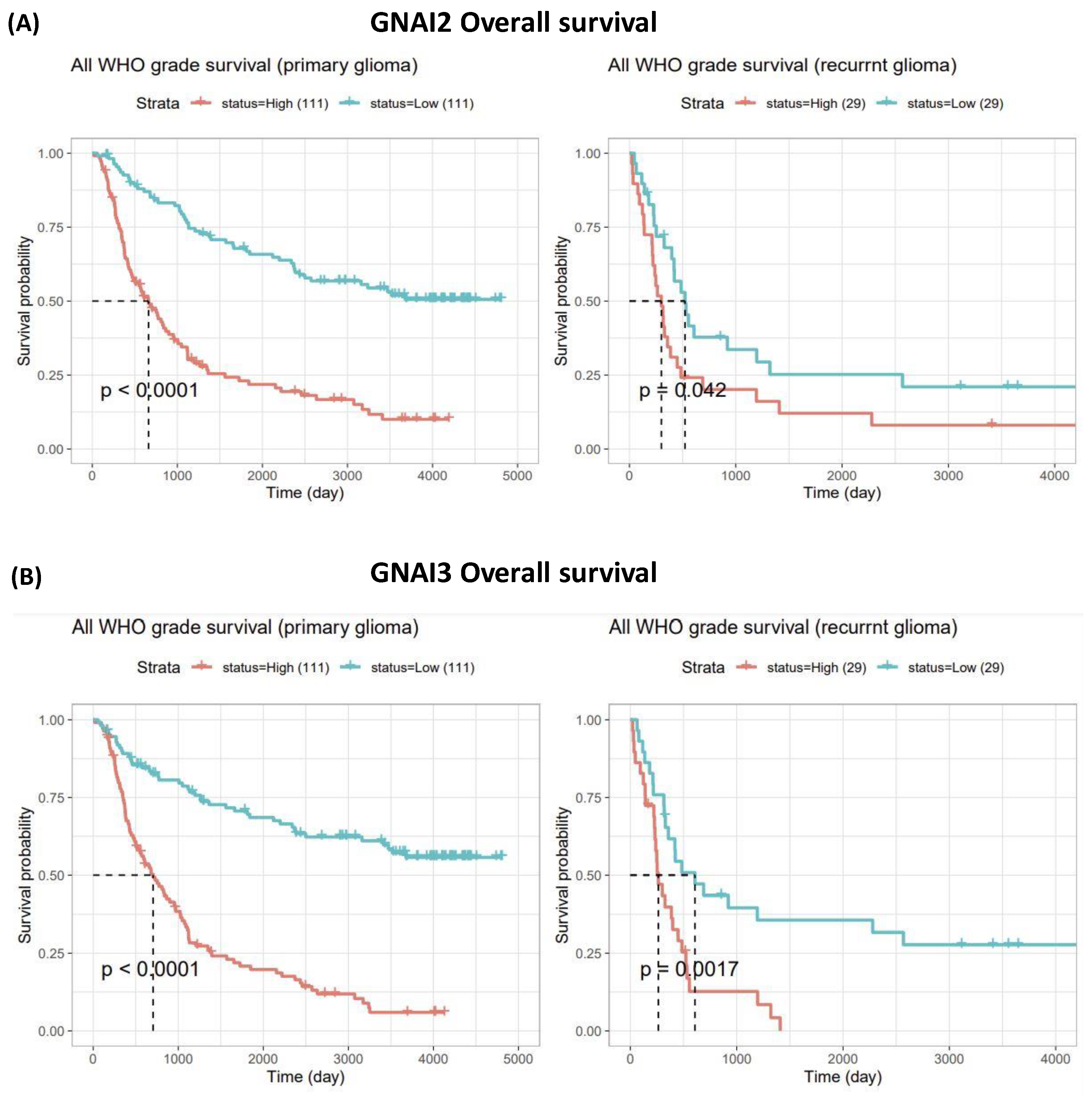
3.2. Expressions of GNAI2 and GNAI3 in IDH WTs Based on WHO Grade II, III, and IV Gliomas and Survival Analysis
3.3. PPI Analysis
3.4. Drug Analysis and the Role of Drugs in GBM Treatment
3.5. Human Protein Atlas (HPA) Analysis Results
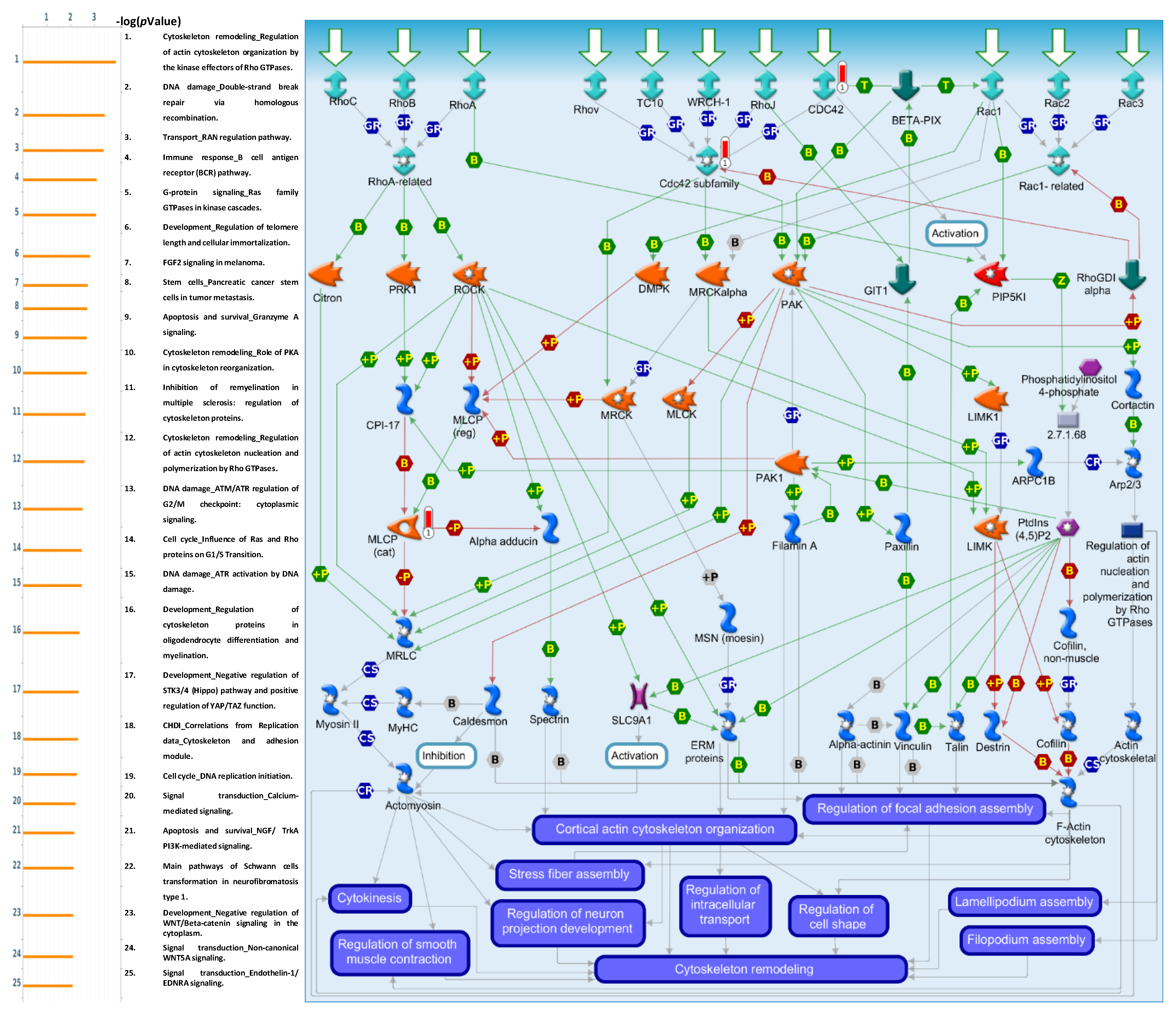
3.6. Pathway Interactions Using GO Tools for GNAI3
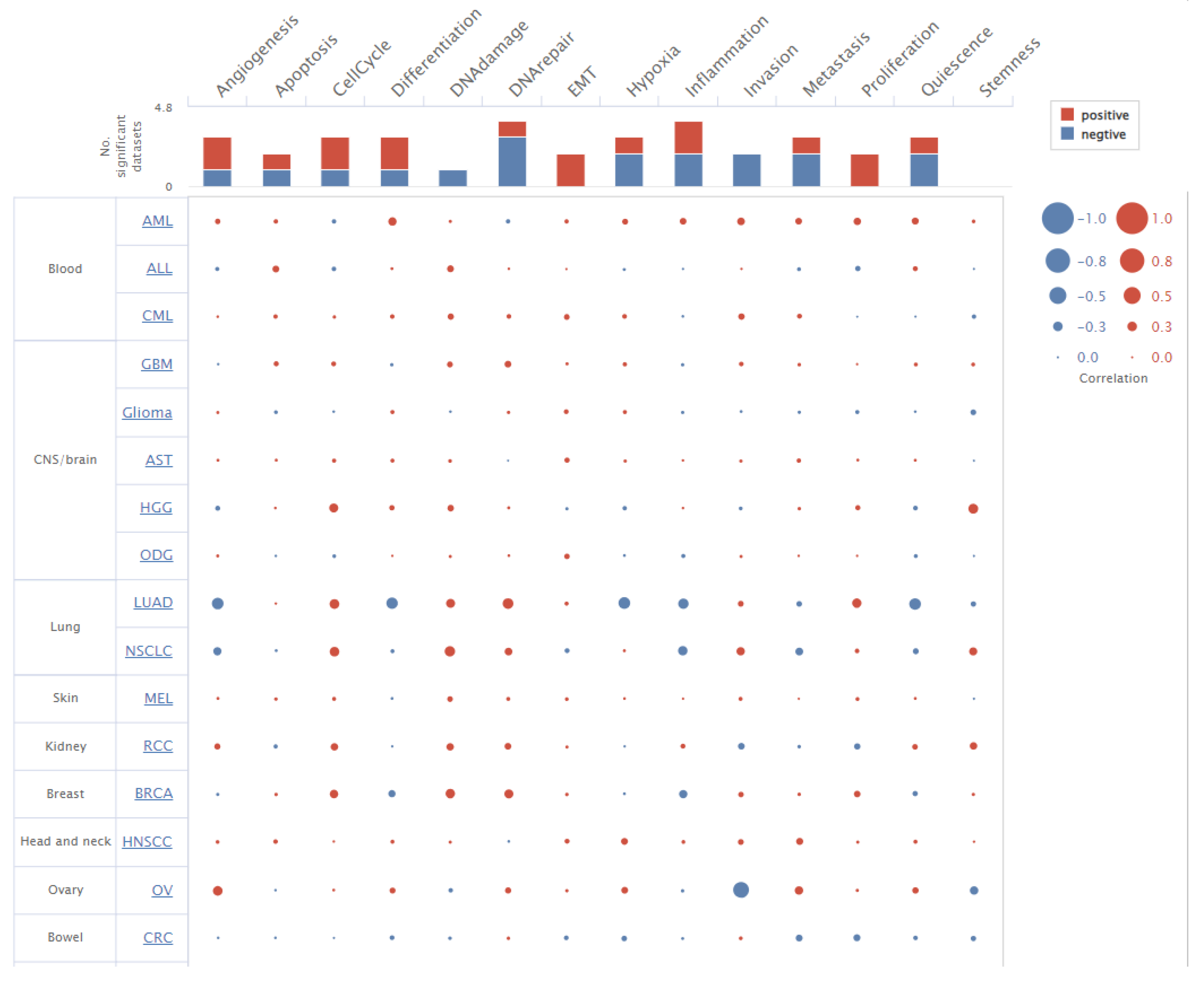
3.7. TIMER Analysis for Immune Infiltration
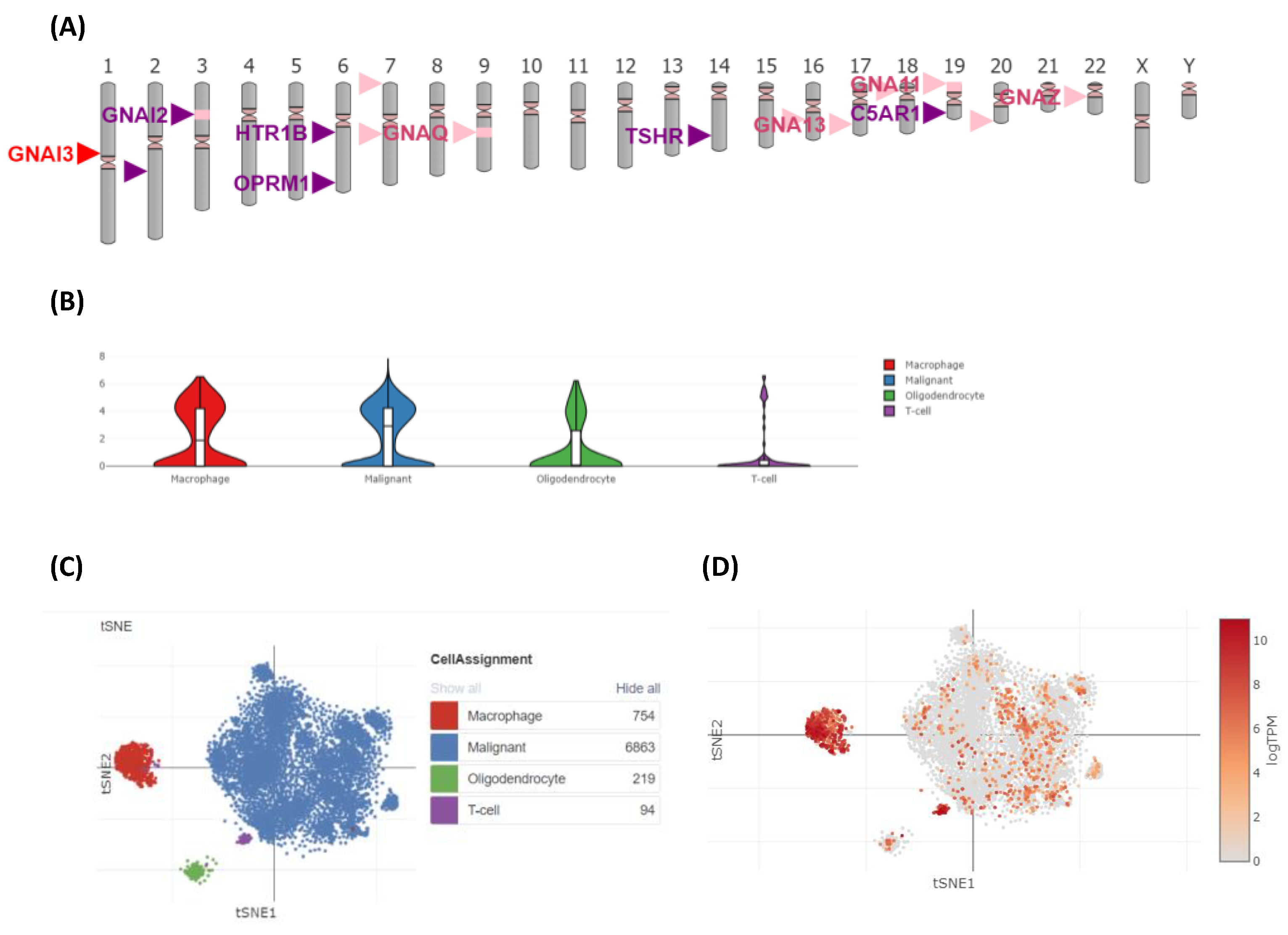
3.8. GNAI3 Expression Using Single-Cell Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinones, A.; Le, A. The multifaceted glioblastoma: From genomic alterations to metabolic adaptations. In The Heterogeneity of Cancer Metabolism; Springer International Publishing: Cham, Switzerland, 2021; pp. 59–76. [Google Scholar]
- Hegde, M.N. A Coursebook on Aphasia and Other Neurogenic Language Disorders; Plural Publishing: San Diego, CA, USA, 2022. [Google Scholar]
- Uddin, M.S.; Al Mamun, A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2022, 83, 100–120. [Google Scholar] [CrossRef] [PubMed]
- Lechpammer, M.; Mahammedi, A.; Krummel, D.A.P.; Sengupta, S. Lessons Learned from Evolving Frameworks in Adult Glioblastoma. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2023; Volume 192, pp. 131–140. [Google Scholar]
- McKinnon, C.; Nandhabalan, M.; Murray, S.A.; Plaha, P. Glioblastoma: Clinical presentation, diagnosis, and management. BMJ 2021, 374, n1560. [Google Scholar] [CrossRef]
- Chen, C.; Ou, X.; Wang, J.; Guo, W.; Ma, X. Radiomics-based machine learning in differentiation between glioblastoma and metastatic brain tumors. Front. Oncol. 2019, 9, 806. [Google Scholar] [CrossRef] [PubMed]
- Bilmin, K.; Kujawska, T.; Grieb, P. Sonodynamic therapy for gliomas. Perspectives and prospects of selective sonosensitization of glioma cells. Cells 2019, 8, 1428. [Google Scholar] [CrossRef]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol. Res. 2021, 171, 105780. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, H.; Gao, X.; Cui, M.; Ma, L.; Zheng, X.; Guan, B.; Ma, X. Surgical treatment of diffuse and multi-lobes involved glioma with the assistance of a multimodal technique. Sci. Rep. 2022, 12, 3343. [Google Scholar] [CrossRef] [PubMed]
- Van Solinge, T.S.; Nieland, L.; Chiocca, E.A.; Broekman, M.L. Advances in local therapy for glioblastoma—Taking the fight to the tumour. Nat. Rev. Neurol. 2022, 18, 221–236. [Google Scholar] [CrossRef]
- Zlochower, A.; Chow, D.S.; Chang, P.; Khatri, D.; Boockvar, J.A.; Filippi, C.G. Deep learning AI applications in the imaging of glioma. Top. Magn. Reson. Imaging 2020, 29, 115–121. [Google Scholar] [CrossRef]
- Nojeem, L.; Shun, M.; Embouma, M.; Inokon, A.; Browndi, I. Uncovering the Complexity of Cell Signaling Pathways Using Systems Biology Approaches. Am. Eurasian J. Sci. Res. 2023, 11, 131–135. [Google Scholar]
- Nůsková, H.; Serebryakova, M.V.; Ferrer-Caelles, A.; Sachsenheimer, T.; Lüchtenborg, C.; Miller, A.K.; Brügger, B.; Kordyukova, L.V.; Teleman, A.A. Stearic acid blunts growth-factor signaling via oleoylation of GNAI proteins. Nat. Commun. 2021, 12, 4590. [Google Scholar] [CrossRef]
- Liccardo, F.; Luini, A.; Di Martino, R. Endomembrane-based signaling by GPCRs and G-proteins. Cells 2022, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; He, J.; Deng, Q.; Chen, J.; Peng, M.; Xiao, J.; Zeng, Y.; Yi, L.; Li, Z.; Tian, R. Neuroglobin inhibits pancreatic cancer proliferation and metastasis by targeting the GNAI1/EGFR/AKT/ERK signaling axis. Biochem. Biophys. Res. Commun. 2023, 664, 108–116. [Google Scholar] [CrossRef]
- Abioja, M.; Logunleko, M.; Majekodunmi, B.; Adekunle, E.; Shittu, O.; Odeyemi, A.; Nwosu, E.; Oke, O.; Iyasere, O.; Abiona, J. Roles of Candidate Genes in the Adaptation of Goats to Heat Stress: A Review. Small Rumin. Res. 2022, 218, 106878. [Google Scholar] [CrossRef]
- Lecoquierre, F.; Duffourd, Y.; Vitobello, A.; Bruel, A.-L.; Urteaga, B.; Coubes, C.; Garret, P.; Nambot, S.; Chevarin, M.; Jouan, T. Variant recurrence in neurodevelopmental disorders: The use of publicly available genomic data identifies clinically relevant pathogenic missense variants. Genet. Med. 2019, 21, 2504–2511. [Google Scholar] [CrossRef]
- Chang, L.-C.; Chiu, H.-M.; Wu, M.-S.; Shen, T.-L. The Role of Small Extracellular Vesicles in the Progression of Colorectal Cancer and Its Clinical Applications. Int. J. Mol. Sci. 2022, 23, 1379. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, K.; Morimoto, N.; Iso, M.; Abe, Y.; Okamura, K.; Nakamura, T.; Matsubara, Y.; Kaname, T. A novel missense variant of the GNAI3 gene and recognisable morphological characteristics of the mandibula in ARCND1. J. Hum. Genet. 2021, 66, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Ekimoto, T.; Gordon, C.T.; Uchijima, Y.; Sugiyama, R.; Kitazawa, T.; Iwase, A.; Kotani, R.; Asai, R.; Pingault, V. Mandibulofacial dysostosis with alopecia results from ET A R gain-of-function mutations via allosteric effects on ligand binding. J. Clin. Investig. 2023, 133, e151536. [Google Scholar] [CrossRef]
- Lian, X. Novel Insights into the Vasodilatory Effects of Relaxins and Supraspinal Sympathetic Control of Murine Peripheral Arteries. Ph.D. Thesis, Charité-Universitätsmedizin Berlin, Berlin, Germany, 2019. [Google Scholar]
- Guda, R.S.; Odegaard, K.E.; Tan, C.; Schaal, V.L.; Yelamanchili, S.V.; Pendyala, G. Integrated systems analysis of mixed neuroglial cultures proteome post oxycodone exposure. Int. J. Mol. Sci. 2021, 22, 6421. [Google Scholar] [CrossRef]
- Song, C.; Pan, S.; Li, D.; Hao, B.; Lu, Z.; Lai, K.; Li, N.; Geng, Q. Comprehensive analysis reveals the potential value of inflammatory response genes in the prognosis, immunity, and drug sensitivity of lung adenocarcinoma. BMC Med. Genom. 2022, 15, 198. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C. Chinese Glioma Genome Atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kao, T.J.; Wu, C.C.; Phan, N.N.; Liu, Y.H.; Ta, H.D.K.; Anuraga, G.; Wu, Y.F.; Lee, K.H.; Chuang, J.Y.; Wang, C.Y. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging 2021, 13, 17970. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Wang, C.Y.; Chiao, C.C.; Phan, N.N.; Li, C.Y.; Sun, Z.D.; Jiang, J.Z.; Hung, J.H.; Chen, Y.L.; Yen, M.C.; Weng, T.Y.; et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am. J. Cancer Res. 2020, 10, 95–113. [Google Scholar]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef]
- Zhang, P.; Xia, Q.; Liu, L.; Li, S.; Dong, L. Current opinion on molecular characterization for GBM classification in guiding clinical diagnosis, prognosis, and therapy. Front. Mol. Biosci. 2020, 7, 562798. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761. [Google Scholar] [CrossRef]
- Tan, R.; Nie, M.; Long, W. The role of B cells in cancer development. Front. Oncol. 2022, 12, 958756. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Anuraga, G.; Chang, C.P.; Weng, T.Y.; Hsu, H.P.; Ta, H.D.K.; Su, P.F.; Chiu, P.H.; Yang, S.J.; Chen, F.W.; et al. Repurposing nitric oxide donating drugs in cancer therapy through immune modulation. J. Exp. Clin. Cancer Res. 2023, 42, 22. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. Jama 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015, 17, iv1–iv62. [Google Scholar] [CrossRef] [PubMed]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Miao, H.; Choi, B.D.; Suryadevara, C.M.; Sanchez-Perez, L.; Yang, S.; De Leon, G.; Sayour, E.J.; McLendon, R.; Herndon, J.E.; Healy, P. EGFRvIII-specific chimeric antigen receptor T cells migrate to and kill tumor deposits infiltrating the brain parenchyma in an invasive xenograft model of glioblastoma. PLoS ONE 2014, 9, e94281. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Wang, X.; Xian, W.; McKeon, F.; Foster, J.; Zhou, J.; Zhang, R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med. Res. Rev. 2021, 41, 961–1021. [Google Scholar] [CrossRef]
- Urbanski, L.; Brugiolo, M.; Park, S.; Angarola, B.L.; Leclair, N.K.; Yurieva, M.; Palmer, P.; Sahu, S.K.; Anczuków, O. MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. 2022, 41, 111704. [Google Scholar] [CrossRef]
- Rasheed, S.A.K.; Subramanyan, L.V.; Lim, W.K.; Udayappan, U.K.; Wang, M.; Casey, P.J. The emerging roles of Gα12/13 proteins on the hallmarks of cancer in solid tumors. Oncogene 2022, 41, 147–158. [Google Scholar] [CrossRef]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune microenvironment of gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. The microenvironmental landscape of brain tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Gómez-Puerta, J.A.; Ramos-Casals, M.; Lozano, F.; Bosch, X. Therapeutic targeting of B cells for rheumatic autoimmune diseases. Pharmacol. Rev. 2011, 63, 127–156. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Ramamoorthi, G.; Albert, G.; Gallen, C.; Beyer, A.; Snyder, C.; Koski, G.; Disis, M.L.; Czerniecki, B.J.; Kodumudi, K. Differentiation and regulation of TH cells: A balancing act for cancer immunotherapy. Front. Immunol. 2021, 12, 669474. [Google Scholar] [CrossRef] [PubMed]
- Ostroumov, D.; Fekete-Drimusz, N.; Saborowski, M.; Kühnel, F.; Woller, N. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell. Mol. Life Sci. 2018, 75, 689–713. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, A.; Yen, M.-C.; Anuraga, G.; Shahzadi, I.; Mazhar, M.W.; Ta, H.D.K.; Xuan, D.T.M.; Dey, S.; Kumar, S.; Santoso, A.W.; et al. Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies. Cancers 2023, 15, 5112. https://doi.org/10.3390/cancers15205112
Raza A, Yen M-C, Anuraga G, Shahzadi I, Mazhar MW, Ta HDK, Xuan DTM, Dey S, Kumar S, Santoso AW, et al. Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies. Cancers. 2023; 15(20):5112. https://doi.org/10.3390/cancers15205112
Chicago/Turabian StyleRaza, Ahmad, Meng-Chi Yen, Gangga Anuraga, Iram Shahzadi, Muhammad Waqar Mazhar, Hoang Dang Khoa Ta, Do Thi Minh Xuan, Sanskriti Dey, Sachin Kumar, Adrian Wangsawijaya Santoso, and et al. 2023. "Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies" Cancers 15, no. 20: 5112. https://doi.org/10.3390/cancers15205112
APA StyleRaza, A., Yen, M.-C., Anuraga, G., Shahzadi, I., Mazhar, M. W., Ta, H. D. K., Xuan, D. T. M., Dey, S., Kumar, S., Santoso, A. W., William, B. T., & Wang, C.-Y. (2023). Comparative Analysis of the GNAI Family Genes in Glioblastoma through Transcriptomics and Single-Cell Technologies. Cancers, 15(20), 5112. https://doi.org/10.3390/cancers15205112








